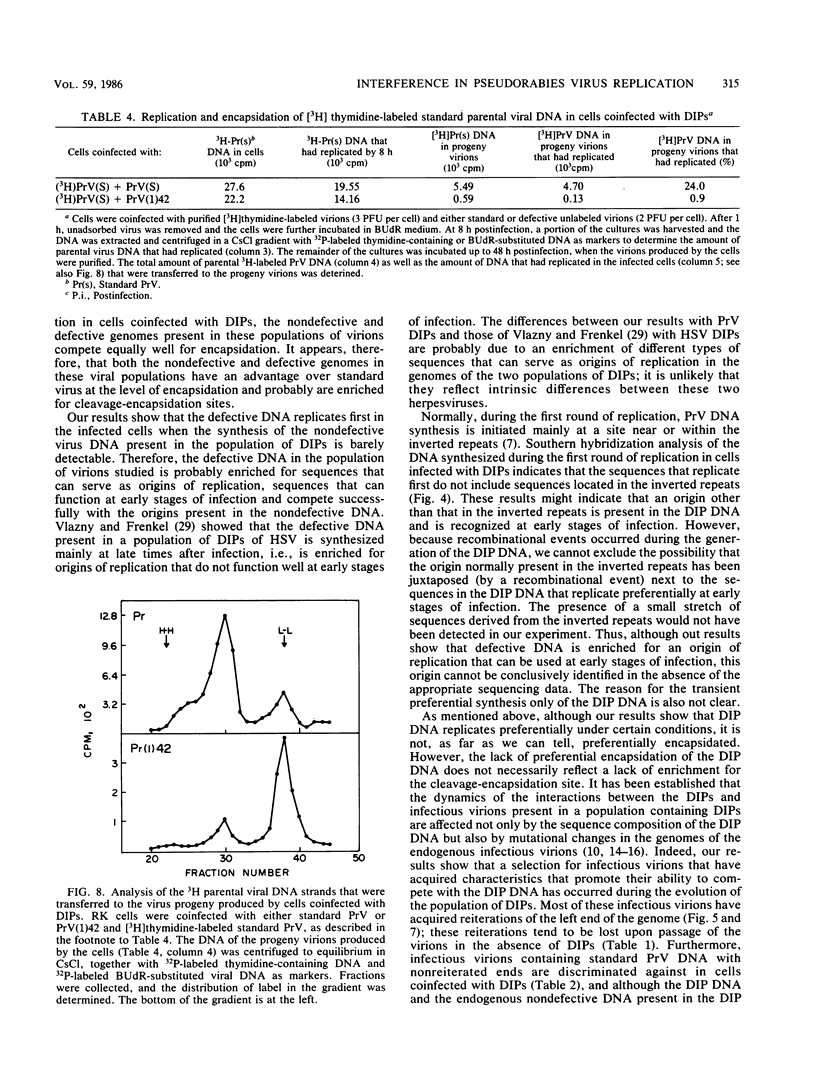
Abstract
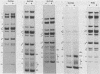
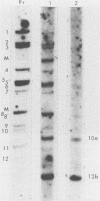
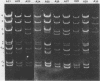
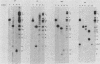
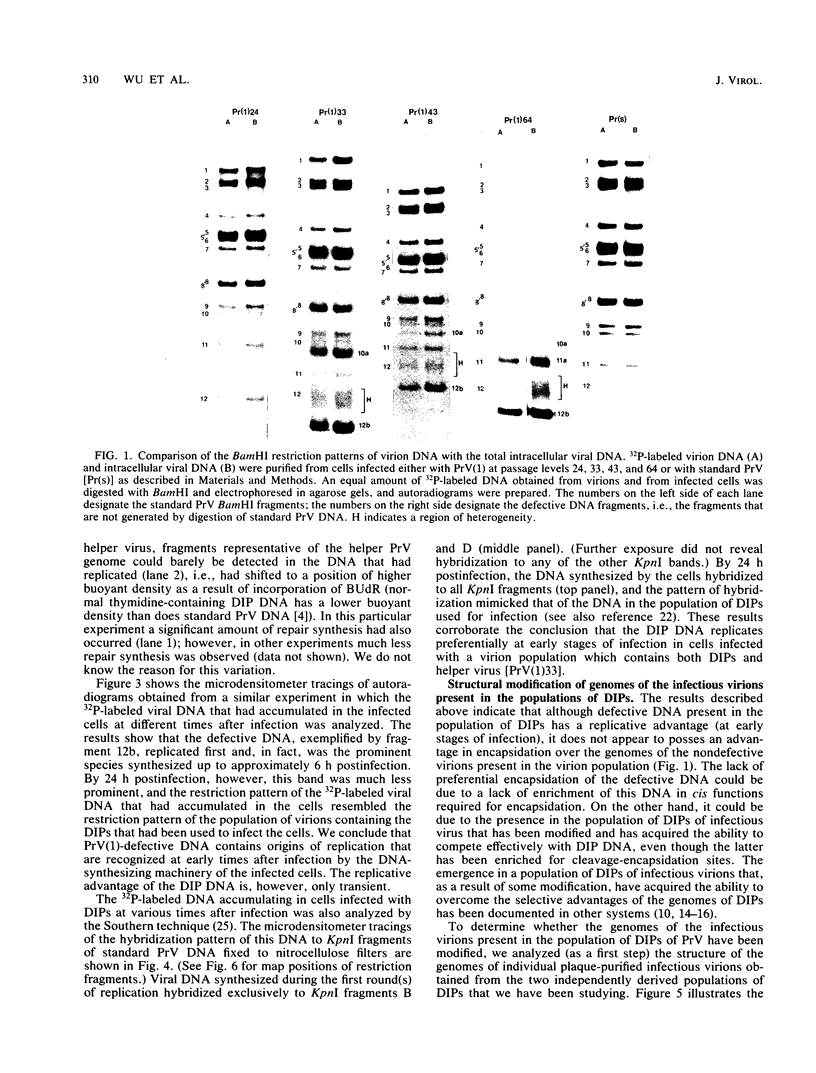
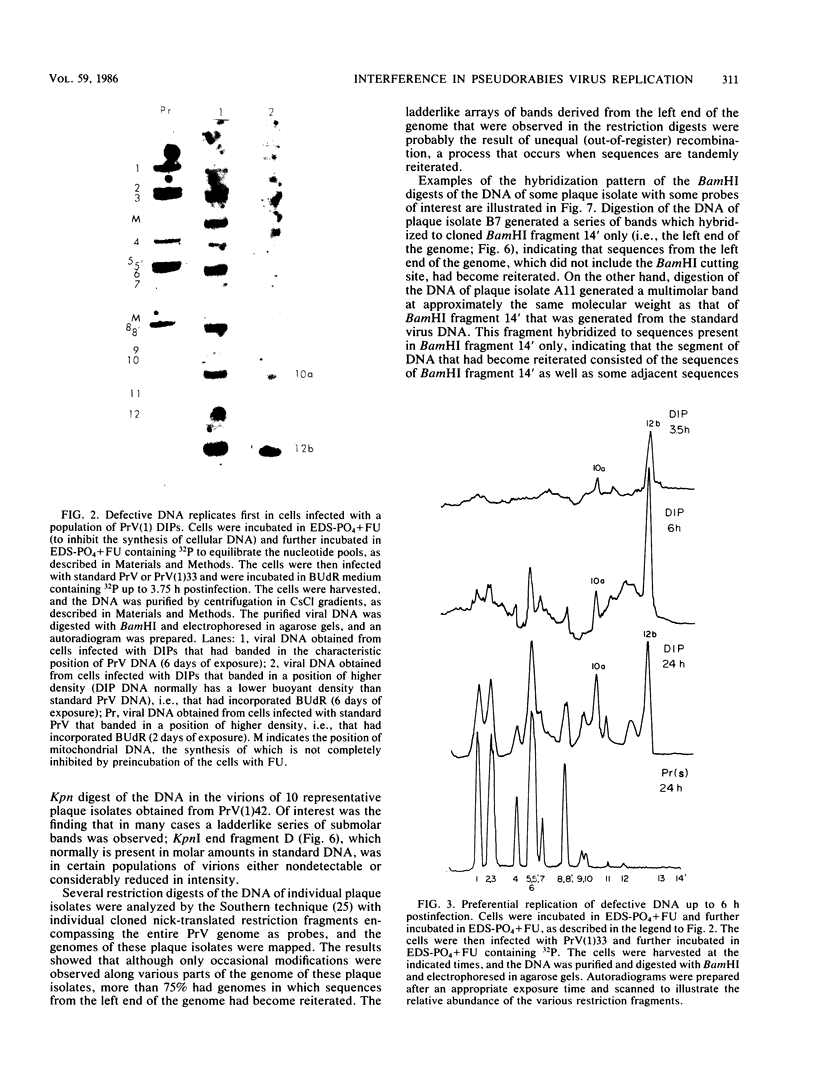
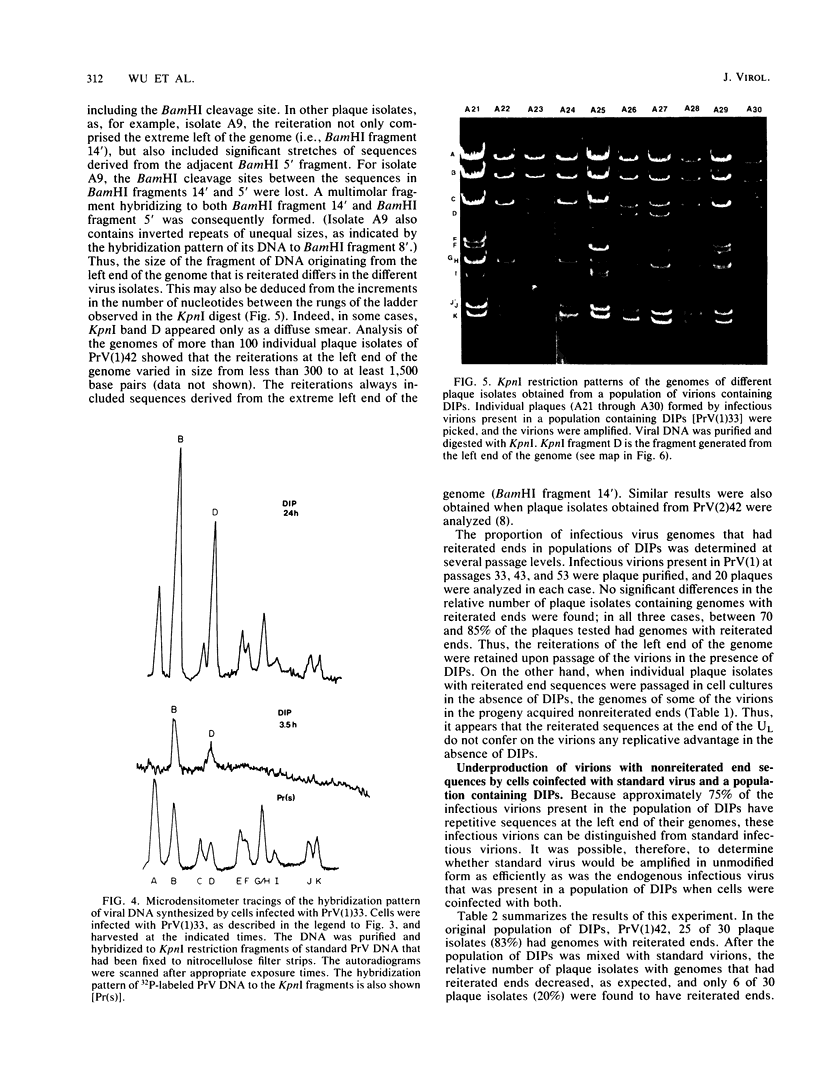
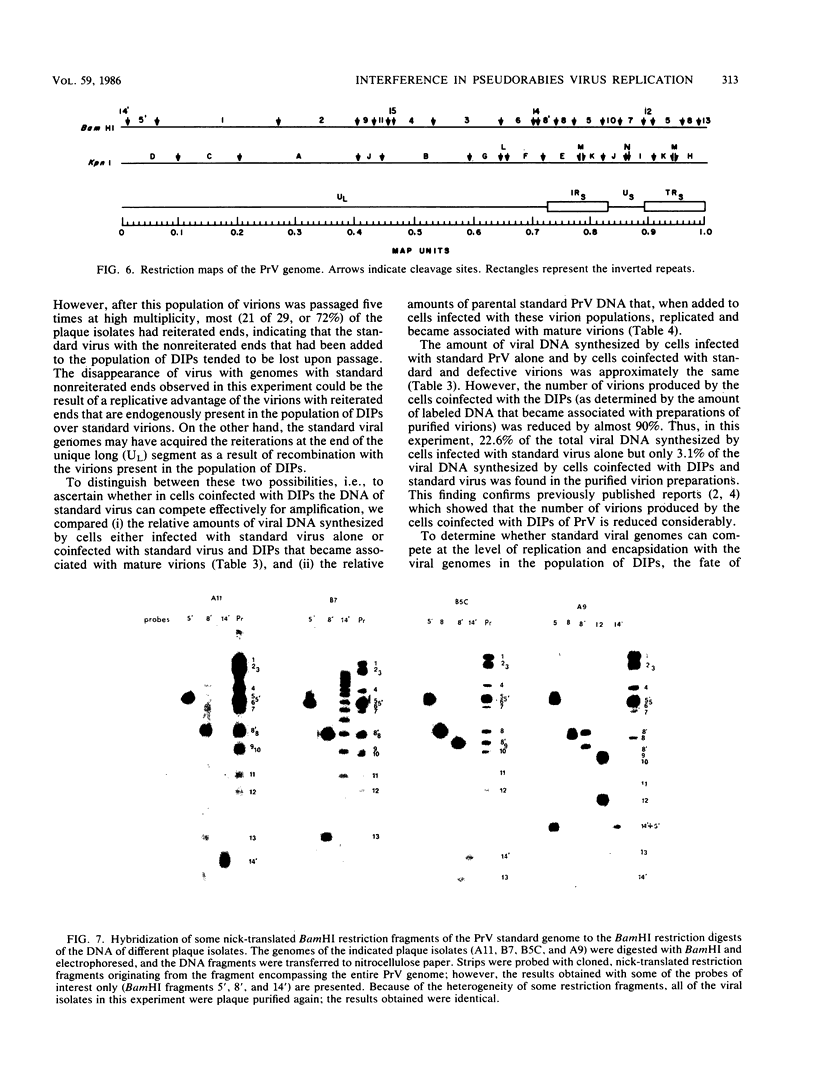
Serial passage of pseudorabies virus (PrV) at high multiplicity yields defective interfering particles (DIPs), but the sharp cyclical increases and decreases in titer of infectious virus that are observed upon continued passage at high multiplicity of most DIPs of other viruses are not observed with DIPs of PrV (T. Ben-Porat and A. S. Kaplan, Virology 72:471-479). We have studied the dynamics of the interactions of the virions present in a population of DIPs to assess the cis functions for which the genomes of the DIPs are enriched. The defective genomes present in one population of DIPs, [PrV(1)42], replicate preferentially over the nondefective genomes present in that virion population at early stages of infection, indicating that the DIP DNA is enriched for sequences that can serve as origins of replication at early stages of infection. This replicative advantage of the DIP DNA is transient and disappears at later stages of infection. The defective DNA does not appear to be encapsidated preferentially over the nondefective DNA present in this virion population, which might indicate that it is not enriched for cleavage-encapsidation sites. However, the nondefective DNA in the DIP virion population has become modified and has