Abstract
Background
Hypermethylation of promoter CpG islands is strongly correlated to transcriptional gene silencing and epigenetic maintenance of the silenced state. As well as its role in tumor development, CpG island methylation contributes to the acquisition of resistance to chemotherapy. Differential Methylation Hybridisation (DMH) is one technique used for genome-wide DNA methylation analysis. The study of such microarray data sets should ideally account for the specific biological features of DNA methylation and the non-symmetrical distribution of the ratios of unmethylated and methylated sequences hybridised on the array. We have therefore developed a novel algorithm tailored to this type of data, Methylation Linear Discriminant Analysis (MLDA).
Results
MLDA was programmed in R (version 2.7.0) and the package is available at CRAN [1]. This approach utilizes linear regression models of non-normalised hybridisation data to define methylation status. Log-transformed signal intensities of unmethylated controls on the microarray are used as a reference. The signal intensities of DNA samples digested with methylation sensitive restriction enzymes and mock digested are then transformed to the likelihood of a locus being methylated using this reference. We tested the ability of MLDA to identify loci differentially methylated as analysed by DMH between cisplatin sensitive and resistant ovarian cancer cell lines. MLDA identified 115 differentially methylated loci and 23 out of 26 of these loci have been independently validated by Methylation Specific PCR and/or bisulphite pyrosequencing.
Conclusion
MLDA has advantages for analyzing methylation data from CpG island microarrays, since there is a clear rational for the definition of methylation status, it uses DMH data without between-group normalisation and is less influenced by cross-hybridisation of loci. The MLDA algorithm successfully identified differentially methylated loci between two classes of samples analysed by DMH using CpG island microarrays.
Background
DNA methylation frequently occurs in mammalian DNA at the 5 position of cytosine in CpG dinucleotides. It has been estimated that over 70% of cytosines of CpG dinucleotides are methylated in the human genome. CpG dinucleotides are under-represented in the genome and methylated CpG dinucleotides predominantly occur within repetitive elements [2]. However, there are CpG rich regions of the genome which generally remain unmethylated [3]. These CpG rich regions are known as CpG islands and are frequently located in the promoter or the first exon regions of approximately 60% of all genes [4]. The unmethylated status of CpG islands is thought to be a prerequisite state to maintain the linked gene in an active transcribed and transcriptional permissive state.
Differential Methylation Hybridisation (DMH) is one of several techniques for examining CpG island methylation at a genome-wide scale that has been applied to the identification of aberrantly methylated gene promoters in various cancers [5-12]. Nouzova et al[13] modified the original method by using digestion with a methylation-dependent enzyme, McrBC. This enzyme only cleaves methylated CpG DNA sequences. Within-sample comparison is applied after competitive hybridisation with McrBC digested DNA and undigested (mock digested) DNA labelled with Cy3 and Cy5. If a locus is unmethylated the signal intensities of Cy3 and Cy5 are equivalent, while if methylated the Cy5/Cy3 (undigested/digested) ratio is greater than one. However, no common reference is generally used in the modified DMH method, and the unequal representation of methylated and unmethylated sequences due to competitive hybridisation may reduce sensitivity and specificity to detect differential methylation.
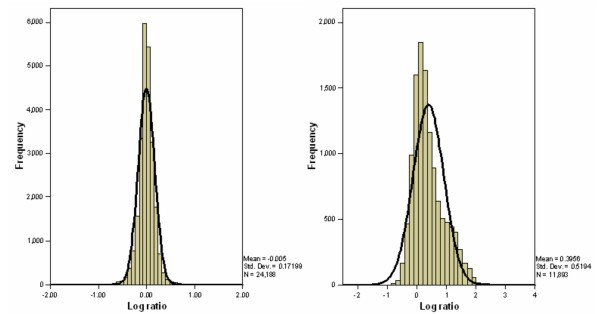
Currently, Significance Analysis of Microarrays (SAM) [14] and Prediction Analysis for Microarrays (PAM) [15] are commonly applied in DNA methylation analysis. Based on the change in hybridisation relative to the standard deviation of repeated measurements, SAM assigns each gene a score that is an extension of the t-statistic. For significant genes with a score over a certain threshold, SAM uses permutations to estimate the false discovery rate (FDR). It has been implemented in many studies of gene expression data [16-21] as well as DMH data, e.g. Wei et al. [22] applied SAM to find the differential methylation of CpG island loci between ovarian caner patient groups with short and long progression-free survival (PFS). However, SAM assumes that the microarray data conform to approximate normality and symmetry, leading to the loss of power in the analysis of DMH data that are inherently skewed due to the biological features of DNA methylation in cancer and competitive hybridisation on DMH arrays (Figure 1).
Figure 1.
Distribution of log-transformed ratio of gene expression data in breast cancer and DMH data in A2780 cell line. The left histogram shows the distribution of log-transformed ratios (cy3/cy5) in gene expression profiling data from a previous study of breast cancer 36] which is symmetric, while the right histogram shows the log-transformed ratios (undigested/digested) of DMH data from the present study which is skewed.
In the modified DMH method, the ratios of raw signal intensities (undigested/digested) greater than 1 reflect the various methylation levels [13]. A ratio cut-off is generally used to identify the hypermethylated loci [7]. However, this is an arbitrary value and does not necessarily accurately reflect the various sources of variation in the experiment. It is therefore desirable to develop an algorithm to more objectively assess the methylation status of loci from DMH data.
PAM is a nearest centroid shrinkage method that identifies those genes that discriminate best between classes. This technique shrinks the class gene centroid towards the overall centroid by a "threshold" amount after standardizing each gene by its within class standard deviation. The "threshold" is identified by cross-validation. This approach was applied in the study by Wei et al. [22] and showed certain power in the identification of differentially methylated loci, but PAM is designed for class prediction rather than class comparison. Although the class predictor used in PAM can reflect the difference between classes, a large number of loci actually differentially methylated between the classes are excluded to improve the accuracy of prediction.
