Abstract
An abundance of protein structures emerging from structural genomics and the Protein Structure Initiative (PSI) are not amenable to ready functional assignment because of a lack of sequence and structural homology to proteins of known function. We describe a high-throughput NMR methodology (FAST-NMR) to annotate the biological function of novel proteins through the structural and sequence analysis of protein-ligand interactions. This is based on basic tenets of biochemistry where proteins with similar functions will have similar active sites and exhibit similar ligand binding interactions, despite global differences in sequence and structure. Protein-ligand interactions are determined through a tiered NMR screen using a library composed of compounds with known biological activity. A rapid co-structure is determined by combining the experimental identification of the ligand-binding site from NMR chemical shift perturbations with the protein-ligand docking program AutoDock. Our CPASS (Comparison of Protein Active Site Structures) software and database is then used to compare this active site with proteins of known function. The methodology is demonstrated using unannotated protein SAV1430 from Staphylococcus aureus.
Keywords: NMR Screening, High-throughput NMR, Protein-Ligand Binding, Function Assignment, Hypothetical Protein, SAV1430
Introduction
The availability of the human genome sequence has just begun to provide a wealth of information on cell biology, development, evolution and physiology that is expanding our understanding of disease and making beneficial contributions to medicine and human health.1 Contributing to this expanding knowledge base is the extensive amount of protein structures emerging from structural genomics and the Protein Structure Initiative (PSI).2
To date, 261 genomes have been completely sequenced with ~1,158 ongoing genome projects3 where 30,000–90,000 proteins are predicted to be from the human genome alone.1 Unfortunately, ~60% of recent structures emerging from structural genomics correspond to folds that provide little insight into function (Fig. 1). The prospect of obtaining functional information for this extensive collection of hypothetical proteins by traditional biochemical approaches presents an extremely overwhelming and daunting task.
Figure 1.
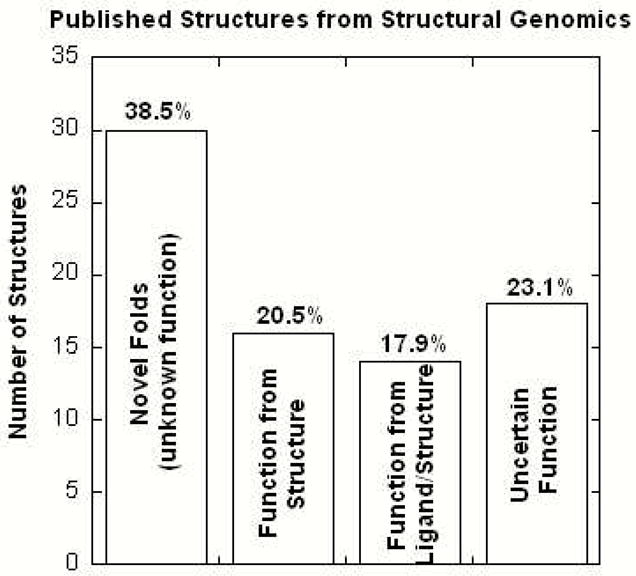
Functional information for hypothetical proteins accumulated from the literature can be classified into four general categories. Functional identification was obtained for ~40% of the proteins where ligand binding played a key role.
A fundamental component to our understanding of the biological role of a protein is through the identification of its functional ligand(s), the identification of its active site or binding site, and the corresponding determination of a structure for this complex. The active sites of proteins interact with unique and specific targets, as is well-documented in numerous metabolic and signaling pathways.4 Promiscuity of binding is inherently detrimental to the overall biological process. The comparison and prediction of putative ligand binding sites from both structural and sequence information has proven to be a valuable approach for functional analysis of proteins.5 Distant evolutionary relationships between enzymes with no sequence similarity have been identified based on the comparison of conserved active site structures.6 Similarly, a number of hypothetical proteins determined from structural genomics have had a function attributed to them by the serendipitous observation of a bound ligand in the resulting structure.7, 8
We have established the Functional Annotation Screening Technology using NMR Spectroscopy (FAST-NMR) to provide initial functional information for proteins originating from structural genomics that lack functional information. By experimentally identifying ligands that have an associated biological function that bind to hypothetical proteins, identifying the protein’s active site from this interaction, and determining a corresponding co-structure of this protein-ligand complex, it is possible to readily infer a potential function for these novel proteins. Functional annotation will emerge from a combined bioinformatics approach that incorporates the identity of the functional ligands that bind the protein, the comparison of the ligand-defined active site identified from FAST-NMR with structures of protein-ligand complexes for proteins of known function using our CPASS software and database9, and other readily available methodologies and software.
Analysis of genome sequences for structural genomics target selection have indicated that the typical protein domain length is ~ 100 residues, where the majority of larger domains fall in the range of >160–300 residues.10 This indicates that a significant percentage of the proteome is amenable to analysis by our FAST-NMR methodology11, where further progress in NMR technology may continue to extend the range of viable protein targets.12 Recent statistical analysis indicates that ~20–40% of protein structures determined from structural genomics may be amenable to analysis by NMR13, 14, further supporting the potential general utility of FAST-NMR. Additional proteins may also become available for analysis when secondary efforts to optimize conditions are applied outside the practical limits imposed by the high-throughput structure determination process.15 This paper describes our development of the FAST-NMR screen and the functional assignment of unannotated protein SAV1430 from Staphylococcus aureus, a typical target of the Northeast Structural Genomics (NESG) consortium.