acquired reiterations of sequences originating from the end of the unique long (UL) region of the genome. Furthermore, both the infectious and defective genomes present in the DIP population compete for encapsidation more effectively than do the genomes of standard PrV. These results indicate that the defective genomes in the population of virions studied are enriched not only for an origin of replication but probably also for sequences necessary for efficient cleavage-encapsidation. Furthermore, the nondefective genomes present in this population of DIPs have also been modified and have acquired the ability to compete with the defective genomes for cleavage-encapsidation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BEN-PORAT T., KAPLAN A. S. The synthesis and fate of pseudorabies virus DNA in infected mammalian cells in the stationary phase of growth. Virology. 1963 Jun;20:310–317. doi: 10.1016/0042-6822(63)90120-x. [DOI] [PubMed] [Google Scholar]
- Barnett J. W., Eppstein D. A., Chan H. W. Class I defective herpes simplex virus DNA as a molecular cloning vehicle in eucaryotic cells. J Virol. 1983 Nov;48(2):384–395. doi: 10.1128/jvi.48.2.384-395.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porat T., Demarchi J. M., Kaplan A. S. Characterization of defective interfering viral particles present in a population of pseudorabies virions. Virology. 1974 Sep;61(1):29–37. doi: 10.1016/0042-6822(74)90239-6. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Kaplan A. S. A comparison of two populations of defective, interfering pseudorabies virus particles. Virology. 