Although normalisation has become a standard procedure for the study of microarray data and is necessary for SAM and PAM analysis, unbalanced shifts in methylation status between class samples in DMH limit the use of between-class normalisation which assumes the changes are roughly symmetric. Thus, the differential methylation can be masked by the over-correction of normalisation and it would be preferable to use a method of analysis that does not require normalisation of the data.
Since PAM and SAM may have limitations for analysing DMH data, we have developed an alternative approach based on the specific features and known biological properties of the arrays used for DMH analysis. The algorithm is named as Methylation Linear Discriminant Analysis (MLDA) and has been applied to identify a set of loci differentially methylated between ovarian cisplatin sensitive and resistant cancer cell lines.
Results
Outline of MLDA
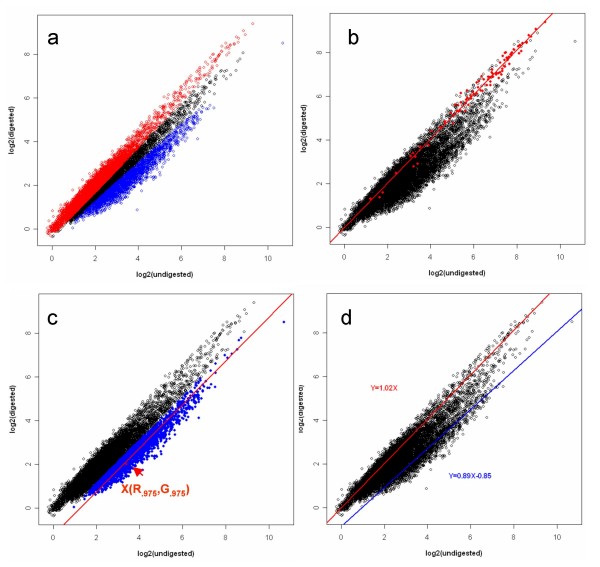
In this study, we have developed a novel approach, named MLDA, for analysing CpG island microarray hybridisation data that allows the identification of differentially methylated loci. MLDA was programmed in R (version 2.7.0) and the package is available at CRAN [1]. This approach uses three relatively simple linear regression models. The first one is constructed by the log-transformed signal intensities of unmethylated features and used as the reference for unmethylation (Figure 2b). The second one is the intermediate model constructed through the point corresponding to the 97.5-quantiles residual below the first linear regression line (Figure 2c). The features with a standardised residual less than 2 from this intermediate model are used to generate the third model which is used as the reference for methylation (Figure 2d). The log likelihood ratio of a locus being methylated is then proportional to the difference between the squared standardised residual from the methylated line and that from the unmethylated line. The log likelihood threshold of zero then provides a more rational basis for distinguishing between methylated and unmethylated loci than a robust undigested/digested ratio of 1.5, as it takes into account the observed variability in the experiment.
Figure 2.
An illustration of unmethylated and methylated model construction in MLDA in A2780 cell line. a: Three patterns can be observed on the scatter plot of log-transformed Cy3 (undigested) against log-transformed Cy5 (digested) intensities. b: The unmethylated model constructed using 94 mitochondrial sequences as a unmethylation reference. c: The intermediate model constructed through the 97.5 quantile residual. The point X is the 97.5 quantile residual. The microarray probes colored in blue (standardised residual to the intermediate model is less than 2) are selected to construct the methylated model. d: Methylated (in blue) and unmethylated (in red) models in A2780 cell line.
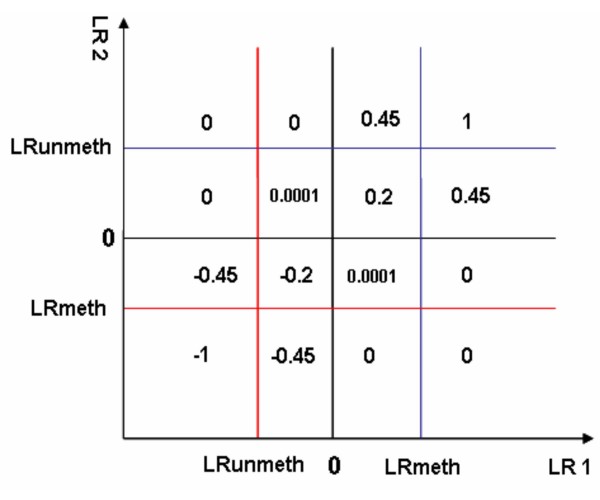
In our approach the consistency and inconsistency rates of log likelihood ratios on dye-swapped/duplicate arrays are used to determine methylation and unmethylation cut-offs, which keep the consistency rate (CR) relatively high (about 140%) and the inconsistency rate (IR) low (about 1%). Each loci is assigned a score based on the cut-offs using the weighted methylation scoring scheme. The feature consistently identified as methylated candidates on dye-swapped/duplicate arrays are scored as 1; similarly unmethylated features are scored as -1; the rest of the feature are assigned a weighted score corresponding to their location on the plot of log-likelihood ratios (Figure 3).
Figure 3.
Weighted scoring scheme. The microarray probes consistently identified as methylated candidates on dye-swap arrays were scored 1; similarly unmethylated microarray probes were scored -1. The rest of the microarray probes were assigned a weighted score based on their location on the plot. LRmeth: log likelihood ratio cut-off for methylated loci; LRunmeth: log likelihood ratio cut-off for unmethylated loci. LR: log likelihood ratio on dye-swapped arrays.
The averaged score for each locus is calculated in each sample class (e.g. resistant or sensitive) and plotted against each other. A robust regression model is then fitted to these data. The standardised residuals from the robust regression model are assumed to follow a normal distribution N(μ, σ2). The outliers of the standardised residuals are identified as the differentially methylated loci between the class samples.
