Abstract
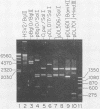
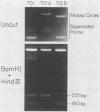
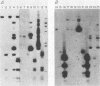
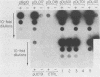
An origin of replication within the long unique sequence of herpes simplex virus type 2 designated oriL2 has been identified in a position homologous to its type 1 counterpart, oriL1, between map coordinates 0.398 and 0.413. The difficulties encountered in previous attempts to clone both oriL2 and oriL1 in an undeleted form were surmounted by minimizing the growth of the host Escherichia coli, using a recBC sbcB E. coli host, and purifying the full-length plasmid from delected forms by using a novel method which exploits the ability of a palindrome-containing plasmid to adopt a cruciform conformation, thereby decreasing its supercoiling. In a previously developed assay for functional origin activity, oriL2 was localized to a 241-base-pair ApaI-SstII fragment. DNA sequence analysis revealed a 136-base pair, almost perfect palindrome. Comparison with oriL1 showed a very high degree of conservation: the two origins differ in only 16 of the 144-base-pair oriL1 palindromic region. Most significantly, the differences between oriL1 and oriL2 mainly occur in pairs so as to generally preserve the potential for intrastrand base pairing. The central region of oriL2 is homologous with the shorter palindromic structures detected in origins located within the repetitive sequences of the short component of herpes simplex virus type 1 or 2.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer W. R. Structure and reactions of closed duplex DNA. Annu Rev Biophys Bioeng. 1978;7:287–313. doi: 10.1146/annurev.bb.07.060178.001443. [DOI] [PubMed] [Google Scholar]
- Behnke D., Malke H., Hartmann M., Walter F. Post-transformational rearrangement of an in vitro reconstructed group-A streptococcal erythromycin resistance plasmid. Plasmid. 1979 Oct;2(4):605–616. doi: 10.1016/0147-619x(79)90058-1. [DOI] [PubMed] [Google Scholar]
- Boissy R., Astell C. R. An Escherichia coli recBCsbcBrecF host permits the deletion-resistant propagation of plasmid clones containing the 5'-terminal palindrome of minute virus of mice. Gene. 1985;35(1-2):179–185. doi: 10.1016/0378-1119(85)90170-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Collins J., Volckaert G., Nevers P. Precise and nearly-precise excision of the symmetrical inverted repeats of Tn5; common features of recA-independent deletion events in Escherichia coli. Gene. 1982 Jul-Aug;19(1):139–146. doi: 10.1016/0378-1119(82)90198-6. [DOI] [PubMed] [Google Scholar]
- Cortini R., Wilkie N. M. Physical maps for HSV type 2 DNA with five restriction endonucleases. J Gen Virol. 1978 May;39(2):259–280. doi: 10.1099/0022-1317-39-2-259. [DOI] [PubMed] [Google Scholar]
- Friedmann A., Shlomai J., Becker Y. Electron microscopy of herpes simplex virus DNA molecules isolated from infected cells by centrifugation in CsCl density gradients. J Gen Virol. 1977 Mar;34(3):507–522. doi: 10.1099/0022-1317-34-3-507. [DOI] [PubMed] [Google Scholar]
- Gray C. P., Kaerner H. C. Sequence of the putative origin of replication in the UL region of herpes simplex virus type 1 ANG DNA. J Gen Virol. 1984 Dec;65(Pt 12):2109–2119. doi: 10.1099/0022-1317-65-12-2109. [DOI] [PubMed] [Google Scholar]
- Greaves D. R., Patient R. K., Lilley D. M. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J Mol Biol. 1985 Oct 5;185(3):461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Hagan C. E., Warren G. J. Viability of palindromic DNA is restored by deletions occurring at low but variable frequency in plasmids of Escherichia coli. Gene. 1983 Oct;24(2-3):317–326. doi: 10.1016/0378-1119(83)90092-6. [DOI] [PubMed] [Google Scholar]
- Hirsch I., Cabral G., Patterson M., Biswal N. Studies on the intracellular replicating DNA of herpes simplex virus type 1. Virology. 1977 Aug;81(1):48–61. doi: 10.1016/0042-6822(77)90057-5. [DOI] [PubMed] [Google Scholar]