Experimental
Functional Chemical Screening Library
To date, our screening library is composed of 414 compounds with known biological activity in a total of 113 mixtures comprising 3–4 compounds.16 Reference NMR spectra are collected for both the individual compounds and the mixtures. These spectra provide NMR assignments for the compounds, while verifying compound solubility, compatibility, and the presence of distinct NMR resonances attributed to each compound in the mixture to avoid deconvolution. Identical chemical shifts, coupling patterns and line-widths between the mixture and individual NMR spectra verify that no interaction is occurring between the compounds and that the compounds are equally soluble and stable in the mixture. All the compounds have been individually weighed, dissolved to a concentration of 20 mM in D2O or D6-DMSO (Sigma Aldrich, Saint Louis, MO), and stored in standard 2 ml 96-well plates at −80°C.
1D NMR Line-Broadening Experiments
The 1D NMR line-broadening samples consist of 100μM of each compound and 25μM protein in a 20mM D-Bis-Tris buffer with 11.1μM TMSP as a NMR reference in 100% D2O at pH 7.0 (uncorrected). The NMR spectra were collected on a Bruker 500 MHz Avance spectrometer equipped with a triple-resonance, Z-axis gradient cryoprobe, a BACS-120 sample changer and Icon NMR software for automated data collection. 1H NMR spectra were collected with 128 transients at 298 K with solvent presaturation of the residual HDO, a sweep-width of 6009 Hz and 32K data points and a total acquisition time of 6 minutes. The NMR spectra were processed automatically using a macro in the ACD/1D NMR manager. The NMR data was Fourier transformed, zero-filled, phased, and baseline corrected. Each spectrum was referenced with the TMSP peak set to 0.0 ppm and peak-picked. Compounds were identified as binding to S. aureus protein SAV1430 by a visual comparison of the free ligand and protein-ligand NMR spectra. The entire functional chemical library composed of 113 mixtures containing 414 compounds was used in the 1D NMR experiments. Software for the automated analysis of 1D NMR line broadening data is currently being developed.17
2D 1H-15N HSQC NMR Experiments
22 2D 1H-15N HSQC were collected with 16 transients at 298 K with a sweep-width of 6009Hz and 1K data points in the direct dimension and 1612Hz and 256 data points in the indirect dimension for a total acquisition time of 2.5 hours. Each NMR sample consists of 100μM protein and 400μM compound in a Bis-Tris buffer in 95% H2O, 5%D2O at pH 7.0 (uncorrected). The spectra were processed using NMRPipe on a Linux Workstation. Chemical shift differences were identified by comparing a reference 2D 1H-15N HSQC spectrum of the free protein to the spectrum of the compound-protein samples. Chemical shift perturbations were then mapped onto the surface of the SAV1430 structure (PDB ID: 1PQX) and visualized using VMD-XPLOR.
Rapid Determination of Protein-Ligand Co-structures
AutoDock was used to obtain co-structures, binding and docking energies for all of the 21 compounds identified to bind SAV1430 from the NMR experiments. Chemical shift changes identified a consensus binding site comprising residues: I6-P10, T14-K16 and I61-V63. A grid was manually defined within AutoDock to encompass this experimentally determined ligand-binding site by adjusting the x, y and z coordinates for the center of the grid box to position the grid in the binding pocket. The grid size is determined by the number of points in the x, y and z dimensions and is just visibly large enough to fit the entire ligand.
CPASS Database and Software for Functional Annotation
Comparison of Protein Active site Structures (CPASS) database and software was developed to aid in the functional annotation of hypothetical proteins by using a protein-ligand co-structure.9 The CPASS database contains ~26,000 ligand-defined active sites identified from structures deposited in the PDB. CPASS returns a similarity score (0–100%) and an interactive 3D graphically display of the structural alignment for each active site comparison. The CPASS software runs on 16-node Beowulf Linux cluster with a simple web-based interface. Each comparison averages ~40s requiring ~18 hrs. to complete a comparison against the entire database.
Bioinformatics Analysis of SAV1430
The SAV1430 PDB structure (http://www.ebi.ac.uk/dali/Interactive.html) was uploaded to the DALI web server to identify homologous 3D structures. The Dali output contains a list of aligned structures and corresponding Z-scores, where a higher Z-score indicates a better 3D fit. Identical structures yield a Z-score of ~30, similar structures a score of ~15, where scores below 2–3 are generally insignificant and mean the structures are dissimilar. Dali analysis of SAV1430 only identified structures with Z-scores <3, which included a number of hypothetical proteins of unknown functions or ferredoxin-like folds.
A web server (http://consurf.tau.ac.il/) interface for the ConSurf software enables a rapid identification of conserved residues by comparison with structural homologues of known functions and generating a phylogenic tree. ConSurf outputs an amino acid conservation score for each residue in the SAV1430 structure, where the highest score represents the highest evolutionary conserved residue. The scores are normalized such that the average score is zero. The ConSurf amino acids with high scores were color-coded onto the SAV1430 surface using VMD-XPLOR.