DMH dataset
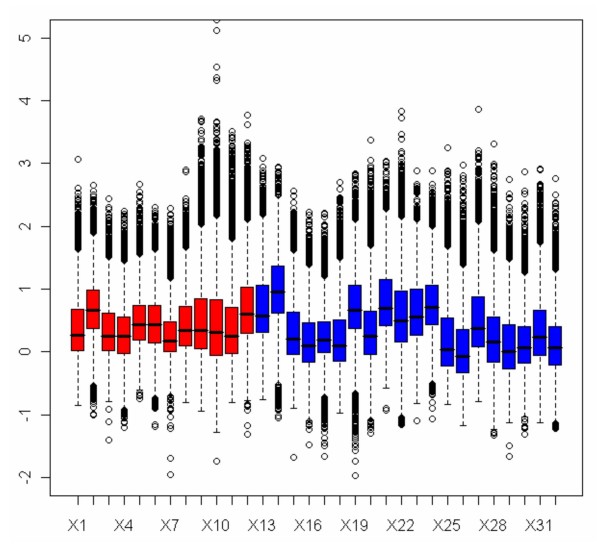
MLDA was applied to identify the CGIs differentially methylated from DMH data derived from sensitive A2780 derivatives (A2780, A2780p3, A2780p5, A2780p6, A2780p13, A2780p14) and isogenically matched, resistant lines [23] derived by multiple exposures to cytotoxic levels of cisplatin and which are 2–5 fold resistant to cisplatin in clonogenic assays (A2780cp70, A2780/MCP1, A2780/MCP2, A2780/MCP3, A2780/MCP4, A2780/MCP5, A2780/MCP6, A2780/MCP7, A2780/MCP8, A2780/MCP9). After background correction, the log-transformed digested and undigested intensities of the 13056 microarray probes show three approximately parallel linear patterns (Figure 2a). The first pattern (digested/undigested is close to 1) represents the unmethylated sequences. The second pattern represents either hemi-methylated sequences or the unmethylated sequences cross-hybridised with the methylated ones on the panel. The third pattern represents the methylated sequences in target DNA. The methylated and unmethylated loci in target DNA can be characterised by a linear regression model for each pattern. As previously mentioned, normalisation may not be appropriate for DMH data, so the log ratios of signal intensities in two classes of samples are not at the same level (Figure 4). Normalisation is not required for MLDA as the determination of the methylation score is based on the data within each experiment.
Figure 4.
Box plot of log ratios of undigested signal intensities against digested signal intensities in 16 cell lines (dye-swapped arrays). The boxes colored in red are the A2780 sensitive cell lines; in blue are the A2780 resistant cell lines. As normalisation is not applied, the center and scale of log ratios for the 16 cell lines are not at the same level.
Mitochondrial DNA is unmethylated [24], therefore, the signal intensities of both channels of microarray probes for mitochondrial sequences are expected to be equal. However, a bi-modal distribution is observed in the log-transformed fluorescence ratios (digested/undigested) of 121 mitochondrial sequences. The first peak represents the unmethylated mitochondrial sequences and the second lower peak is assumed to be the mitochondrial sequences cross-hybridised with other methylated sequences on the panel. Thus, we selected 94 of 121 mitochondrial sequences that were consistently unmethylated through all the cell lines and used them as the unmethylation reference in target DNA.
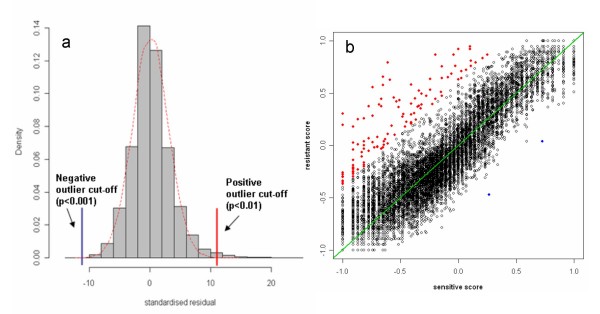
The parameters of those two models in all 16 cell lines were estimated (Table 1). The slope of the unmethylated regression line constructed by 94 mitochondrial sequences is indeed close to 1. After computing the log-likelihood ratios, the methylation and unmethylation cut-offs and associated IRs and CRs were determined from the dye-swapped array pairs (details in Method section). As shown in Figure 5, IR tends to rise with the increase of CR slowly, but starts to increase dramatically when the CR goes above 140%, at which point IR is generally about 1%. We have therefore used CR > 140% and IR < 1% as the criteria for determining the methylation and unmethylation cut-offs. Each locus was scored using the weighted scoring scheme based on those cut-offs. The averaged scores in 6 cisplatin-sensitive cell lines and 10 cisplatin-resistant cell lines were used to construct a robust regression model. Figure 6a shows that the standardised residuals (residual/σ) from the robust regression model roughly follow a normal distribution. The positive and negative outliers are determined as described in Method section.
Table 1.
