Abstract
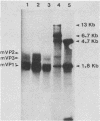
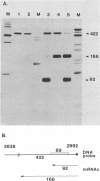
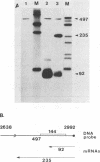
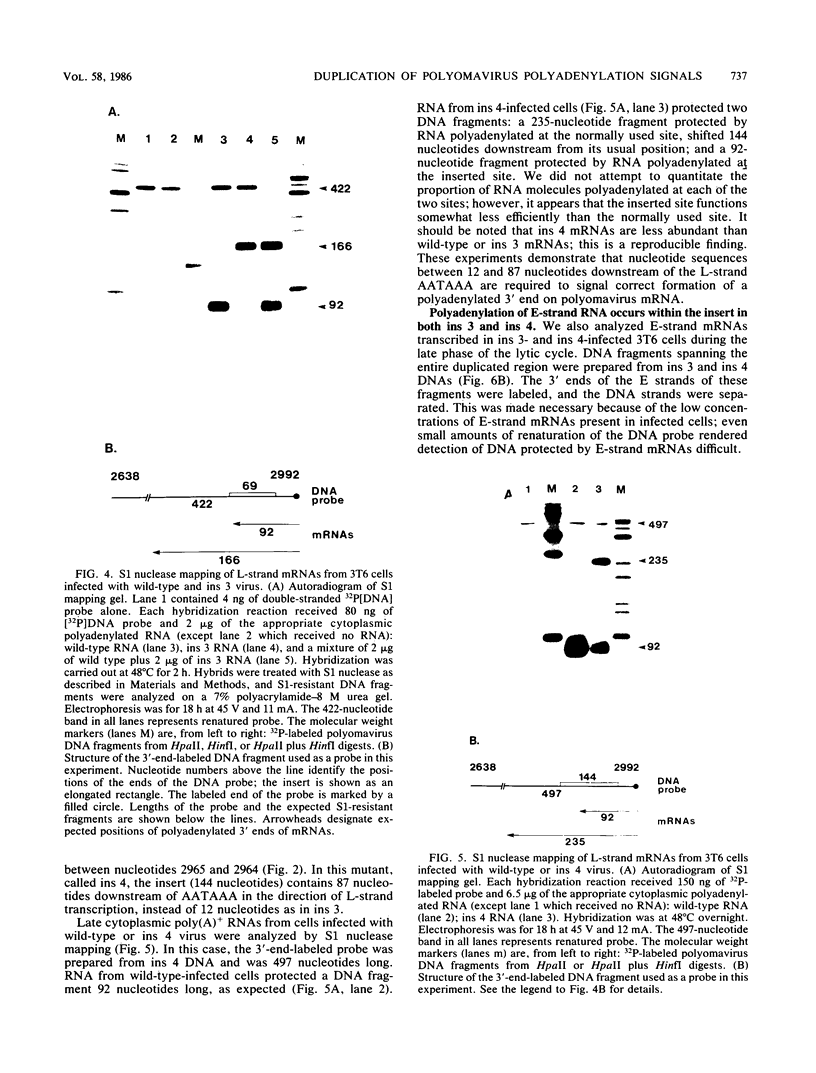
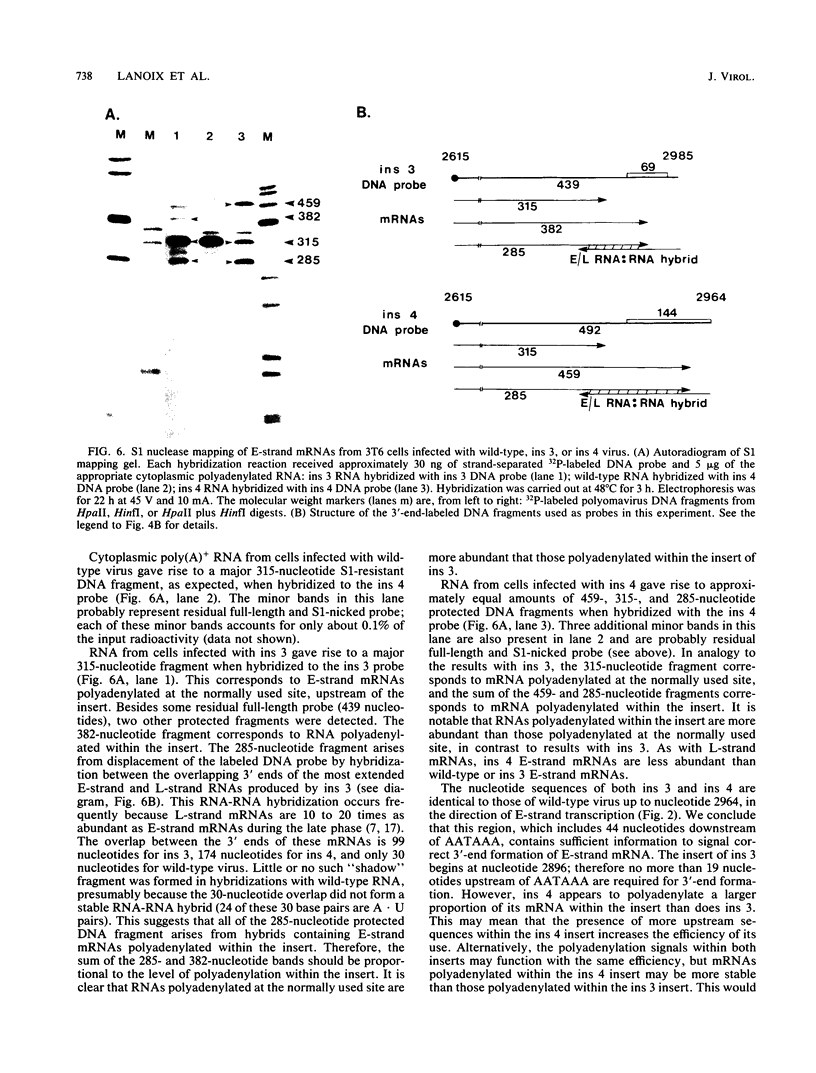
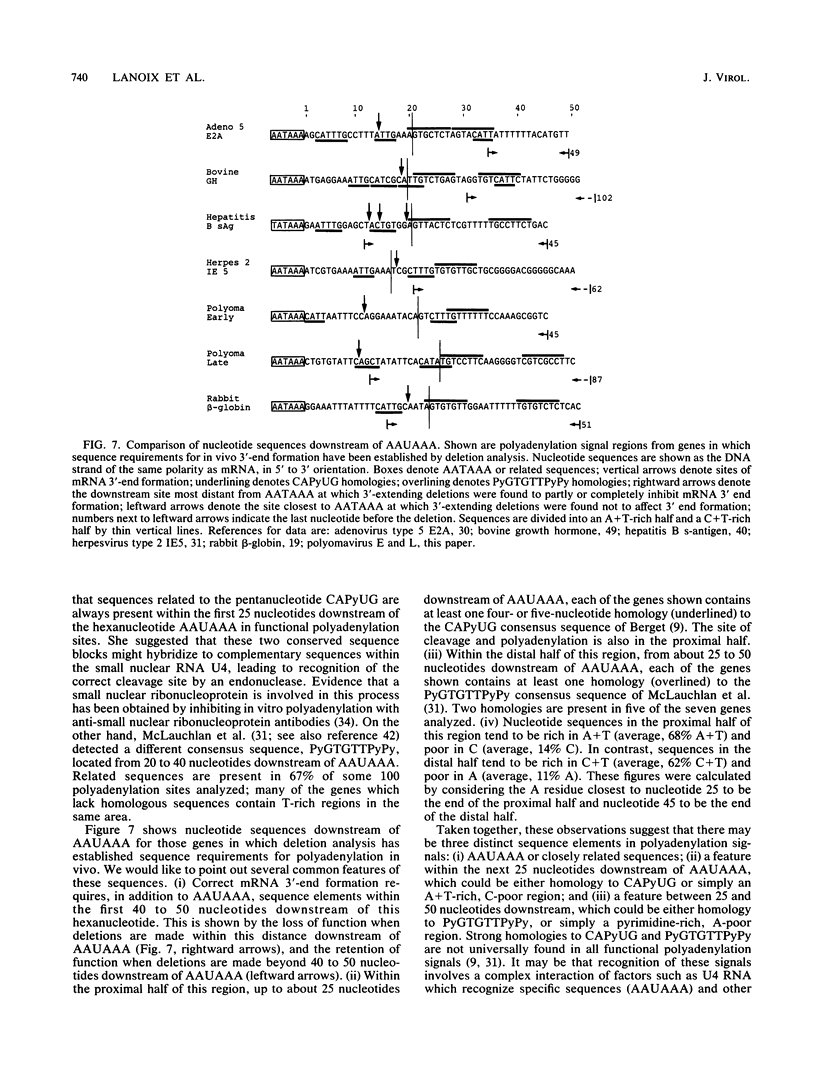
We constructed viable insertion mutants of polyomavirus that contain duplications of the nucleotide sequences surrounding the polyadenylation sites for both E- and L-strand RNAs. Our results showed that formation of poly(A)+ 3'termini of L-strand mRNAs requires sequence elements located between 12 and 87 nucleotides downstream of AAUAAA. No more than 19 nucleotides upstream and 44 nucleotides downstream of AAUAAA are required for polyadenylation of E-strand mRNAs. Our results and those of others suggest that there are three distinct sequence elements required for mRNA 3' end formation: AAUAAA and two downstream elements. An insertion mutant containing two adjacent functional polyadenylation signals produced E-strand and L-strand mRNAs with 3' ends at both sites. However, the overall level of polyadenylation of L-strand RNAs was not increased over the low (10 to 25%) levels seen with wild-type virus. Neither was the efficiency of termination of L-strand transcription increased in mutant virus-infected cells. We conclude that factors required for both polyadenylation and transcription termination are limiting in polyomavirus-infected mouse cells.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Acheson N. H. Efficiency of processing of viral RNA during the early and late phases of productive infection by polyoma virus. J Virol. 1981 Feb;37(2):628–635. doi: 10.1128/jvi.37.2.628-635.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson N. H. Kinetics and efficiency of polyadenylation of late polyomavirus nuclear RNA: generation of oligomeric polyadenylated RNAs and their processing into mRNA. Mol Cell Biol. 1984 Apr;4(4):722–729. doi: 10.1128/mcb.4.4.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson N. H., Miéville F. Extent of transcription of the E strand of polyoma virus DNA during the early phase of productive infection. J Virol. 1978 Dec;28(3):885–894. doi: 10.1128/jvi.28.3.885-894.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson N. H. Polyoma virus giant RNAs contain tandem repeats of the nucleotide sequence of the entire viral genome. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4754–4758. doi: 10.1073/pnas.75.10.