Abstract
An ESI-MS/MS strategy employing the thymine-selective KMnO4 oxidation reaction to detect conformational changes and ligand binding sites in non-covalent DNA/drug complexes is reported. ESI-MS/MS is used to detect specific mass shifts of the DNA ions that are associated with the oxidation of thymines. This KMnO4 oxidation/ESI-MS/MS approach is an alternative to conventional gel-based oxidation methods and affords excellent sensitivity while eliminating the reliance on radiolabelled DNA. Comparison of single strand versus duplex DNA indicates that the duplexes exhibit a significant resistance to the reaction, thus confirming that the oxidation process is favored for unwound or single strand regions of DNA. DNA complexes containing different drugs including echinomycin, actinomycin-D, ethidium bromide, Hoechst 33342 and cis-C1 were subjected to the oxidation reaction. Echinomycin, a ligand with a bisintercalative binding mode, was found to induce the greatest KMnO4 reactivity, while Hoechst 33342, a minor groove binder, caused no increase in the oxidation of DNA. The oxidation of echinomycin/DNA containing duplexes with different sequences and lengths was also assessed. Duplexes with thymines closer to the terminal ends of the duplex demonstrated a greater increase in the degree of oxidation than those with thymines in the middle of the sequence. CAD and IRMPD experiments were used to determine the site of oxidation based on oligonucleotide fragmentation patterns.
Introduction
Electrospray ionization mass spectrometry (ESI-MS) has been established as a useful tool for the analysis of non-covalent complexes formed between small molecule drugs and DNA.1-3 The advantages of using ESI-MS in this capacity are low sample consumption coupled with fast analysis time, which make the technique adaptable to high throughput screening techniques. In early reports, the binding of well-studied, commercially available drugs to duplex DNA was examined, with results indicating that the binding trends observed by ESI-MS such as binding stoichiometries, sequence selectivities, binding affinities and complex stabilities, can be correlated to known solution binding behavior.4-10 Recent accounts have extended the use of the technique to look at more novel types of ligand/DNA complexes such as those containing bisintercalators,11 ligands that are metal-mediated,12, 13 and quadruplex DNA,14-21 further demonstrating the capabilities of the method.
While the success of using ESI-MS to screen binding affinities and stoichiometries has been established, one of the drawbacks of the technique is the lack of structural information that can be directly obtained; drug/DNA complexes are observed, but information about the drug binding site or alterations in the DNA structure due to drug binding can not be easily determined. Chemical probes that modify nucleic acid substrates in a structure-dependant manner22,23 coupled with ESI-MS/MS analysis represent a promising technique for the structural analysis of DNA and RNA. Solvent accessibility probes (such as dimethylsufate (DMS), kethoxal, and 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide metho-p-toluenesulfonate (CMCT)) that modify nucleotides that are not base-paired or involved in binding interactions with proteins or ligands have been used with Fourier transform mass spectrometry (FTMS) to examine structural features of RNA24-26 and RNA/protein27 complexes. In these previous studies, tandem mass spectrometry or chemical digestion techniques were used to sequence the RNA and identify the site of chemical modification, thereby introducing an additional analytical tool for the analysis of RNA and RNA/ligand complexes.24-27
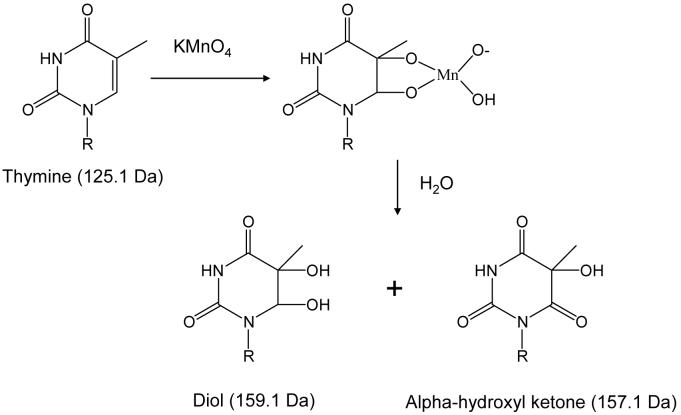
In the present study, we have exploited the use of chemical probes with ESI-MS analysis by developing a technique that utilizes KMnO4 to detect conformational changes in duplex DNA and identify the ligand binding site in non-covalent drug/DNA complexes. Under neutral conditions, KMnO4 oxidizes thymine and, to a much lesser extent, cytosine by attacking the double bond of the nucleobase to produce a mixture of the diol and an alpha-hydroxylketone products as demonstrated in Scheme 1.22,23,28 Each oxidized base results in a mass shifts of +34 for the diol and +32 Da for the alpha-hydroxylketone, which can easily be detected by mass spectrometry. Thymines in unstacked, single strand-like DNA are susceptible to reaction with KMnO4 while double stranded DNA is resistant to the oxidation.29 As a result, KMnO4 has been used to detect conformational changes upon ligand30,31 and protein binding,32-35 determine the base pair composition,36-37 uncover the specific structure of DNA (Z-DNA, hairpins, curvatures, parallel stranded helices, etc.),38-41 and ascertain conformational changes of mismatched DNA, in conjunction with gel electrophoresis analysis.42,43
Scheme 1.
