Abstract
Summary: We present MetaRoute, an efficient search algorithm based on atom mapping rules and path weighting schemes that returns relevant or textbook-like routes between a source and a product metabolite within seconds for genome-scale networks. Its speed allows the algorithm to be used interactively through a web interface to visualize relevant routes and local networks for one or multiple organisms based on data from KEGG.
Availability: http://www-bs.informatik.uni-tuebingen.de/Services/MetaRoute.
Contact: blum@informatik.uni-tuebingen.de
Supplementary information: Supplementary details are available at http://www-bs.informatik.uni-tuebingen.de/Services/MetaRoute
1 INTRODUCTION
The computational prediction of all biologically relevant or novel alternative routes in metabolic networks has numerous applications in systems biology, for example, the design of tracer or knockout experiments.
Related approaches include graph theory techniques where the metabolic network is mapped onto a graph and (shortest) path algorithms compute the metabolic routes (Arita, 2000; Croes et al., 2005; Rahman et al., 2005). The network expansion approach (Handorf and Ebenhöh, 2007) iteratively adds reactions and their products from a given network to a set of initial source compounds. Routes between source and product metabolites are extracted using a shortest path approach. Others have applied convex analysis to the stoichiometric matrix of the network to calculate elementary flux modes (Schuster et al., 1999). However, a major problem is still the computational effort caused by the combinatorical explosion of the number of possible routes or flux modes in a metabolic network at genome scale (Planes and Beasley, 2008).
In this work, we present MetaRoute, a web-based application for efficient (and thus interactive) search and navigation through genome-scale metabolic networks combined with an easy-to-use visualization of the search results. Compared with other graph-based tools (Croes et al., 2005; Klukas and Schreiber, 2007; Rahman et al., 2005) MetaRoute offers interactive speed, cross-species comparison and the dynamic retrieval of local networks. Another related web tool is MetaPath Online (Handorf and Ebenhöh, 2007), an implementation of the network expansion approach that delivers information on which products can be synthesized in principle from a given set of seed compounds by the calculation of an expanded network. Additionally, the shortest route to a particular product can be extracted from such a network. This is different from MetaRoute where up to 500 different routes (between a source and a product) of increasing weight and size can be calculated and combined into a local network. This allows a systematic enumeration and a very compact representation of alternative routes. By enforcing biochemical constraints, MetaRoute strongly favors ‘textbook pathways’ over less relevant pathways.
2 METHODS
The search engine of MetaRoute is an extended version of a graph-based approach which we recently reported (Blum and Kohlbacher, 2008). Several extensions and novel concepts (explained in the Supplementary Material) improve both the search time and quality of the result. The basic algorithm uses atom mapping rules (Arita, 2003) and path weighting schemes to search for relevant paths in a directed graph representing the metabolic network. The weighted path search is constrained to trace only feasible paths transferring atoms from the source to the product metabolite. Efficient implementation and effective weighting strategies enable the interactive use of the algorithm through a web server.
Atom mapping rules were calculated as described in Blum and Kohlbacher (2008). These precalculated atom mapping rules are integrated into the graph representation of the metabolic networks and the path-finding algorithm. We use a modified version of Eppstein's k-shortest path algorithm (Eppstein, 1998), which efficiently computes the first k-shortest (or lightest) paths between two given nodes in a directed graph. The edges in our graph represent the intermediary compounds of two succeeding reactions in turn represented by the nodes of the graph. For each reaction there can be multiple nodes where each node represents a distinct educt/product pair. Compared with previous works using weighted graphs (Blum and Kohlbacher, 2008; Croes et al., 2005), a novel weighting scheme, called the combined weight, can be used to rank the routes which are extracted by Eppstein's algorithm. The combined weight for each directed edge is equal to the number of reactions in the network in which the represented compound participates as substrate (the connectivity) plus the context weight of the entering reaction node. The context weight of a reaction node is the sum of the transformed connectivities of the compounds present in the reaction but not represented by the node. The transformation makes sure that rarely occuring compounds receive a higher weight and pool metabolites a lower weight in the context of the reaction node. The combined weighting is more suited for routes passing the core metabolism or routes like purine biosynthesis which involve ‘hub compounds’ like ADP as main intermediates.
MetaRoute is built upon BNDB (Küntzer et al., 2007) and BN++ (Küntzer et al., 2006; Sirava et al., 2002) and uses the metabolic data imported from KEGG (Kanehisa, 1996). All reactions (∼7000) and the compounds involved therein were used to create a metabolic ‘super network’. Organism-specific networks are constructed by removing reactions catalyzed by enzymes absent in that organism.
We use the graph visualization software Graphviz (http://www.graphviz.org) for drawing the search results.
3 APPLICATIONS
The main application of MetaRoute is the exploration of genome-scale metabolic networks by the search for relevant routes transforming a source metabolite into a product.
Before starting a search, several search settings can be defined by the user. However, only three (source metabolite, product metabolite, network) are necessary. The remaining settings are pre-defined using default values which can be changed by experienced users. The user can enter the name of the metabolite which will be used as source. However, it is also possible to leave the field empty and to search for metabolic routes with an unspecified source but that produce a given product metabolite. The product metabolite field is analogously used. It is possible to search for relevant routes in the metabolic network of one organism of interest or in the combined network of multiple organisms, which is interesting for cross-species comparison. Furthermore, the user can search in the ‘super network’ containing all KEGG reactions as well as in an external or user-defined network.