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akusjärvi G., Persson H. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature. 1981 Jul 30;292(5822):420–426. doi: 10.1038/292420a0. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Shavit R., Laub O., Aloni Y. The frequencies of transcription from the E- and L-strands of polyoma DNA. J Gen Virol. 1978 May;39(2):357–360. doi: 10.1099/0022-1317-39-2-357. [DOI] [PubMed] [Google Scholar]
- Berget S. M. Are U4 small nuclear ribonucleoproteins involved in polyadenylation? Nature. 1984 May 10;309(5964):179–182. doi: 10.1038/309179a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Carmichael G. G., McMaster G. K. The analysis of nucleic acids in gels using glyoxal and acridine orange. Methods Enzymol. 1980;65(1):380–391. doi: 10.1016/s0076-6879(80)65049-6. [DOI] [PubMed] [Google Scholar]
- Chiu N. H., Radonovich M. F., Thoren M. M., Salzman N. P. Selective degradation of newly synthesized nonmessenger simian virus 40 transcripts. J Virol. 1978 Nov;28(2):590–599. doi: 10.1128/jvi.28.2.590-599.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron B., Falck-Pedersen E., Salditt-Georgieff M., Darnell J. E., Jr Transcription termination occurs within a 1000 base pair region downstream from the poly(A) site of the mouse beta-globin (major) gene. Nucleic Acids Res. 1984 Nov 26;12(22):8723–8731. doi: 10.1093/nar/12.22.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Santangelo G. M. Analysis in Cos-1 cells of processing and polyadenylation signals by using derivatives of the herpes simplex virus type 1 thymidine kinase gene. Mol Cell Biol. 1983 Feb;3(2):267–279. doi: 10.1128/mcb.3.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck-Pedersen E., Logan J., Shenk T., Darnell J. E., Jr Transcription termination within the E1A gene of adenovirus induced by insertion of the mouse beta-major globin terminator element. Cell. 1985 Apr;40(4):897–905. doi: 10.1016/0092-8674(85)90349-6. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Kamen R. Strand-specific transcription of polyoma virus DNA late during productive infection. J Mol Biol. 1977 Sep 15;115(2):237–242. doi: 10.1016/0022-2836(77)90099-7. [DOI] [PubMed] [Google Scholar]