1976 Jul 15;72(2):471–479. doi: 10.1016/0042-6822(76)90175-6. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Rixon F. J. Replication of herpesvirus DNA. IV: analysis of concatemers. Virology. 1979 Apr 15;94(1):61–70. doi: 10.1016/0042-6822(79)90438-0. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Stehn B., Kaplan A. S. Fate of parental herpesvirus DNA. Virology. 1976 Jun;71(2):412–422. doi: 10.1016/0042-6822(76)90369-x. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Veach R. A. Origin of replication of the DNA of a herpesvirus (pseudorabies). Proc Natl Acad Sci U S A. 1980 Jan;77(1):172–175. doi: 10.1073/pnas.77.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EAGLE H. Amino acid metabolism in mammalian cell cultures. Science. 1959 Aug 21;130(3373):432–437. doi: 10.1126/science.130.3373.432. [DOI] [PubMed] [Google Scholar]
- Enea V., Zinder N. D. Interference resistant mutants of phage f1. Virology. 1982 Oct 15;122(1):222–226. doi: 10.1016/0042-6822(82)90395-6. [DOI] [PubMed] [Google Scholar]
- Frenkel N., Jacob R. J., Honess R. W., Hayward G. S., Locker H., Roizman B. Anatomy of herpes simplex virus DNA. III. Characterization of defective DNA molecules and biological properties of virus populations containing them. J Virol. 1975 Jul;16(1):153–167. doi: 10.1128/jvi.16.1.153-167.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkeĺ N., Locker H., Batterson W., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. VI. Defective DNA originates from the S component. J Virol. 1976 Nov;20(2):527–531. doi: 10.1128/jvi.20.2.527-531.