Parameters of linear models in MLDA for 16 cell lines in DMH dataset I
| Unmethylation linear regression model | ||||||||
| cell line | intercetp (α) | slope (β) | σ | R2 | interceptds (α) | slopeds (β) | σds | R2ds |
| A2780 | -0.0003 | 1.0122 | 0.1727 | 0.9829 | 0.0005 | 1.0574 | 0.1897 | 0.978 |
| A2780p3 | -0.0003 | 1.0343 | 0.1212 | 0.9897 | 0.0018 | 1.1065 | 0.1425 | 0.9882 |
| A2780p5 | -0.0003 | 1.0138 | 0.1684 | 0.984 | -0.0002 | 1.0728 | 0.1425 | 0.9883 |
| A2780p6 | 0.0002 | 0.9914 | 0.1605 | 0.9778 | -0.0001 | 1.0012 | 0.1638 | 0.9747 |
| A2780p13 | -0.0012 | 1.024 | 0.1628 | 0.9786 | -0.0005 | 1.0744 | 0.1436 | 0.9852 |
| A2780p14 | -0.0022 | 1.0499 | 0.1523 | 0.9809 | -0.0009 | 1.034 | 0.2069 | 0.9691 |
| A2780cp70 | 0.0013 | 0.9604 | 0.2532 | 0.9524 | -0.0002 | 1.0119 | 0.2402 | 0.9479 |
| MCP1 | 0.0002 | 0.9946 | 0.145 | 0.9803 | -0.0023 | 1.112 | 0.1452 | 0.9836 |
| MCP2 | 0.0000 | 0.998 | 0.137 | 0.9727 | -0.0028 | 1.0475 | 0.1719 | 0.9653 |
| MCP3 | 0.0004 | 0.9932 | 0.2253 | 0.9183 | -0.0023 | 1.0517 | 0.2795 | 0.8978 |
| MCP4 | 0.0006 | 0.9838 | 0.1739 | 0.9718 | -0.0028 | 1.077 | 0.1947 | 0.9751 |
| MCP5 | 0.0009 | 0.9857 | 0.2464 | 0.9639 | -0.0008 | 1.017 | 0.2166 | 0.9692 |
| MCP6 | -0.0022 | 1.0352 | 0.122 | 0.9751 | -0.0068 | 1.1283 | 0.154 | 0.9752 |
| MCP7 | -0.0005 | 1.0079 | 0.1379 | 0.9791 | -0.0045 | 1.1529 | 0.1588 | 0.9764 |
| MCP8 | -0.0028 | 1.0578 | 0.1903 | 0.9431 | -0.0068 | 1.1193 | 0.1885 | 0.9575 |
| MCP9 | -0.0017 | 1.0331 | 0.1834 | 0.9614 | -0.0091 | 1.1538 | 0.1691 | 0.9674 |
| Methylation linear regression model | ||||||||
| cell line | intercetp (α) | slope (β) | σ | R2 | interceptds (α) | slopeds (β) | σds | R2ds |
| A2780 | -0.8839 | 0.8917 | 0.1491 | 0.9438 | -0.8055 | 0.9086 | 0.1706 | 0.926 |
| A2780p3 | -1.1672 | 0.9797 | 0.1518 | 0.9553 | -0.7414 | 0.9774 | 0.179 | 0.9438 |
| A2780p5 | -0.8991 | 0.8978 | 0.1515 | 0.9476 | -0.9246 | 0.9766 | 0.1673 | 0.9523 |
| A2780p6 | -0.9455 | 0.9562 | 0.1838 | 0.9378 | -1.1995 | 0.9641 | 0.1788 | 0.9324 |
| A2780p13 | -1.8918 | 0.9807 | 0.2535 | 0.8962 | -1.8049 | 0.9652 | 0.2936 | 0.8512 |
| A2780p14 | -1.5637 | 0.9142 | 0.2549 | 0.8857 | -1.4468 | 0.9066 | 0.2128 | 0.8837 |
| A2780cp70 | -1.0317 | 0.8501 | 0.1581 | 0.9115 | -1.3074 | 0.8967 | 0.1541 | 0.9265 |
| MCP1 | -1.199 | 0.9781 | 0.1692 | 0.9467 | -1.0935 | 1.0384 | 0.1775 | 0.9525 |
| MCP2 | -0.8037 | 0.9292 | 0.1486 | 0.9557 | -0.9738 | 0.9381 | 0.2176 | 0.8848 |
| MCP3 | -1.1244 | 0.9151 | 0.1755 | 0.9482 | -0.9303 | 0.9205 | 0.2599 | 0.8098 |
| MCP4 | -1.4326 | 0.9171 | 0.1418 | 0.966 | -1.6205 | 0.961 | 0.2348 | 0.8323 |
| MCP5 | -1.1187 | 0.9425 | 0.1839 | 0.9404 | -1.2007 | 0.9546 | 0.1757 | 0.9295 |
| MCP6 | -1.246 | 0.925 | 0.1966 | 0.9294 | -1.2182 | 0.9826 | 0.2248 | 0.8989 |
| MCP7 | -1.8972 | 0.9909 | 0.1977 | 0.9442 | -1.4894 | 1.0139 | 0.2458 | 0.8886 |
| MCP8 | -1.0219 | 0.9905 | 0.1975 | 0.9468 | -0.4735 | 0.9421 | 0.228 | 0.8761 |
| MCP9 | -1.3399 | 0.9837 | 0.2073 | 0.9352 | -1.1497 | 1.0078 | 0.1967 | 0.9115 |
ds: dye swap
σ: standard deviation
R2: coefficient of determination
Figure 5.
CR against IR in 16 cell lines. X axis is the consistency rate (CR) and y axis is the inconsistency rate (IR). IR tends to rise with the increase of CR slowly, but starts to increase dramatically when the CR goes above 140%, at which point the inconsistency rate is generally about 1%. Not all cell lines could reach this point e.g. MCP3.
Figure 6.
Outliers identifications. a: Distribution of the observed (histogram) standardised residuals and the theoretical distribution based on the fitted model (dashed smooth line in red). The red and blue solid line are the positive and negative cut-offs, respectively. b: Scatter plot of sensitive scores against resistant scores in A2780 series cell lines. The hypermethylated loci are colored in red and hypomethylated loci are in blue. The robust regression model is Y = 0.9956X + 0.0019.
Finally, 115 loci were identified as candidates differentially methylated between A2780 sensitive and these resistant cell lines (additional file 1). Noticeably, 113 of 115 loci (p = 8.8 × 10-3, outlier detection test [25]) were hypermethylated, but only 2 loci (p < 0.001, outlier detection test) lost methylation in the resistant cell lines (Figure 6b). This is consistent with the unbalanced shift in DMH data and indicates cisplatin treatment of cells selects preferentially for hypermethylation of loci, rather than hypomethylation in these tumor cells.