Rosetta Stone analysis (http://www.igs.cnrs-mrs.fr/FusionDB/main.html) of SAV1430 was done using FusionDB, a database of bacterial and archael gene fusion events. Submitting the protein sequence for SAV1430 indicated a gene fusion for proteins SAV1430 and SAV0936 based on hypothetical proteins Q92GV4 and the Q9ZCQ2 in Rickettsia conorii and Rickettsia proazeki, respectively.18 The quality of the predicted fusion event is measured by the separation index where a value >0.6 is indicative of a real gene fusion event. A separation index score of 0.79 was determined for the predicted fusion of genes SAV1430 and SAV0936.
BLAST, FASTA and ClustalW were used with the SAV1430 and SAV0936 sequences to identify bacterial proteins with homology to both SAV1430 and SAV0936. A prevalence of hypothetical proteins were identified from the sequence alignments, but a mixture of small (~80aa) and large (~190aa) NifU-like proteins corresponding to the C-terminal domain of the multi-domain NifU protein were also observed. Specifically, the sequence alignment of SAV1430 and SAV0936 against the C-terminal NifU domain from Brucella melitensis yielded 30% and 47% sequence identity, respectively.
Results and Discussion
Description of the FAST-NMR methodology
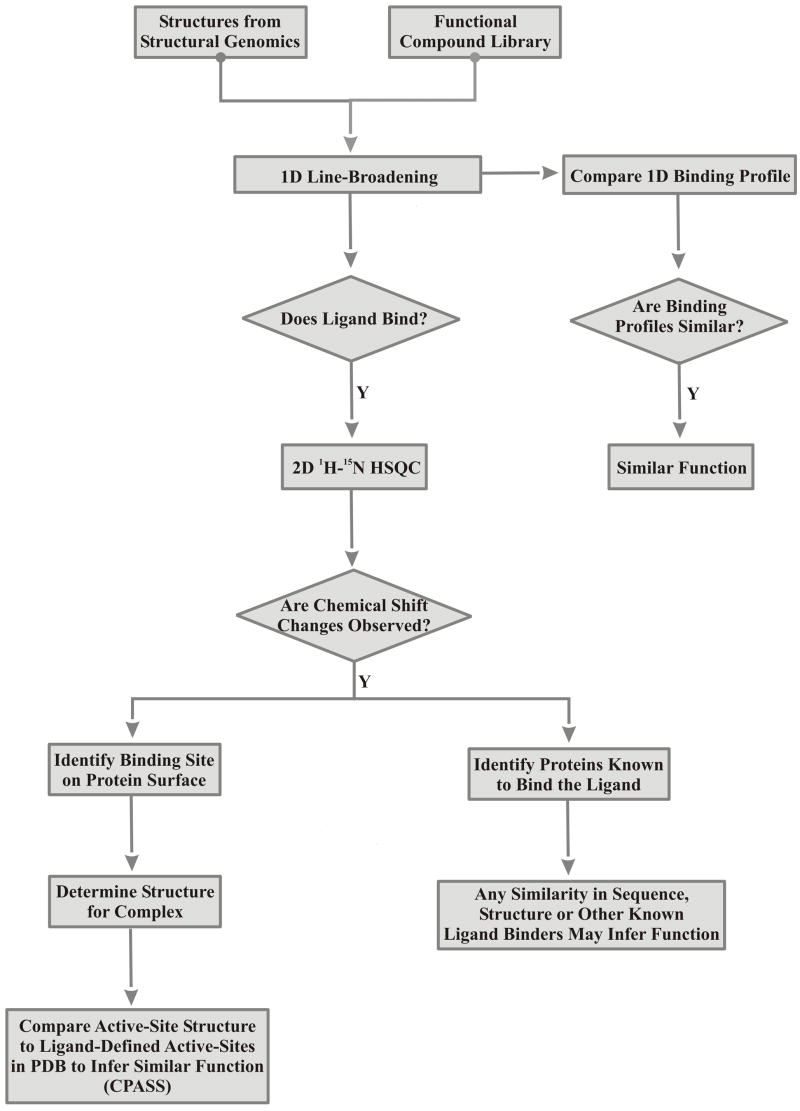
The overall protocol for the FAST-NMR assay is illustrated in Fig. 2. There are three major components to the process: (i) identify functional ligands that bind the protein, (ii) use the ligands to determine protein-ligand co-structures and (iii) use the co-structures with bioinformatics to infer function. An NMR based screen is used to determine which ligands from a chemical library bind a hypothetical protein that has been identified from structural genomics. The structure and NMR assignments for the hypothetical protein are available prior to initiating the screen. The chemical library that is screened in the FAST-NMR assay is composed of small molecules with defined functional and biological activity.16 This library is composed of aminoacids, carbohydrates, co-factors, fatty-acids, hormones, inhibitors, known drugs, metabolites, neurotransmitters, nucleotides, substrates and vitamins.
Figure 2.
Flow Chart for the FAST-NMR screen.
The tiered NMR screening assay minimizes valuable resources, such as protein samples and NMR instrument time, while increasing throughput. To further improve efficiency, ligands are screened as mixtures of 3–4 compounds based on a statistical analysis to optimize NMR based screens.19 The mixtures were designed to avoid deconvolution and maximize structural and functional diversity.