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B. E., Newcomb W. W., O'Callaghan D. J. Biological and biochemical properties of defective interfering particles of equine herpesvirus type 1. Virology. 1979 Jan 30;92(2):495–506. doi: 10.1016/0042-6822(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Horiuchi K. Co-evolution of a filamentous bacteriophage and its defective interfering particles. J Mol Biol. 1983 Sep 15;169(2):389–407. doi: 10.1016/s0022-2836(83)80057-6. [DOI] [PubMed] [Google Scholar]
- Horodyski F. M., Holland J. J. Viruses isolated from cells persistently infected with vesicular stomatitis virus show altered interactions with defective interfering particles. J Virol. 1980 Nov;36(2):627–631. doi: 10.1128/jvi.36.2.627-631.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson S., Pfau C. J. Viral pathogenesis and resistance to defective interfering particles. Nature. 1980 Jan 17;283(5744):311–313. doi: 10.1038/283311a0. [DOI] [PubMed] [Google Scholar]
- KAPLAN A. S., BEN-PORAT T. The action of 5-fluorouracil on the nucleic acid metabolism of pseudorabies virus-infected and noninfected rabbit kidney cells. Virology. 1961 Jan;13:78–92. doi: 10.1016/0042-6822(61)90034-4. [DOI] [PubMed] [Google Scholar]
- KAPLAN A. S. STUDIES ON THE REPLICATING POOL OF VIRAL DNA IN CELLS INFECTED WITH PSEUDORABIES VIRUS. Virology. 1964 Sep;24:19–25. doi: 10.1016/0042-6822(64)90143-6. [DOI] [PubMed] [Google Scholar]
- KAPLAN A. S., VATTER A. E. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959 Apr;7(4):394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- Ladin B. F., Ihara S., Hampl H., Ben-Porat T. Pathway of assembly of herpesvirus capsids: an analysis using DNA+ temperature-sensitive mutants of pseudorabies virus. Virology. 