- Horii Z., Clark A. J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973 Oct 25;80(2):327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- Leach D. R., Stahl F. W. Viability of lambda phages carrying a perfect palindrome in the absence of recombination nucleases. 1983 Sep 29-Oct 5Nature. 305(5933):448–451. doi: 10.1038/305448a0. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. In vivo consequences of plasmid topology. Nature. 1981 Jul 23;292(5821):380–382. doi: 10.1038/292380a0. [DOI] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol. 1982 Apr 5;156(2):229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie M. J., McGeoch D. J. DNA sequence analysis of an immediate-early gene region of the herpes simplex virus type 1 genome (map coordinates 0.950 to 0.978). J Gen Virol. 1982 Sep;62(Pt 1):1–15. doi: 10.1099/0022-1317-62-1-1. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader W. F., Edlind T. D., Huettermann A., Sauer H. W. Cloning of Physarum actin sequences in an exonuclease-deficient bacterial host. Proc Natl Acad Sci U S A. 1985 May;82(9):2698–2702. doi: 10.1073/pnas.82.9.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Rao R. N., Rogers S. G. Plasmid pKC7: a vector containing ten restriction endonuclease sites suitable for cloning DNA segments. Gene. 1979 Sep;7(1):79–82. doi: 10.1016/0378-1119(79)90044-1. [DOI] [PubMed] [Google Scholar]
- Sadler J. R., Betz J. L., Tecklenburg M., Goeddel D. V., Yansura D. G., Caruthers M. H. Cloning of chemically synthesized lactose operators. II. EcoRI-linkered operators. Gene. 1978 May;3(3):211–232. doi: 10.1016/0378-1119(78)90033-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E., Francke B. Organization of herpes simplex virus type 1 deoxyribonucleic acid during replication probed in living cells with 4,5',8-trimethylpsoralen. Biochemistry. 1982 Aug 31;21(18):4484–4490. doi: 10.1021/bi00261a045. [DOI] [PubMed] [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci U S A. 1985 Feb;82(3):694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D. Localization of an origin of DNA replication within the TRS/IRS repeated region of the herpes simplex virus type 1 genome. EMBO J. 1982;1(7):863–867. doi: 10.1002/j.1460-2075.1982.tb01261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., McMonagle E. C. Characterization of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983 Oct 30;130(2):427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- Stow N. D. Mutagenesis of a herpes simplex virus origin of DNA replication and its effect on viral interference. J Gen Virol. 1985 Jan;66(Pt 1):31–42. doi: 10.1099/0022-1317-66-1-31. [DOI] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walboomers J. M., Schegget J. T. A new method for the isolation of herpes simplex virus type 2 DNA. Virology. 1976 Oct 1;74(1):256–258. doi: 10.1016/0042-6822(76)90151-3. [DOI] [PubMed] [Google Scholar]
- Watson R. J., Vande Woude G. F. DNA sequence of an immediate-early gene (IEmRNA-5) of herpes simplex virus type I. Nucleic Acids Res. 1982 Feb 11;10(3):979–991. doi: 10.1093/nar/10.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Lee K. J., Sabourin D. J., Schaffer P. A. Genetic analysis of temperature-sensitive mutants which define the gene for the major herpes simplex virus type 1 DNA-binding protein. J Virol. 1983 Jan;45(1):354–366. doi: 10.1128/jvi.45.1.354-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S. K., Spadaro A., Schaffer J. E., Murray A. W., Maxam A. M., Schaffer P. A. Cloning, sequencing, and functional analysis of oriL, a herpes simplex virus type 1 origin of DNA synthesis. Mol Cell Biol. 1985 May;5(5):930–942. doi: 10.1128/mcb.5.5.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitton J. L., Clements J. B. Replication origins and a sequence involved in coordinate induction of the immediate-early gene family are conserved in an intergenic region of herpes simplex virus. Nucleic Acids Res. 1984 Feb 24;12(4):2061–2079. doi: 10.1093/nar/12.4.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman A. R., Wolfe L. B., Botstein D. Propagation of some human DNA sequences in bacteriophage lambda vectors requires mutant Escherichia coli hosts. Proc Natl Acad Sci U S A. 1985 May;82(9):2880–2884. doi: 10.1073/pnas.82.9.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]