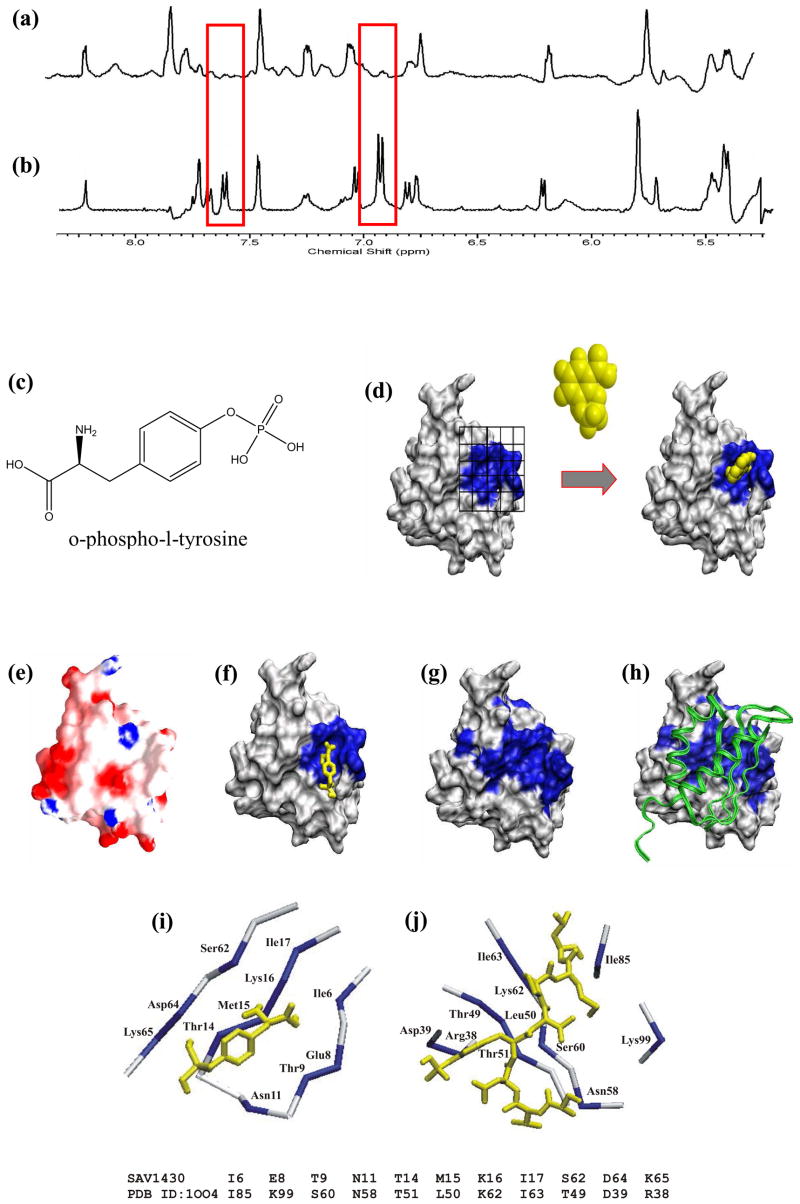
The first one-dimensional (1D) 1H line-broadening NMR experiment is most suitable for screening a larger number of compounds using minimal resources while providing preliminary binding information. A protein-ligand binding interaction is identified by a change in line-width or complete disappearance of the ligands 1H NMR resonances in the presence of the protein (Fig. 3a–b). Effectively, a bound ligand acquires the broad NMR line-widths of the large MW protein.
Figure 3.
FAST-NMR analysis of SAV1430. Example of 1D NMR line-broadening spectra obtained in the SAV1430 screen. Expanded NMR region corresponding to the aromatic residues (a) in the presence of SAV1430 and (b) the free mixture, illustrating the binding of SAV1430 by changes in observed NMR line-widths. SAV1430 surface where residues that incur a chemical shift change are darkened. (c) Chemical structure of O-phospho-l-tyrosine shown to bind SAV1430. (d) Illustration of the use of the NMR defined ligand binding site to direct a protein-ligand co-structure determination with AutoDock36. (e) GRASP37 electropotential surface where positive and negative regions are blue and red, respectively. (f) NMR-based ligand-defined SAV1430 binding site where residues that incur a chemical shift change upon binding phosphotyrosine are colored blue. (g) Mapping of functionally conserved residues (colored blue) identified by ConSurf 38 on the SAV1430 surface. (h) Molecular model of SAV1430- SE0630 (structural homolog of SAV0936) complex determined by Hex39. SE0630 is shown as a ribbon. Sequence and structural alignment of (i) SAV1430-pTyr binding site to the (j) Src SH2 domain’s phosphotyrosine peptide binding site (PDB ID: 1004). The sequence alignment is shown below the structure where the aligned residues are colored blue.
The second 2D 1H-15N HSQC NMR experiment is only conducted on positive “hits” from the first NMR experiment, because it is more resource intensive. This experiment monitors NMR chemical shifts for the backbone amide resonances for each amino acid residue in the protein, which has been previously assigned during the determination of the protein’s NMR structure.20 These backbone amide chemical shifts are sensitive to the local environment and change proportionally to the residues’ proximity to a bound ligand. The residues that incur a chemical shift change in the presence of the bound ligand can be mapped onto the protein surface to identify the ligand binding site (Fig. 3d). Non-specific binders will exhibit a random distribution or complete lack of chemical shift changes.