Validation of differential methylation
To confirm the differential methylation of loci identified in this study, we experimentally tested the methylation of 26 loci by methylation-specific PCR (MSP) and/or pyrosequencing of bisulphite modified DNA [26] in sensitive A2780 derivatives and cisplatin resistant derivatives. Twenty-three out of the 26 loci have been confirmed as differentially methylated (additional file 1). It should be noted that MSP and pyrosequencing only examine methylation at a limited number of CpG sites of the sequence present on the DMH analysis. It is possible that the loci which were not confirmed as differentially methylated are methylated at other CpG sites which are detected by DMH but not targeted by MSP and/or pyrosequencing primers and so 23 out of 26 loci confirmed as differentially methylated is a minimum estimation.
To compare the results from MLDA, SAM and PAM, we analysed the DMH dataset by all three methods. MLDA identified 115 loci (113 hypermethylated and 2 hypomethylated loci, misclassification error < 0.001), SAM identified 152 loci (149 hypermethylated and 3 hypomethylated loci, misclassification error = 0.227, FDR = 6.17 × 10-3), and PAM found 24 hypermethylated loci (misclassification error = 0.084, FDR < 0.001) in the resistant cell lines. Twenty-four loci identified by all three methods are listed in Table 2.
Table 2.
24 loci identified by MLDA, PAM and SAM as differentially methylated candidates in the comparison between A2780 cisplatin sensitive and cisplatin multiple-selected resistant cell lines.
| microarray ID | status | validation | MLDA rank* | PAM rank | SAM rank | CGI*** | gene symbol** | GenBank Accession | Chromosome |
| 66_G_6 | hypermethylated | Yes | 1 | 1 | 1 | Yes | |||
| 121_D_9 | hypermethylated | Yes | 2 | 5 | 6 | Yes | CRABP1 | NM_004378 | 15 |
| 39_E_1 | hypermethylated | ND | 3 | 2 | 2 | No | |||
| 122_D_9 | hypermethylated | No | 4 | 11 | 11 | Yes | SOX12 | NM_006943 | 20 |
| 123_D_9 | hypermethylated | No | 5 | 10 | 10 | Yes | SOX12 | NM_006943 | 20 |
| 51_H_8 | hypermethylated | Yes | 6 | 18 | 19 | No | FEZF2 | NM_018008 | 3 |
| 58_A_1 | hypermethylated | ND | 7 | 22 | 22 | Yes | |||
| 80_H_5 | hypermethylated | ND | 8 | 14 | 16 | No | |||
| 21_A_11 | hypermethylated | Yes | 9 | 17 | 18 | Yes | NTN4 | NM_021229 | 12 |
| 38_D_7 | hypermethylated | Yes | 11 | 23 | 24 | Yes | AGBL2 | NM_024783 | 11 |
| 40_E_1 | hypermethylated | ND | 12 | 19 | 17 | No | |||
| 18_A_7 | hypermethylated | ND | 13 | 20 | 20 | No | EDIL3 | NM_005711 | 5 |
| 55_F_8 | hypermethylated | Yes | 14 | 9 | 9 | Yes | BC127881 | BC127881 | 7 |
| 122_B_1 | hypermethylated | ND | 15 | 16 | 14 | No | |||
| 109_A_6 | hypermethylated | No | 18 | 12 | 13 | Yes | |||
| 41_D_9 | hypermethylated | Yes | 22 | 8 | 8 | Yes | WNT1 | NM_005430 | 12 |
| 42_D_9 | hypermethylated | ND | 23 | 4 | 4 | No | |||
| 119_A_6 | hypermethylated | Yes | 24 | 6 | 5 | Yes | NR2E1 | NM_003269 | 6 |
| 63_A_8 | hypermethylated | ND | 26 | 15 | 15 | No | |||
| 6_D_4 | hypermethylated | Yes | 31 | 3 | 3 | Yes | LMX1A | NM_177398 | 1 |
| 17_H_9 | hypermethylated | Yes | 34 | 13 | 12 | Yes | HRASLS3 | NM_006290 | 6 |
| 5_D_4 | hypermethylated | Yes | 35 | 7 | 7 | Yes | LMX1A | NM_177398 | 1 |
| 24_D_3 | hypermethylated | Yes | 75 | 24 | 23 | Yes | SP5 | NM_001003845 | 2 |
| 122_G_1 | hypermethylated | ND | 101 | 21 | 21 | Yes |
ND: not done. Yes: validated. No: not validated
MLDA rank*: the rank of standardised residuals to the robust regression line constructed by the averaged sensitive scores against averaged resistant scores
Gene symbol**: only the gene of which transcription start site (TSS) is within 5 kb span of the loci
CGI***: CpG island defined by Gardiner-Garden and Frommer [31].
Discussion
Hypermethylation of promoter CpG islands is strongly correlated to transcriptional gene silencing and epigenetic maintenance of the silenced state and is a potential rich source of biomarkers of cancer. Differential Methylation Hybridisation (DMH) is one technique used for genome-wide DNA methylation analysis. The study of such microarray data sets should ideally account for the specific biological features of DNA methylation and the non-symmetrical distribution of the ratios of unmethylated and methylated sequences hybridised on the array. We have therefore developed a novel algorithm tailored to this type of data, Methylation Linear Discriminant Analysis (MLDA). MLDA utilises log likelihood ratios representing the relative probability that loci are methylated instead of log ratios of signal intensities used in previous studies [6-10,27]. Validation of 23/26 identified loci using independent methods of methylation analysis shows that MLDA can robustly identify differential methylated loci between ovarian cancer sensitive and resistant cell lines without requiring the data to be normalised.
Although a log likelihood ratio above zero means that the locus tends to be methylated, we did not use zero as the cut-off to determine the number of methylated and unmethylated sequences, as the existence of cross-hybridisation and measurement errors in the DMH assay makes this unreliable. To increase the precision of the methylation classification, we used the inconsistency (IR) and consistency (CR) rates between the dye-swap arrays to determine likelihood ratio cut-offs for methylation and unmethylation and assigned each locus a methylation score based on the position relative to these cut-offs. As shown in Figure 5, not all cell lines can reach the point that CR is around 140% and IR is about 1%. IR and CR need to be carefully selected as the methylation scores of loci are consequently influenced by the change of IR and CR. We also observed a lower CR (about 120%) and a higher IR (about 2%) in another CpG island array using DMH (data not shown), therefore, further examination of what factors influence the achievable CR and IR rates may improve the utility of the MLDA approach.