- Ford J. P., Hsu M. T. Transcription pattern of in vivo-labeled late simian virus 40 RNA: equimolar transcription beyond the mRNA 3' terminus. J Virol. 1978 Dec;28(3):795–801. doi: 10.1128/jvi.28.3.795-801.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Wellauer P. K., Cribbs D. L., Schibler U. Termination of transcription in the mouse alpha-amylase gene Amy-2a occurs at multiple sites downstream of the polyadenylation site. Cell. 1984 Oct;38(3):737–744. doi: 10.1016/0092-8674(84)90269-1. [DOI] [PubMed] [Google Scholar]
- Heiser W. C., Eckhart W. Polyoma virus early and late mRNAs in productively infected mouse 3T6 cells. J Virol. 1982 Oct;44(1):175–188. doi: 10.1128/jvi.44.1.175-188.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann R., Neugebauer K., Pirkl E., Zentgraf H., Schaller H. Conversion of bacteriophage fd into an efficient single-stranded DNA vector system. Mol Gen Genet. 1980 Jan;177(2):231–242. doi: 10.1007/BF00267434. [DOI] [PubMed] [Google Scholar]
- Higgs D. R., Goodbourn S. E., Lamb J., Clegg J. B., Weatherall D. J., Proudfoot N. J. Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature. 1983 Nov 24;306(5941):398–400. doi: 10.1038/306398a0. [DOI] [PubMed] [Google Scholar]
- Hofer E., Darnell J. E., Jr The primary transcription unit of the mouse beta-major globin gene. Cell. 1981 Feb;23(2):585–593. doi: 10.1016/0092-8674(81)90154-9. [DOI] [PubMed] [Google Scholar]
- Kahn M., Kolter R., Thomas C., Figurski D., Meyer R., Remaut E., Helinski D. R. Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol. 1979;68:268–280. doi: 10.1016/0076-6879(79)68019-9. [DOI] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J. Topography of the three late mRNA's of polyoma virus which encode the virion proteins. J Virol. 1980 Feb;33(2):637–651. doi: 10.1128/jvi.33.2.637-651.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legon S., Flavell A. J., Cowie A., Kamen R. Amplification in the leader sequence of late polyoma virus mRNAs. Cell. 1979 Feb;16(2):373–388. doi: 10.1016/0092-8674(79)90013-8. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Sharp P. A., Gefter M. L. Rna synthesis in isolated nuclei processing of adenovirus serotype 2 late messenger rna precursors. J Mol Biol. 1982 Aug 25;159(4):581–599. doi: 10.1016/0022-2836(82)90102-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T. J., Stephens D. L., Mertz J. E. Kinetics of accumulation and processing of simian virus 40 RNA in Xenopus laevis oocytes injected with simian virus 40 DNA. Mol Cell Biol. 1982 Dec;2(12):1581–1594. doi: 10.1128/mcb.2.12.