A protein-ligand co-structure is rapidly generated by combining this experimentally determined ligand binding site with AutoDock. A grid (Fig. 3d) is used to direct AutoDock to dock the ligand in this NMR binding site and determine the lowest energy conformation for the complex. Thus, AutoDock permits an initial structure of the protein-ligand complex to be calculated in minutes. The AutoDock derived co-structure provides a complete and detailed description of the ligand binding site. The chemical-shift perturbation mapping of the ligand binding site is generally incomplete because of chemical shift overlap. Moreover, a lack of a perturbation in the NH NMR resonances may occur, because the primary interaction between the ligand and the amino-acid residue is with the side-chain instead of the backbone. Also, amino-acid residues not in direct contact with the ligand may incur a chemical shift perturbation because of local structural changes. Additionally, the NMR chemical shift perturbations only identify residues involved in an interaction with the ligand. The AutoDock co-structure also identifies relative distances between the ligand and the amino-acid residues.
The Comparison of Protein Active Site Structures (CPASS) database and software enables the comparison of experimentally identified ligand-binding sites to infer biological function.9 Proteins that exhibit a high similarity in the structural characteristics of their active sites would suggest a corresponding similarity in function. This is simply a further refinement of generally accepted paradigms where global sequence and structure homology are routinely used to annotate function.21 The AutoDock structure of the protein-ligand complex is used to generate a ligand-defined active site, which comprises any residue in the protein that contains at least one atom that is ≤ 6Å from any atom in the ligand. The entire PDB database has been similarly analyzed where each protein that contains a bound ligand had its corresponding ligand-defined active site extracted as part of the CPASS database. The ligand-defined active site identified from the FAST-NMR assay using AutoDock is then compared with the CPASS database using the CPASS software. CPASS iteratively compares the ligand-defined active site with the database to maximize both the sequence and structural alignments and reports a similarity score ranging from 0–100%. Protein(s) of known function that exhibit a high similarity with the unannotated protein’s active site would suggest potential functions for the unknown protein. It is important to note that the alignment of the ligand-defined active sites does not include the ligands, but relative distances between the ligands and the active site residues are used in the CPASS scoring function. The identity of the ligand(s) that bind the unannotated protein in conjunction with other available bioinformatics tools provide additional insight into possible functions for the unannotated protein.
Validation of FAST-NMR Using Staphylococcus aureus Protein SAV1430
Staphylococcus aureus (S. aureus) protein SAV1430 is an example of a protein of unknown biological function that is a typical target of NESG. Dali analysis suggests SAV1430 has a topology similar to ferredoxin-like folds; however, the Z-score of < 3 is not significant.22 The only significant sequence homology was to hypothetical proteins. Taken together, bioinformatic analysis alone failed to assign a reliable function to SAV1430. The NMR resonance assignments and corresponding solution structure for SAV1430 were determined by the effort at NESG.23, 24
The 1D NMR line-broadening experiments using our entire functional chemical library identified 21 compounds that bind SAV430 (Fig. 3a–b). The compounds exhibit similar structural features that are consistent with binding in an optimized active site for SAV1430 and suggestive of the natural ligand. The compounds are generally similar to O-phospho-L-tyrosine (pTyr). The structures tend to contain one or more aromatic rings that overlap with the phenyl ring in pTyr with one or more hydrophilic substituents that either overlaps with the amino acid or phosphate group in pTyr (e.g. 2-amino-4-methylphenol, nicotinic acid and acetylsalicylic acid).
These 21 compounds were further evaluated for SAV1430 binding in the second tiered 2D 1H-15N HSQC NMR experiments. Specifically, pTyr was identified as one of the compounds that exhibited higher chemical shift perturbations through a visual inspection of histogram plots of the chemical shift perturbations for the 21 compounds (Fig. 3c). Analysis of the 2D 1H-15N HSQC NMR spectra implies a distinct, consensus binding site that comprises residues I6-P10, T14-K16 and I61-V63. Based on these NMR results, protein-ligand co-structures have been generated using AutoDock targeting the identified binding pocket (Fig. 3d–f). This binding site corresponds to a shallow cleft on the SAV1430 surface surrounded by relatively flat structural features. A GRASP representation of the electropotential surface indicates that most of the ligand binding site is hydrophobic in nature (Fig. 3e). A small positive patch (Lys 16) and negative patch (Asp 64) are part of or proximal to the ligand binding site. The weak ligand binding interactions with SAV1430 in conjunction with the indistinct and generally flat hydrophobic structural characteristics of the consensus ligand binding site is strongly suggestive of a protein-protein interaction site.
ConSurf was used to identify residues in the SAV1430 structure that are conserved with homologous proteins with a known structure. These conserved residues are predicted to be functionally important to the biological activity of SAV1430 and were mapped onto the protein’s surface (Fig. 3g). The experimentally determined SAV1430 ligand binding site is consistent with the functionally conserved residues identified by ConSurf supporting the biological relevance of this region of SAV1430. Furthermore, the FAST-NMR defined ligand binding site corresponds to the most prominent structural feature (binding cleft) within the larger surface area described by ConSurf.
CPASS analysis of the SAV1430-pTyr binding site identified a Src SH2 domain complexed with a phosphotyrosine containing peptide, along with other SH2 domains, as a significant hit (37% similarity). Visual comparison of the two ligand binding sites clearly indicates a similarity in local structure, primarily a flat β-sheet, and sequence composition (Fig. 3i-j). SH2 domains are typically part of multidomain proteins involved in cell signaling and form a protein-protein complex with a kinase after phosphorylation of a tyrosine.25 Phosphorylation of Ser, Thr and Tyr are also common mechanisms for regulating protein activity in bacteria.26, 27 The similarity in the characteristics of the SAV1430 and Src SH2 ligand binding sites and the fact that SAV1430 binds pTyr further supports the general proposal that SAV1430 functions by forming a protein-protein complex.