General pathway of permanganate oxidation of thymine nucleobases. The mass of thymine and the resulting diol and alpha-hydroxyl ketone products are given in parenthesis.
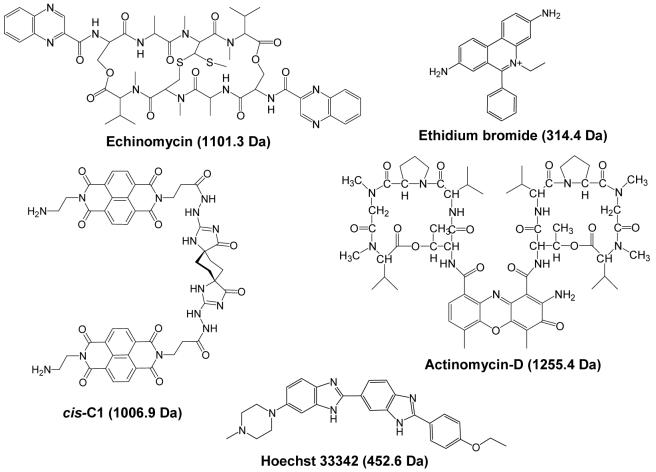
In past studies using the KMnO4 reaction to examine conformational changes upon small molecule binding, it was determined that the permanganate ion reacts with thymines around the ligand binding site as a result of the unwinding and extension of the duplex upon intercalator binding.30,31 Therefore, ligand binding sites can be elucidated after determining the positions of the oxidized thymines. The site of oxidation is determined using a piperidine heat treatment to cleave the DNA at the site of the oxidation, followed by polyacrylamide gel electrophoresis (PAGE) analysis to sequence the fragments.30,31 We have developed a streamlined technique to oxidize drug/DNA complexes in solution, analyze the extent of oxidation using ESI-MS and determine the site of oxidation using two tandem mass spectrometry techniques: collisional activated dissociation (CAD)44 and infrared multiphoton dissociation (IRMPD).45,46 Our technique simplifies the experiment, affords an alternative to the more time consuming PAGE analysis, and eliminates the need for radiolabeled DNA, while also offering detailed binding site and sequence information. We establish the technique by examining the oxidation of the duplex DNA of different sizes and sequences as well as drug/DNA complexes containing echinomycin, ethidium bromide, actinomycin-D, Hoechst 33342, and cis-C1 (structures shown in Figure 1).
Figure 1.
Structures of ligands used in this study with molecular weights given in parenthesis.
EXPERIMENTAL SECTION
Materials
Single strand oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA) and synthesized as ammonium salts on the 1 μmole scale and purified by reverse phase HPLC. Actinomycin-D, ethidium bromide, Hoechst 33342, and KMnO4 were purchased from Thermo Fisher Scientific (Waltham, MA) and used without further purification. Echinomycin was purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification and cis-C1 was prepared as previously described.47 All solvents were of HPLC grade purity.
KMnO4 Reactions
Prior to the KMnO4 oxidation reactions, stock solutions of each single strand oligo were prepared at 2 mM in 250 mM ammonium acetate buffer. Solutions containing complementary single strands, each at 1 mM concentration were prepared in 250 mM ammonium acetate and annealed by heating the solution to 90 °C, followed by cooling to room temperature over a period of 7 hours. Stock solutions of the drug molecules were prepared at 1 mM concentration in water for actinomycin-D, Hoechst 33342 and cis-C1, and in methanol for echinomycin and ethidium bromide.