1982 Jan 30;116(2):544–561. doi: 10.1016/0042-6822(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rixon F. J., Ben-Porat T. Structural evolution of the DNA of pseudorabies-defective viral particles. Virology. 1979 Aug;97(1):151–163. doi: 10.1016/0042-6822(79)90381-7. [DOI] [PubMed] [Google Scholar]
- Rixon F. J., Feldman L. T., Ben-Porat T. Expression of the genome of defective interfering pseudorabies virions in the presence or absence of helper functions provided by standard virus. J Gen Virol. 1980 Jan;46(1):119–138. doi: 10.1099/0022-1317-46-1-119. [DOI] [PubMed] [Google Scholar]
- Rubenstein A. S., Kaplan A. S. Electron microscopic studies of the DNA of defective and standard pseudorabies virions. Virology. 1975 Aug;66(2):385–392. doi: 10.1016/0042-6822(75)90211-1. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: a new eucaryotic defective-virus cloning-amplifying vector. Cell. 1982 Aug;30(1):295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci U S A. 1985 Feb;82(3):694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann B., Zentgraf H., Ott A., Schröder C. H. Synthesis and packaging of herpes simplex virus DNA in the course of virus passages at high multiplicity. Intervirology. 1978;10(4):228–240. doi: 10.1159/000148986. [DOI] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. A., Harper L., Ben-Porat T. cis Functions involved in replication and cleavage-encapsidation of pseudorabies virus. J Virol. 1986 Aug;59(2):318–327. doi: 10.1128/jvi.59.2.318-327.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]