Data on methylation status for 121 mitochondrial derived sequences were available in this study. Mitochondrial sequences would be expected to be unmethylated. We used 94 mitochondrial sequences to construct unmethylated linear model at the beginning of the study, and indeed, 93 of 121 mitochondrial loci were defined as unmethylated and 25 loci being of uncertain methylation status by MLDA. However, three mitochondrial loci were identified as hypermethylated candidates in the resistant ovarian carcinoma cell lines by both MLDA and SAM. One explanation of this discrepancy is that all these three loci have more than one BLAT hit indicating the existence of homology with nuclear DNA sequences, raising the possibility of hybridisation with these nuclear DNA sequences which may be differentially methylated. As shown in Figure 2a, the loci in the middle pattern represent either hemi-methylated sequences or the unmethylated sequences cross-hybridised with the methylated ones on the panel. No specific allowance is made for these intermediate points in analysis by SAM and PAM, whereas MLDA attempts specifically to down-weight these points in the identification of the methylation regression line. By giving a lower weighted score (close to 0) (Figure 3) to those loci, MLDA reduces the influence of cross-hybridisation among this group of sequences. Of course cross-hybridisation may also occur in the loci in the other two patterns (methylated and unmethylated patterns), but it is not possible for any mathematical approach to identify this.
The misclassification error of MLDA based on the methylation score is much lower than that for either SAM or PAM based on the log ratios, indicating the potential of MLDA methylation scores to be used as a reliable discriminator between classes of samples.
Conclusion
We have developed a novel method, named MLDA, for genome-wide DNA methylation studies. MLDA can transform the signal intensities to log-likelihood ratios through three linear regression models. Using this approach MLDA allows determination of the methylation status of a locus based on dye-swapped/duplicate arrays. The method has been applied to assess the methylation status of each locus and identified 115 loci that exhibit differential methylation between A2780 sensitive and resistant cell lines. A minimum of 23 out of 26 loci have been confirmed by independent methods as differentially methylated.
Methods
First, all intensity values were log transformed. A multiplicative background correction was applied to correct signal intensities for the background noise in each array. After background correction, the log-transformed digested and undigested intensities show three approximately parallel linear patterns (Figure 2a). The first pattern (digested/undigested is close to 1) represents the unmethylated sequences. The second pattern represents either hemi-methylated sequences or the unmethylated sequences cross-hybridised with the methylated ones on the panel. The third pattern represents the methylated sequences in target DNA. The methylated and unmethylated loci in target DNA can be characterised by a linear regression model for each pattern. The distance of each spot to the methylated and unmethylated lines respectively can then be estimated by standardised residuals. The log likelihood ratio of a locus being methylated is then proportional to the difference between the squared standardised residual from the methylated line and that from the unmethylated one. The algorithm based around this regression approach is named Methylation Linear Discriminant Analysis (MLDA) and was programmed in R version 2.7.0.
Log-likelihood ratio transformation
a An univariate linear regression model was constructed for the unmethylated probes (e.g. mitochondrial derived features) using formula (1) where α is the intercept, β is the slope of the model, and ξ is the error representing the unpredicted or unexplained variation in the model (Figure 2b). The parameters of regression line were estimated by the method of least squares (formula 2 and 3).
| Gi = α + βRi + ξi i = 1,2,3.......k | (1) |
| (2) |
| (3) |
k is the number of unmethylated controls on DMH array. Gi and Ri are the logarithmic-transformed digested and undigested intensities of microarray probes for mitochondrial sequences, respectively. and are the averaged logarithmic-transformed undigested and digested intensities of the k unmethylated controls.
b. The scale estimate σmito associated with the error term in the linear regression model was estimated from the residuals from the observed k points to the fitted line. The most extreme 10% of residuals was omitted from either end of the distribution to minimise the impact of extreme residuals on this estimate.
c. The standardised residuals of all the microarray probes to the unmethylation regression line were calculated as formula (4).
| (4) |
d. The point corresponding to the 97.5-quantiles residual below the unmethylation line is represented as X (R.975, G.975). The intermediate linear model (Figure 2c) was constructed through point X with a slope assumed to be 1 and the intercept estimated as formula (5).
| (5) |
e. The standardised residuals of all the microarray probes to the line with slope 1 and intercept estimated from (5) were calculated as formula (6). The variance of the residuals to the intermediate model was assumed to be similar as that in the mitochondrial model.
| (6) |
f. The microarray probes with standardised residuals less than 2 were included for later robust regression analysis. The line estimated from this regression analysis represents the methylation regression line (Figure 2d).
g. The scale estimate σmeth of the methylation regression line was estimated using only those microarray probes below the line, with the most extreme 5% removed.
h. The standardised residuals of all the microarray probes to the methylated regression line were calculated as formula (7). The log likelihood ratio (LR) of all the microarray probes was estimated by formula (8) for further analysis.