Solutions containing a DNA duplex or single strand and were prepared at 40 μM in 50 μL of 90 mM ammonium acetate. Where indicated, a drug was added at 120 μM and the solution was allowed to equilibrate for 30 min. to allow for binding. To initiate oxidation 5 μL of a 5 mM KMnO4 solution prepared in water was added, and the reaction mixture was incubated for 4-30 min. at room temperature. After the desired incubation time, KMnO4 was immediately removed from the solution using a Pierce PepClean C18 spin column (Rockford, IL). The DNA was eluted from the column using 40 μL of 50% acetonitrile solution and then diluted to 100 μL so that the final solution contained 50 mM ammonium acetate.
Mass spectrometry
Analytical solutions were directly infused into a ThermoFinnigan LCQ Duo mass spectrometer (San Jose, CA) at 3 μL/min using a Harvard Apparatus PHD 2000 syringe pump (Holliston, MA). Negative ions were produced using an ESI voltage of 3.5 kV and a heated capillary temperature of 110 °C. Nitrogen sheath and auxiliary gas flows of 40 and 20 arbitrary units, respectively were used to aide the desolvation of the ions. Spectra were acquired using an ionization time of 50 - 100 ms and by summing 300 scans. For CAD experiments the desired precursor ion was isolated in the trap using the default activation time of 30 ms and a qz value of 0.25. The collision energy was increased until the abundance of the precursor ion was reduced to ∼10% relative abundance. The base pressure of the trap was nominally 1 × 10−5 torr.
IRMPD experiments were undertaken on a modified ThermoFinnigan LCQ Deca XP mass spectrometer equipped with model 48-5 Synrad 50 W continuous wave laser (Mukilteo, WA) that has been described previously.46 Briefly, qz values of 0.1 to 0.2 were used and the laser was triggered during the activation portion of the scan function. Activation times of 1 to 3 ms at 50 W laser power were used to achieve dissociation. The base pressure of the trap was nominally 2.8 × 10−5 torr which corresponds to a helium pressure of 1 mTorr.
RESULTS AND DISCUSSION
For development, optimization and validation of this new KMnO4 oxidation/ESI-MS/MS strategy, first the oxidation of single strand and then duplex DNA is evaluated. The oxidation of drug/duplex DNA complexes with varying duplex sequences, drug molecules, and duplex length are subsequently described in order to map the range of utility of the new method. The use of CAD and IRMPD to identify the position of the oxidized thymines is then demonstrated. The duplex sequences (listed in Table 1) used in this study were selected using different criteria. Ds1 and ds5 are general sequences that have been used by our group in previous ESI-MS studies of ligand/DNA complexes and were used in the present study to maintain consistency. The sequences of these duplexes were chosen to contain A/T and G/C tracts of alternating length to examine general sequence selectivity of the ligands. The sequence of one strand is G-rich while the other is C-rich to allow the masses of the complementary single strands to be easily distinguished. The A/T bases in these sequences are contained in the middle of the sequence to ensure the duplexes efficiently anneal and do not unwind during the electrospray process. The sequence for ds2 was selected based on the sequence fragment of a 265-mer DNA sequence used in a past study by Bailly et al. to determine if the sites of oxidation observed in the ESI-MS study were consistent with those of the gel-based study.31 The sequences for ds3 and ds4 were selected to be more random and not contain A/T or G/C tracts like ds1 and ds5, but were known to form complexes with echinomycin based on previous pilot studies. The sequence for ds4 contains two repeats of the sequence found in ds3 bracketed with a G/C rich to ensure extensive annealing and gas-phase stability.
Table 1.
Percent oxidation values of the duplexes and duplex/ligand complexes.a
| [ds]b | [ds + L] | [ds + 2L] | [ds + 3L] | |
|---|---|---|---|---|
| ds1/echinomycinc | 15 | 68 | 76 | 88 |
| ds2/echinomycind | 55 | 74 | 76 | 79 |
| ds3/echinomycine | 66 | 100 | 100 | nd |
| ds4/echinomycinf | 39 | 60 | 74 | nd |
| ds5/echinomycing | 20 | 59 | 73 | 76 |
| ds5/actinomycin-Dg | 20 | 21 | nd | nd |
| ds1/ethidium bromidec | 15 | 46h | nd | nd |
| ds2/ethidium bromided | 55 | 59h | nd | nd |
| ds5/cis-C1g | 20 | 42 | nd | nd |
| ds1/Hoechst 33342c | 15 | 22 | nd | nd |
All values ± 5.