Bioinformatics Analysis of FAST-NMR Results of SAV1430
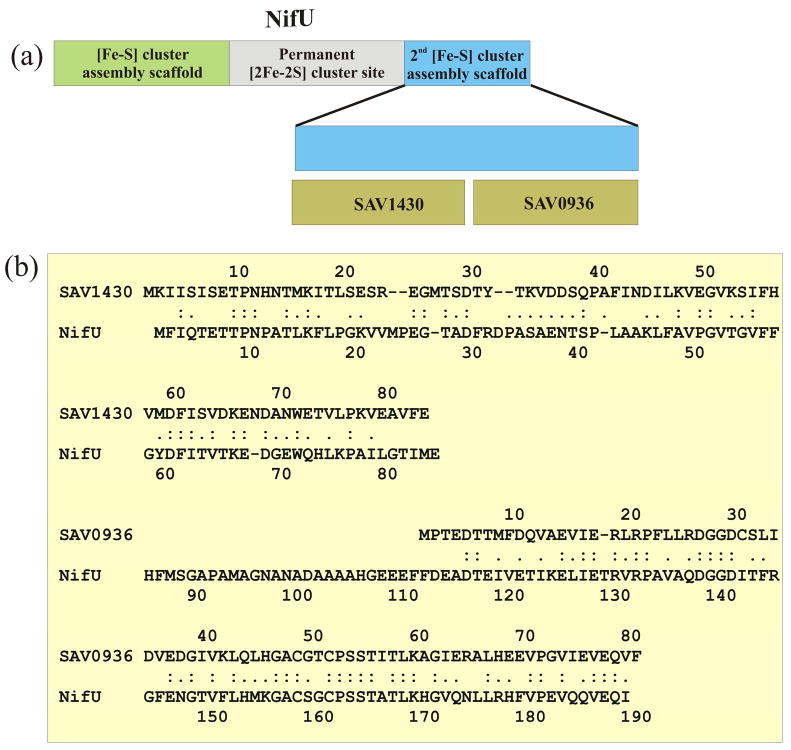
The NIF (nitrogen fixation), ISC (iron-sulfur cluster) and SUF (sulfur) [Fe-S] cluster assembly networks are essential for the viability of bacteria.28, 29 The assembly of [Fe-S] clusters is a complex, poorly understood process involving multiple proteins. [Fe-S] clusters are utilized in >100 proteins for a variety of functions. NifU is a multi-protein complex within the NIF [Fe-S] cluster assembly network that is involved in the overall biosynthesis of FeMo-cofactor. The current viewpoint is that NifU provides a scaffold for the assembly of transient [2Fe–2S] units for the formation of a [Fe–S] cluster in nitrogenase, an important component of biological nitrogen fixation. The [Fe-S] assembly scaffold is located in the N-terminus of the NifU homodimer structure, the central region of the protein contains a permanent [2FE-2S] cluster and the C-terminus appears to contain a second transient [Fe-S] assembly scaffold. NifU functions as a homodimer, where a number of proteins have been identified as components of the FeMo-cofactor biosynthesis pathway. NifU has been shown to form a macromolecular complex with NifS.30 Also, the identification of two transient [Fe-S] cluster assembly scaffolds within NifU suggests target specificity between NifU and the other components of the FeMo-cofactor biosynthesis pathway. The assembly of macromolecular complexes appears to be an important component of the FeMo-cofactor biosynthesis pathway.
SAV1430 is homologous to the N-terminal domain of the S. cerivisae NIFU protein. Rosetta Stone analysis reveals that the C-terminal domain of S. cervivsae NIFU is also found in a distant part of the S. aureus genome as gene SAV0936. This analysis suggests that SAV0936 may be a binding partner of SAV1430. In order to validate the FAST-NMR active site prediction we were able to use a homolog of SAV0936, the Staphylococcus epidermidis protein SE063031, 32 (95% identity) to perform in-silico docking experiments. An unbiased model of a SAV1430-SE0630 complex was generated using Hex (Fig. 3h). Hex determines a protein-protein complex by identifying complementary binding sites based on surface shape and electrostatic charge and potential distributions. SE0630 was preferentially docked to SAV1430 in the same binding site determined by the NMR ligand binding studies. This further supports a protein-protein binding interaction as part of the biological activity of SAV1430.
SAV0936 exhibits 47% sequence identity with the N-terminal region of the C-terminal NifU domain (Fig. 4). SAV0936 contains a surface exposed CXXC loop motif that aligns with conserved cysteines in NifU and have been identified as [Fe-S] ligands, enriched in Ser and Thr residues (typical of regulatory phosphorylation sites) and required for growth.33 A more exhaustive sequence analysis of SAV1430, based on the results with SAV0936, indicates the protein shares ~30% sequence identity with the C-terminal region of the C-terminal domain of the NifU multidomain structure (Fig. 4). The observed ferredoxin-like fold for SAV1430 is consistent with the electron transfer activity in the [Fe-S] cluster assembly pathway.34 But, the lack of any cysteine residues in the SAV1430 sequence and an inability to bind iron indicate SAV1430 would not play a direct role in the electron transfer activity. SAV1430 and SAV0936 sequences account for the majority of the full length NifU domain. These results imply that SAV1430 would interact with SAV0936 to form a complex that exhibits similar activity as the full length NifU domain.