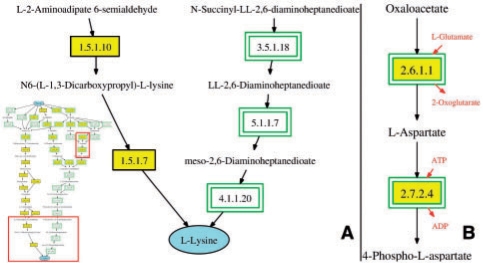
As an example, we can search for lysine-producing routes (pyruvate as source) in the combined network of yeast and Eschenchia coli. This is interesting because lysine biosynthesis is very different in each organism. All routes found can be visualized simultaneously in a local network showing only the main metabolites which were traced in the path-finding algorithm. If multiple organisms are selected (as in our example), the user can assign them to two groups which are differently highlighted for species comparisons. The resulting network for our search example, including the species highlighting, is shown in Figure 1A. Another option is to visualize each route found in a separate window (Fig. 1B). Image maps offer direct links to the KEGG database, but the user also immediately gets more information in a small popup box simply by moving the mouse over the compounds and reactions.
Fig. 1.
An example cut-out of the local network view is shown on the left. Enzymatic reactions present in yeast are drawn in yellow boxes and those in E.coli in green double borders. On the right, an example cut-out of the single route view is shown. Here, the side metabolites are also drawn.
After the first run, the user can refine the search by the addition of arbitrary constraints (forbidden or required compounds and reactions). Reactions can be defined by experienced users as irreversible. All reactions are set reversible by default. The reason is that detailed knowledge of thermodynamic data and physiological conditions is required in order to decide whether a reaction is irreversible. Search results can be exported in Systems Biology Markup Language (SBML) for further processing or analysis.
Besides its network exploration capabilities, MetaRoute can be used to complement related approaches. For example, Metatool (Kamp and Schuster, 2006), which is based on the elementary flux mode approach, allows sophisticated pathway analysis. However, the approach represents a computationally hard problem and cannot be used for genome-scale networks. Furthermore, a predefined distinction between internal and external (pool) metabolites is necessary. Using MetaRoute, the user can select a local network of moderate size and export it in the Metatool format including an automated definition of external compounds.
4 CONCLUSIONS
MetaRoute is a user-friendly tool for exploring genome-scale metabolic networks. It integrates efficient search for relevant routes with interactive network navigation and visualization. Due to the weighting scheme and the application of atom mapping rules, the resulting routes are close to textbook-like routes. Further developments of MetaRoute will try to further improve these pathways by including thermodynamic information, information on enzyme subcellular location and pathway regulation.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Arita M. Metabolic reconstruction using shortest paths. Simulat. Pract. Theory. 2000;8:109–125. [Google Scholar]
- Arita M. In silico atomic tracing by substrate-product relationships in Escherichia coli intermediary metabolism. Genome Res. 2003;13:2455–2466. doi: 10.1101/gr.1212003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum T, Kohlbacher O. Using atom mapping rules for an improved detection of relevant routes in weighted metabolic networks. J. Comput. Biol. 2008;15:565–576. doi: 10.1089/cmb.2008.0044. [DOI] [PubMed] [Google Scholar]
- Croes D, et al. Metabolic PathFinding: inferring relevant pathways in biochemical networks. Nucleic Acids Res. 2005;33:W326–W330. doi: 10.1093/nar/gki437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppstein D. Finding the k shortest paths. SIAM J. Comput. 1998;28:652–673. [Google Scholar]
- Handorf T, Ebenhöh O. MetaPath Online: a web server implementation of the network expansion algorithm. Nucleic Acids Res. 2007;35:W613–W618. doi: 10.1093/nar/gkm287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp A, Schuster S. Metatool 5.0: fast and flexible elementary modes analysis. Bioinformatics. 2006;22:1930–1931. doi: 10.1093/bioinformatics/btl267. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. Toward pathway engineering: a new database of genetic and molecular pathways. Sci. Technol. Jpn. 1996;59:34–38. [Google Scholar]
- Klukas C, Schreiber F. Dynamic exploration and editing of KEGG pathway diagrams. Bioinformatics. 2007;23:344–350. doi: 10.1093/bioinformatics/btl611. [DOI] [PubMed] [Google Scholar]
- Küntzer, et al. BN++– a biological information system. J. Integr. Bioinform. 2006;3:34. [Google Scholar]
- Küntzer, et al. BNDB – the biochemical network database. BMC Bioinformatics. 2007;8:367. doi: 10.1186/1471-2105-8-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planes FJ, Beasley JE. A critical examination of stoichiometric and path-finding approaches to metabolic pathways. Brief. Bioinform. 2008 doi: 10.1093/bib/bbn018. [Epub ahead of print, April 24, 2008] [DOI] [PubMed] [Google Scholar]
- Rahman SA, et al. Metabolic pathway analysis web service (Pathway Hunter Tool at CUBIC) Bioinformatics. 2005;21:1189–1193. doi: 10.1093/bioinformatics/bti116. [DOI] [PubMed] [Google Scholar]
- Schuster S, et al. Detection of elementary flux modes in biochemical networks: a promising tool for pathway analysis and metabolic engineering. Trends Biotechnol. 1999;17:53–60. doi: 10.1016/s0167-7799(98)01290-6. [DOI] [PubMed] [Google Scholar]
- Sirava, et al. BioMiner – modeling, analyzing, and visualizing biochemical pathways and networks. Bioinformatics. 2002;18:S219–S230. doi: 10.1093/bioinformatics/18.suppl_2.s219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.