Values for the duplex alone were determined from the oxidation of solutions containing the duplex without ligand.
ds1 = d(GCGGATATATGGCG/CGCCATATATCCGC)
ds2 = d(GTAGAGTCGACCTG/CAGGTCGACTCTAC)
ds3= d(GCAGTGA/TCACTGC)
ds4= d(GGACAGTGAGGGCAGTGAGGG/CCCTCACTGCCCTCACTGTCC)
ds5 =(GCGGGGATGGGGCG/CGCCCCATCCCCGC) was used in the experiment.
Because the KMnO4 oxidation caused a loss of ethidium bromide from the duplex, the values for ethidium bromide were determined based oxidation adducts formed on the free duplex.
Oxidation of single strand DNA
To confirm that the oxidation protocol, subsequent work-up, and mass spectrometric analysis allows effective monitoring and detection of the oxidation of thymines in DNA, the KMnO4 reaction was first carried out on solutions containing the single strand d(GCGGATATATGGCG), which contains three possible sites for thymine oxidation. Figure 2a shows the ESI mass spectrum of a solution containing the single strand prior to reaction with KMnO4. To obtain this spectrum, the reaction and sample purification procedures were followed as described in the Experimental section, except the KMnO4 was not added to the solution. The 3- and 4- charge states of the single strand are present with very low abundance sodium adducts. The spectral region around [ss]4− (m/z 1070 to 1115) is shown in the inset of Figure 2a, and the sodium adduct ions are labeled with asterisks. The corresponding spectrum of the single strand after a 5 min. reaction with KMnO4 is shown in Figure 2b. Compared to Figure 2a, new ions are present in Figure 2b and the m/z values of the new ions are shifted by multiples of +34 Da compared to the m/z of the single strand ions. The m/z values of the new ions are consistent with the formation of up to three oxidized thymines on the single strand ions via the reaction shown in Scheme 1. The ions in Figure 2 that correspond to single strand DNA containing the oxidized thymine(s) are labeled with “▼”, and the number of oxidation adducts is indicated by the number in parenthesis. The formation of up to three adducts is observed and is expected since three thymines are present in the single stand. The spectral enlargements of the region around [ss]4− also demonstrate that peaks pertaining to sodium adducts (again labeled with asterisks) can be distinguished from those containing oxidation adducts (labeled with black triangles)
Figure 2.
ESI-mass spectra showing solutions containing the single strand d(GCGGATATATGGCG) (ss1) (a) before reaction with KMnO4, (b) after 4 min. reaction with KMnO4, and (c) after 30 min. reaction with KMnO4. Spectral enlargements of the region around m/z 1070 to 1115 are shown in the inset. Sodium adducts are labeled with an asterisk. Ions containing oxidized thymines are labeled with ▼, with the number in parenthesis indicating the number of oxidation adducts.
After 30 min. reaction with KMnO4, the oxidation of the single strand is more extensive, as demonstrated by the spectrum shown in Figure 2c. Up to three oxidation adducts are still observed, and the abundances of the ions containing the oxidation adducts relative to the single strand ions containing no adducts are significantly increased in Figure 2c compared to Figure 2b. These results indicate that at longer incubation times, more extensive oxidation of the single strand DNA occurs. These initial results confirm that the oxidation process, as well as the extent of oxidation as a function of time, can be monitored readily by ESI-MS.
Oxidation of duplex DNA
After observing significant oxidation of single strand DNA, a similar series of experiments were performed with a duplex to compare the degree of thymine oxidation. The duplex d(GCGGATATATGGCG/CGCCATATATCCGC) (ds1) was used and contains the same single strand used in the oxidation experiments described above, then annealed with its complementary strand. The spectrum of the duplex prior to oxidation is shown in Figure 3a. The dominant charge state of the duplex is 5-, and the region around this ion (m/z 1700-1730) is enlarged and shown in the inset. After 4 min. reaction with KMnO4, two low abundance ions are detected with mass shifts that are consistent with the formation of duplexes containing one or two oxidized thymines (Figure 3b). The abundance of these adducts relative to the unoxidized [ds]5− ion is significantly lower than the abundance of the oxidation adducts formed after the KMnO4 reaction with the single strand oligonucleotide d(GCGGATATATGGCG) (Figure 2b). After 30 min. reaction with the permanganate ion, the relative abundances of the oxidation adducts of the duplex do not significantly change, as demonstrated by the spectrum shown in Figure 3c. Furthermore, no more than two oxidation adducts are formed even though there are six thymines in the duplex sequence. It is also interesting to note that increasing the reaction time from 4 to 30 min. resulted in more extensive oxidation of the single strand (Figure 2b versus 2c), while the degree of oxidation of the duplex did not change. The striking differences between the spectra obtained for the oxidation of single strand (Figure 2) and duplex DNA (Figure 3) demonstrates the resistance of duplex DNA to KMnO4 oxidation. This resistance stems from the stacking of the nucleobases which sterically hinders the access and attack of the permanganate ion on the double bond of thymine.28 Based on the results shown in Figures 1 and 2, a reaction period of 20 min. was selected for all subsequent KMnO4 reactions since this time allowed sufficient reaction of thymines on single strand DNA, while the extent of oxidation of the duplex did not change substantially at even longer times.