| (7) |
| LR = SR2mito - SR2meth | (8) |
Determination of log likelihood ratio cut-offs
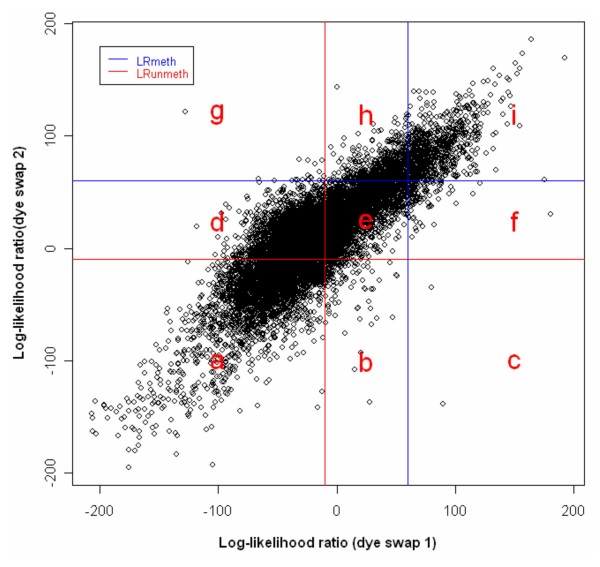
Two inconsistency rates (IRmeth and IRunmeth) and two consistency rates (CRmeth and CRunmeth) between dye-swap arrays were used to determine the log like likelihood ratio threshold. IRmeth (formula 9) represents the rate of the microarray probes identified as methylated in one array but as unmethylated in the other one, while IRunmeth (formula 10) is the rate of the microarray probes identified as unmethylated in one array but as methylated in the other one. CRmeth (formula 11) and CRunmeth (formula 12) are the rates for the spots identified as methylated (CRmeth) and unmethylated (CRunmeth) in both dye-swap arrays (Figure 7).
Figure 7.
Determination of methylation and unmethylation cut-offs of likelihood ratios on dye-swapped arrays. LRmeth: log likelihood ratio cut-off for methylated spots; LRunmeth: log likelihood ratio cut-off for unmethylated spots.
Table 3.
Classification of loci
| Unmethylated | uncertain | methylated | |
| Unmethylated | a | b | c |
| Uncertain | d | e | f |
| Methylated | g | h | i |
| (9) |
| (10) |
| (11) |
| (12) |
b The log likelihood ratio thresholds (LRmeth and LRunmeth) for methylated and unmethylated microarray probes, which kept the IR rates low (at or close to 1%) and the CR rates high (at or close to 140%), were used as the cut-offs for methylated and unmethylated loci. IR tends to rise with the increase of CR slowly, but starts to increase dramatically when the CR goes above 140%, at which point the inconsistency rate is generally about 1%. We have therefore used CR > 140% and IR < 1% as the criteria for determining the methylation cut-offs.
Identification of robust regression outliers
Each microarray probe was scored based on the cut-offs of likelihood ratios for methylation and unmethylation on dye-swap arrays using the weighted methylation scoring scheme shown in Figure 3. The microarray probes consistently identified as methylated candidates on dye-swap arrays were scored of 1; similarly unmethylated microarray probes were scored of -1. The rest of the microarray probes were assigned a weighted score based on their location on the plot.
A robust regression model [28] was constructed with the averaged scores in one class of samples as the explanatory variable, and the corresponding scores in the other class of samples as the dependent variable. The degree of trimming was determined according to Barnett et al. [29] when estimating the variance of residuals to the robust linear regression model.
It was assumed that the standardised residuals (SRs) from the robust regression line followed a normal distribution N(μ, σ2). μ and σ were estimated excluding outliers using the MAD-Median Rule [30]. The p value for each SR cut-off was calculated as described by Simon et al [25]. This p-value reflects the probability of observing a group of more extreme residuals from the fitted normal distribution. Microarray probes were identified as outliers if their SRs were larger than the cut-off for which the p-value was less than 0.01.
Estimation of misclassification rate
The misclassification rate was estimated by drawing bootstrap samples 500 times with replacement from the two classes (sensitive and resistant) and carrying out hierarchical clustering based on the loci identified as differentially methylated using weighted scores for MLDA and log ratios without between-group normalisation for SAM and PAM, respectively. Clustering was carried out using Euclidean distance as the distance metric, and clusters were agglomerated using the average linkage criterion. The clustering tree was cut into two groups and the number of misclassified cell lines was counted. The misclassification rate was obtained from the averaged number of misclassified samples in 500 bootstraps divided by the total number of samples.
SAM and PAM analysis
The raw signal intensities of each channel were subtracted by the median signal intensities of corresponding channel of controls on HCGI12K array. After this correction, SAM in samr package and PAM in pamr package were applied using log ratios (digested/undigested) in R version 2.7.0. Between-group normalisation was not used in SAM and PAM to avoid over-correction masking the differential methylation.
Authors' contributions
WD conducted the statistical analysis and algorithm development supervised by JP and RB. The DMH data was produced by JMT in collaboration with TH and PY. RB, JP and KV conceived the study. JMT, JG and CZ conducted validation by MSP or pyrosequencing in RB's lab. WD, JP and RB prepared the manuscript with review by all authors. Funding was obtained by RB and TH.
Supplementary Material
115 differential methylated candidates identified by MLDA in A2780 series cell lines.
Acknowledgments
Acknowledgements
This work was supported by a CR-UK programme grant to RB, NIH R21 grant CA110475 to RB and TH, and an Ovarian Cancer Action project grant to RB, JMT and JG.
Contributor Information
Wei Dai, Email: w.dai@imperial.ac.uk.
Jens M Teodoridis, Email: j.teodoridis@imperial.ac.uk.
Janet Graham, Email: j.graham@imperial.ac.uk.
Constanze Zeller, Email: c.zeller@imperial.ac.uk.
Tim HM Huang, Email: tim.huang@osumc.adu.
Pearlly Yan, Email: pearlly.yan@osumc.edu.
J Keith Vass, Email: j.k.vass@dundee.ac.uk.
Robert Brown, Email: b.brown@imperial.ac.uk.
Jim Paul, Email: j.paul@clinmed.gla.ac.uk.