Figure 4.
(a) Cartoon illustration of the NifU domain structure and the sequence overlap with SAV1430 and SAV0936. (b) Sequence alignment of the C-terminal NifU domain from Brucella melitensis with SAV1430 (30% identity) and SAV0936 (47% identity).
The apparent requirement for prokaryotic SAV1430 to form a protein-protein complex, instead of an intact single domain observed in eukaryotic organisms, may suggest a protein assembly or regulation mechanism for SAV1430 in S. aureus. The proper assembly of the SAV1430-SAV0936 complex may be required for activity of the NifU-like domain or to provide a mechanism to regulate the [Fe-S] cluster assembly pathway. Potentially, phosphorylation of SAV0936 in the CXXC loop motif may regulate the SAV1430-SAV0936 complex formation in a manner similar to SH2 domains. Precedence for staphylococcus utilizing heterodimeric proteins to accomplish what other bacteria accomplish using monomeric proteins can be found in the enzyme serine dehydratase. In most bacteria, a single gene encodes the serine dehydratase; however, in staphylococci two genes (sdhA and sdhB) encode serine dehydratase.35
Conclusion
Our analysis of hypothetical S. aureus protein SAV1430 in the FAST-NMR screen suggests SAV1430 is part of a multi-protein complex within the [Fe-S] cluster assembly network, where SAV1430 may interact with SAV0936 to form a complex that exhibits similar activity as the intact C-terminus domain of NifU. Formation of the SAV1430-SAV0936 complex may be regulated in a manner similar to SH2 domains through the phosphorylation of SAV0936. It is also plausible that the SAV1430-SAV0936 complex may be used to regulate the [Fe-S] cluster assembly network. It is important to stress that the only information available for hypothetical protein SAV1430 at the beginning of the FAST-NMR screen was a novel fold and a sequence similar to other hypothetical proteins. Although the results of the FAST-NMR screen are not definitive, it provides important direction for further exploration of the biological function of SAV1430, SAV0936 and other similar hypothetical proteins. The success of structural genomics has resulted in hundreds of unannotated protein structures being deposited in the PDB. Inevitably, the inherent value of data generated by genomics sequencing and structural efforts is significantly reduced without new methodologies to provide annotation information.
Acknowledgments
This work was supported by grants from the Protein Structure Initiative of the National Institutes of Health (U54 GM074958), Nebraska Tobacco Settlement Biomedical Research Development Funds, Nebraska EPSCoR, and an Eloise Gerry Fellowship from Sigma Delta Epsilon. The research was performed in facilities renovated with support from NIH (RR015468-01).
References
- 1.Venter C, Adams MD, et al. Science. 2001;297:1304. [Google Scholar]
- 2.Burley SK. Nat Struct Biol. 2000;7:932. doi: 10.1038/80697. [DOI] [PubMed] [Google Scholar]
- 3.Frishman D, Mokrejs M, Kosykh D, Kastenmuller G, Kolesov G, Zubrzycki I, Gruber C, Geier B, Kaps A, Albermann K, Volz A, Wagner C, Fellenberg M, Heumann K, Mewes HW. Nucleic Acids Research. 2003;31:207. doi: 10.1093/nar/gkg005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. Nucleic Acids Research. 2004;32:D277. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell SJ, Gold ND, Jackson RM, Westhead DR. Current Opinion in Structural Biology. 2003;13:389. doi: 10.1016/s0959-440x(03)00075-7. [DOI] [PubMed] [Google Scholar]
- 6.Hasson MS, Schlichting I, Moulai J, Taylor K, Barrett W, Kenyon GL, Babbitt PC, Gerlt JA, Petsko GA, Ringe D. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:10396. doi: 10.1073/pnas.95.18.10396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsons JF, Lim K, Tempczyk A, Krajewski W, Eisenstein E, Herzberg O. Proteins. 2002;46:393. doi: 10.1002/prot.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eswaramoorthy S, Gerchman S, Graziano V, Kycia H, Studier FW, Swaminathan S. Acta Crystallographica Section D: Biological Crystallography. 2003;59:127. doi: 10.1107/s0907444902018012. [DOI] [PubMed] [Google Scholar]
- 9.Powers R, Copeland J, Germer K, Mercier KA, Ramanathan V, Revesz P. Proteins. 2006;65:124. doi: 10.1002/prot.21092. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Hegyi H, Acton TB, Montelione GT, Rost B. Proteins. 2004;56:188. doi: 10.1002/prot.20012. [DOI] [PubMed] [Google Scholar]
- 11.Kanelis V, Forman-Kay JD, Kay LE. IUBMB Life. 2001;52:291. doi: 10.1080/152165401317291147. [DOI] [PubMed] [Google Scholar]
- 12.Tugarinov V, Kay LE. ChemBioChem. 2005;6:1567. doi: 10.1002/cbic.200500110. [DOI] [PubMed] [Google Scholar]