Figure 3.
ESI-mass spectra showing solutions containing the duplex d(GCGGATATATGGCG/CGCCATATATCCGC) (a) before reaction with KMnO4, (b) after 4 min. reaction with KMnO4, and (c) after 30 min. reaction with KMnO4. Spectral enlargements of the region around m/z 1700 - 1730 are shown in the inset. Sodium adducts are labeled with an asterisk. Ions containing oxidized thymines are labeled with ▼, with the number in parenthesis indicating the number of oxidation adducts. The d(GCGGATATATGGCG) single strand is abbreviated as ss1, while d(CGCCATATATCCGC) is ss2.
Oxidation of complexes containing echinomycin
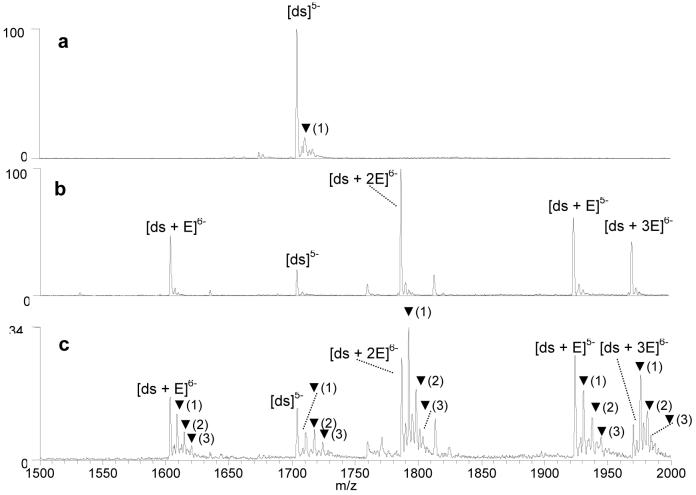
The KMnO4 oxidation of drug/DNA duplex complexes was also examined by ESI-MS to assess the ability to identify or predict structural changes of DNA upon ligand binding based on the observed changes in the oxidation patterns. Drug binding sites also can be determined from the oxidation pattern since the thymines that encompass the drug binding site are most susceptible to oxidation.30,31 Previous PAGE-based studies have found that intercalators induce changes to the DNA conformation, and in doing so, greatly increases the reactivity of thymines to the permanganate ions.30,31 This effect is especially pronounced for echinomycin (Figure 1), a bis-intercalator antibiotic that binds to duplex DNA via the intercalation of two quinoxaline chromophores at CpG sites, with the central bicyclic peptide residing in the minor groove. Intercalation of echinomycin causes considerable unwinding and distortion of the DNA duplex.48-51 In the present study, the duplex d(GCGGATATATGGCG/CGCCATATATCCGC) (ds1) was selected for experiments with echinomycin since this sequence contains multiple CpG ligand binding sites. Prior to examining the oxidation of the echinomycin/duplex complexes, the oxidation of the duplex in the absence of the ligand was assessed as a control. The results are shown in Figure 4a, and they are virtually the same as those shown in Figure 3b and 3c for the same duplex, respectively.
Figure 4.
ESI-mass spectra showing solutions containing the duplex d(GCGGATATATGGCG/CGCCATATATCCGC) (a) after 20 min, reaction with KMnO4, (b) with echinomycin (E), prior to reaction with KMnO4, and (c) with echinomycin (E), after 20 min. reaction with KMnO4. Ions containing oxidized thymines are labeled with ▼, with the number in parenthesis indicating the number of oxidation adducts.