References
- http://cran.r-project.org/
- Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet. 1989;83:181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, Huang TH. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- Wei SH, Chen CM, Strathdee G, Harnsomburana J, Shyu CR, Rahmatpanah F, Shi H, Ng SW, Yan PS, Nephew KP, Brown R, Huang TH. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin Cancer Res. 2002;8:2246–2252. [PubMed] [Google Scholar]
- Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Huang TH. Applications of CpG island microarrays for high-throughput analysis of DNA methylation. J Nutr. 2002;132:2430S–2434S. doi: 10.1093/jn/132.8.2430S. [DOI] [PubMed] [Google Scholar]
- Yan PS, Efferth T, Chen HL, Lin J, Rodel F, Fuzesi L, Huang TH. Use of CpG island microarrays to identify colorectal tumors with a high degree of concurrent methylation. Methods. 2002;27:162–169. doi: 10.1016/S1046-2023(02)00070-1. [DOI] [PubMed] [Google Scholar]
- Yan PS, Wei SH, Huang TH. Differential methylation hybridization using CpG island arrays. Methods Mol Biol. 2002;200:87–100. doi: 10.1385/1-59259-182-5:087. [DOI] [PubMed] [Google Scholar]
- Rahmatpanah FB, Carstens S, Guo J, Sjahputera O, Taylor KH, Duff D, Shi H, Davis JW, Hooshmand SI, Chitma-Matsiga R, Caldwell CW. Differential DNA methylation patterns of small B-cell lymphoma subclasses with different clinical behavior. Leukemia. 2006;20:1855–1862. doi: 10.1038/sj.leu.2404345. [DOI] [PubMed] [Google Scholar]
- Zighelboim I, Goodfellow PJ, Schmidt AP, Walls KC, Mallon MA, Mutch DG, Yan PS, Huang TH, Powell MA. Differential methylation hybridization array of endometrial cancers reveals two novel cancer-specific methylation markers. Clin Cancer Res. 2007;13:2882–2889. doi: 10.1158/1078-0432.CCR-06-2367. [DOI] [PubMed] [Google Scholar]
- Nouzova M, Holtan N, Oshiro MM, Isett RB, Munoz-Rodriguez JL, List AF, Narro ML, Miller SJ, Merchant NC, Futscher BW. Epigenomic changes during leukemia cell differentiation: analysis of histone acetylation and cytosine methylation using CpG island microarrays. J Pharmacol Exp Ther. 2004;311:968–981. doi: 10.1124/jpet.104.072488. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamey CA, Glendining KA, Kreiman G, Kang ND, Wang KH, Fassler R, Sawatari A, Tonegawa S, Sur M. Differential gene expression between sensory neocortical areas: potential roles for Ten_m3 and Bcl6 in patterning visual and somatosensory pathways. Cereb Cortex. 2008;18:53–66. doi: 10.1093/cercor/bhm031. [DOI] [PubMed] [Google Scholar]
- Flavell JR, Baumforth KR, Wood VH, Davies GL, Wei W, Reynolds GM, Morgan S, Boyce A, Kelly GL, Young LS, Murray PG. Down-regulation of the TGF-beta target gene, PTPRK, by the Epstein-Barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin lymphoma cells. Blood. 2008;111:292–301. doi: 10.1182/blood-2006-11-059881. [DOI] [PubMed] [Google Scholar]
- Lau SK, Boutros PC, Pintilie M, Blackhall FH, Zhu CQ, Strumpf D, Johnston MR, Darling G, Keshavjee S, Waddell TK, Liu N, Lau D, Penn LZ, Shepherd FA, Jurisica I, Der SD, Tsao MS. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol. 2007;25:5562–5569. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Li C, Giacomini CP, Salari K, Huang S, Wang P, Ferrari M, Hernandez-Boussard T, Brooks JD, Pollack JR. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–8510. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
- Goold R, Hubank M, Hunt A, Holton J, Menon RP, Revesz T, Pandolfo M, Matilla-Duenas A. Down-regulation of the dopamine receptor D2 in mice lacking ataxin 1. Hum Mol Genet. 2007;16:2122–2134. doi: 10.1093/hmg/ddm162. [DOI] [PubMed] [Google Scholar]
- Cheishvili D, Maayan C, Smith Y, Ast G, Razin A. IKAP/hELP1 deficiency in the cerebrum of familial dysautonomia patients results in down regulation of genes involved in oligodendrocyte differentiation and in myelination. Hum Mol Genet. 2007;16:2097–2104. doi: 10.1093/hmg/ddm157. [DOI] [PubMed] [Google Scholar]
- Wei SH, Balch C, Paik HH, Kim YS, Baldwin RL, Liyanarachchi S, Li L, Wang Z, Wan JC, Davuluri RV, Karlan BY, Gifford G, Brown R, Kim S, Huang TH, Nephew KP. Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res. 2006;12:2788–2794. doi: 10.1158/1078-0432.CCR-05-1551. [DOI] [PubMed] [Google Scholar]
- Anthoney DA, McIlwrath AJ, Gallagher WM, Edlin AR, Brown R. Microsatellite instability, apoptosis, and loss of p53 function in drug-resistant tumor cells. Cancer Res. 1996;56:1374–1381. [PubMed] [Google Scholar]
- Maekawa M, Taniguchi T, Higashi H, Sugimura H, Sugano K, Kanno T. Methylation of mitochondrial DNA is not a useful marker for cancer detection. Clin Chem. 2004;50:1480–1481. doi: 10.1373/clinchem.2004.035139. [DOI] [PubMed] [Google Scholar]
- Simon RM, Korn EL, McShane LM, Radmacher MD, Wright GW, Zhao Y. Statistics for Biology and Health. New York , Springer; 2003. Design and Analysis of DNA Microarray Investigations. [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SH, Brown R, Huang TH. Aberrant DNA methylation in ovarian cancer: is there an epigenetic predisposition to drug response? Ann N Y Acad Sci. 2003;983:243–250. doi: 10.1111/j.1749-6632.2003.tb05979.x. [DOI] [PubMed] [Google Scholar]
- Tukey JW. Exploratory Data Analysis. Rading Massachusetts , Addison-Wesley; 1977. [Google Scholar]
- Wilcox RR. Robust Regression. In: Holland BA, editor. Robust Esitmation and Hypothesis Testing. London , Elsevier Academic Press; 2005. [Google Scholar]
- Wilcox RR. Estimating Measures of Location and Scale. In: Holland BA, editor. Robust Esitmation and Hypothesis Testing. London , Elsevier Academic Press; 2005. [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
115 differential methylated candidates identified by MLDA in A2780 series cell lines.