- 13.Snyder DA, Chen Y, Denissova NG, Acton T, Aramini JM, Ciano M, Karlin R, Liu J, Manor P, Rajan PA, Rossi P, Swapna GVT, Xiao R, Rost B, Hunt J, Montelione GT. Journal of the American Chemical Society. 2005;127:16505. doi: 10.1021/ja053564h. [DOI] [PubMed] [Google Scholar]
- 14.Yee AA, Savchenko A, Ignachenko A, Lukin J, Xu X, Skarina T, Evdokimova E, Liu CS, Semesi A, Guido V, Edwards AM, Arrowsmith CH. Journal of the American Chemical Society. 2005;127:16512. doi: 10.1021/ja053565+. [DOI] [PubMed] [Google Scholar]
- 15.Liu ZJ, Shah AK, Habel JE, Ng JD, Kataeva I, Xu H, Horanyi P, Yang H, Chang J, Zhao M, Huang L, Chang S, Tempel W, Chen L, Zhou W, Lee D, Lin D, Zhang H, Newton MG, Rose J, Wang BC. Journal of Structural and Functional Genomics. 2005;6:121. doi: 10.1007/s10969-005-5242-x. [DOI] [PubMed] [Google Scholar]
- 16.Mercier KA, Germer K, Powers R. Combinatorial Chemistry and High Throughput Screening. 2006;9:515. doi: 10.2174/138620706777935342. [DOI] [PubMed] [Google Scholar]
- 17.Ramanathan V, Mercier K, Powers R, Revesz P. Using Databases and Computational Techniques to Infer the Function of Novel Proteins. 2005 IEEE International Conference on Electro Information Technology; Lincoln, NE. [Google Scholar]
- 18.Renesto P, Ogata H, Audic S, Claverie JM, Raoult D. FEMS Microbiology Reviews. 2005;29:99. doi: 10.1016/j.femsre.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Mercier KA, Powers R. Journal of Biomolecular NMR. 2005;31:243. doi: 10.1007/s10858-005-0948-4. [DOI] [PubMed] [Google Scholar]
- 20.Ferentz AE, Wagner G. Quarterly Reviews of Biophysics. 2000;33:29. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 21.Whisstock JC, Lesk AM. Quarterly Reviews of Biophysics. 2003;36:307. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]
- 22.Dietmann S, Park J, Notredame C, Heger A, Lappe M, Holm L. Nucleic Acids Research. 2001;29:55. doi: 10.1093/nar/29.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baran MC, Aramini JM, Huang YJ, Xiao R, Acton TB, Shih L-y, Montelione GT. Solution Structure Determination of the Staphylococcus Aureus Hypothetical Protein SAV1430. Northeast Structure Consortium target ZR18. Department of Biochemistry, University of Wisconson-Madison; 2003. BMRB accession number 5844. [Google Scholar]
- 24.Baran MC, Aramini JM, Xiao R, Huang YJ, Acton TB, Shih L, Montelione GT. Solution Structure of the Hypothetical Staphylococcus Aureus protein SAV1430. Northest Strucutral Genomics Consortium target ZR18. 2003. PDBID: 1PQX. [Google Scholar]
- 25.Marengere LEM, Pawson T. Journal of Cell Science, Supplement. 1994;18:97. doi: 10.1242/jcs.1994.supplement_18.14. [DOI] [PubMed] [Google Scholar]
- 26.Kennelly PJ, Potts M. Journal of Bacteriology. 1996;178:4759. doi: 10.1128/jb.178.16.4759-4764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alzari PM. Structure (Cambridge, MA, United States) 2004;12:1923. doi: 10.1016/j.str.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Schilke B, Voisine C, Beinert H, Craig E. Proceedings of the National Academy of Sciences of the United States of America. Vol. 96. 1999. p. 10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson JW, Agar JN, Johnson MK, Maier RJ. Biochemistry. 2000;39:16213. doi: 10.1021/bi001744s. [DOI] [PubMed] [Google Scholar]
- 30.Yuvaniyama P, Agar JN, Cash VL, Johnson MK, Dean DR. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:599. doi: 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baran MC, Huang YP, Acton T, Xiao R, Montelione GT. Solution Structure Of The Staphylococcus Epidermis Protein SE0936. Northest Strucutral Genomics Consortium Target SeR8. Department of Biochemistry, University of Wisconson-Madison; 2004. BMRB accession number 6355. [Google Scholar]
- 32.Baran MC, Huang YP, Acton T, Xiao R, Montelione GT. Solution Structure Of The Staphylococcus Epidermidis Protein SE0630. Northest Strucutral Genomics Consortium Target SeR8. 2004. PDB-ID: 1XHJ. [Google Scholar]
- 33.Dos Santos PC, Smith AD, Frazzon J, Cash VL, Johnson MK, Dean DR. Journal of Biological Chemistry. 2004;279:19705. doi: 10.1074/jbc.M400278200. [DOI] [PubMed] [Google Scholar]
- 34.Bertini I, Luchinat C, Provenzani A, Rosato A, Vasos PR. Proteins. 2001;46:110. doi: 10.1002/prot.10009. [DOI] [PubMed] [Google Scholar]
- 35.Ogawa H. Trends in Comparative Biochemistry & Physiology. 2000;6:1. [Google Scholar]
- 36.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Journal of Computational Chemistry. 1998;19:1639. [Google Scholar]
- 37.Petrey D, Honig B. Methods in Enzymology. 2003;374:492. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- 38.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. Bioinformatics. 2003;19:163. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 39.Ritchie DW. Proteins. 2003;52:98. doi: 10.1002/prot.10379. [DOI] [PubMed] [Google Scholar]