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Moore C. L., Sharp P. A. Site-specific polyadenylation in a cell-free reaction. Cell. 1984 Mar;36(3):581–591. doi: 10.1016/0092-8674(84)90337-4. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Darnell J. E., Jr Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978 Dec;15(4):1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Nevins J. R., Wilson M. C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981 Mar 12;290(5802):113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Ray D. S., Schekman R. W. Replication of bacteriophage M13. I. Sedimentation analysis of crude lysates of M13-infected bacteria. Biochim Biophys Acta. 1969 Apr 22;179(2):398–407. [PubMed] [Google Scholar]
- Simonsen C. C., Levinson A. D. Analysis of processing and polyadenylation signals of the hepatitis B virus surface antigen gene by using simian virus 40-hepatitis B virus chimeric plasmids. Mol Cell Biol. 1983 Dec;3(12):2250–2258. doi: 10.1128/mcb.3.12.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Taya Y., Devos R., Tavernier J., Cheroutre H., Engler G., Fiers W. Cloning and structure of the human immune interferon-gamma chromosomal gene. EMBO J. 1982;1(8):953–958. doi: 10.1002/j.1460-2075.1982.tb01277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]
- Tosi M., Young R. A., Hagenbüchle O., Schibler U. Multiple polyadenylation sites in a mouse alpha-amylase gene. Nucleic Acids Res. 1981 May 25;9(10):2313–2323. doi: 10.1093/nar/9.10.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Kamen R. Structure of polyoma virus late nuclear RNA. J Mol Biol. 1981 May 25;148(3):273–301. doi: 10.1016/0022-2836(81)90539-8. [DOI] [PubMed] [Google Scholar]
- Tseng R. W., Acheson N. H. Use of a novel S1 nuclease RNA-mapping technique to measure efficiency of transcription termination on polyomavirus DNA. Mol Cell Biol. 1986 May;6(5):1624–1632. doi: 10.1128/mcb.6.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M., Stephenson P. Role of the conserved AAUAAA sequence: four AAUAAA point mutants prevent messenger RNA 3' end formation. Science. 1984 Nov 30;226(4678):1045–1051. doi: 10.1126/science.6208611. [DOI] [PubMed] [Google Scholar]
- Woychik R. P., Lyons R. H., Post L., Rottman F. M. Requirement for the 3' flanking region of the bovine growth hormone gene for accurate polyadenylylation. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3944–3948. doi: 10.1073/pnas.81.13.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]