Figure 4b shows the ESI mass spectrum of a solution containing the duplex with echinomycin (abbreviated as “E”) prior to KMnO4 reaction. Echinomycin/duplex complexes are observed with drug/DNA binding stoichiometries ranging from 1:1 to 3:1 in the 6- charge state, confirming that echinomycin/duplex complexes are readily formed in solution and maintained upon transport to the gas phase. Then the KMnO4 reaction was undertaken for twenty minutes on a solution containing echinomycin/duplex complexes, and the resulting mass spectrum is shown in Figure 4c. Compared to the oxidation of duplex alone (Figure 4a), the degree of oxidation is significantly increased for the echinomycin/duplex complexes. Interestingly, the degree of oxidation changes with the echinomycin/duplex binding stoichiometry; as the number of bound ligands increases, the abundance of the ions containing oxidation adducts increases relative to the unoxidized ions. This is demonstrated by comparing the relative abundances of the oxidation adducts associated with the 1:1, 2:1, and 3:1 echinomycin/duplex complexes in the 6- charge state. It is expected that the unwinding, elongation and hence the distortion of the duplex should increase with the number of intercalator ligands bound to the duplex, thus making thymines in the duplex more susceptible to reaction with KMnO4.51
To allow the semi-quantitative comparison of the degree of oxidation of DNA complexes, we calculate the percent oxidation of a given DNA complex, M, using the following equation:
where A[M] is the ion abundance of the single strand, duplex or drug/DNA complex designated in the subscript bracket, A[M + O] corresponds to each complex containing one or more oxidation adducts, and n is the maximum number of oxidation adducts associated with the particular complex [M]. The complexes containing multiple oxidation adducts are weighted more heavily in the equation to account for the greater extent of oxidation. As an example, for the spectra shown in Figure 4 the percent oxidation of the duplex in the absence of echinomycin is 15%, while the oxidation values of the corresponding echinomycin/DNA complexes are 68% for the 1:1 complex, 76% for the 2:1 complex, and 88% for the 3:1 complex (all in the 6- charge state). The increase in the extent of oxidation with each additional echinomycin molecule bound to the duplex is reflected in the percent oxidation values. It is interesting to note that the percent oxidation of the single strand d(GCGGATATATGGCG) shown in Figure 2c was calculated to be 91%, which is very similar to the oxidation value obtained for the 3:1 echinomycin/duplex DNA complexes. Clearly the multiple intercalation of echinomycin causes a dramatic distortion of double stranded DNA.
Effect of DNA sequence
The KMnO4 oxidation of complexes containing echinomycin with another duplex, d(GTAGAGTCGACCTG/CAGGTCGACTCTAC) (ds2) was also examined (spectra not shown). The sequence of this duplex was selected from a fragment of a 265-mer DNA sequence used in a past study by Bailly et al. which reported the binding of echinomycin by DNase I, methidium propyl EDTA (MPE)·FeII , diethyl pyrocarbonate (DEPC), and KMnO4.31 This former study identified multiple binding sites along the DNA fragment including one strong site at the sequence 5'-AGTCGACCT-3' which is thus incorporated in our sequence, ds2. The KMnO4 reaction with this duplex in the absence of echinomycin was undertaken, and the percent oxidation for the duplex was 55%. This is a higher degree of oxidation than the value determined for ds1 in the absence of echinomycin (15%). While both duplexes contain an equal number of thymines, we speculate that the higher degree of oxidation of ds2 results from the specific location of the thymines. All of the thymines in ds1 are located in the center of the duplex and are flanked by G/C base pairs, while in ds2, there are three thymines closer to the termini of the duplex. Therefore, a slight unwinding of duplex ds2 in solution, a process more likely to occur near the ends of the duplex, may make the thymines more susceptible to permanganate oxidation even in the absence of an intercalating ligand.54
While the variation in the degree of thymine oxidation in different DNA sequences is interesting, the change in the percent oxidation upon ligand binding is more important to the goals of this study. Echinomycin was found to form abundant complexes with ds2 with 1:1 and 2:1 binding stoichiometries (spectra not shown). After reaction with KMnO4, the formation of one and, to a lesser degree, two oxidation adducts were formed for each complex. As summarized in Table 1, the percent oxidation values of the 1:1, 2:1, and 3:1 echinomycin/duplex complexes are 74%, 76%, and 79%, respectively. While the overall increase in the percent oxidation values for the complexes containing echinomycin with ds2 was not as great as the increase for the echinomycin/ds1 complexes described in the previous section, the increase was still significant. These results confirm that the oxidation of DNA upon echinomycin binding24 can be determined by the ESI-MS technique. As will be discussed in the sections below, the specific sites of thymine oxidation on the duplex can be determined by CAD, and they correlate with the findings of the past gel electrophoresis study.31
Effect of duplex length
The effect of duplex length on the KMnO4 oxidation reaction was also examined. Our previous ESI-MS studies of drug/DNA complexes typically employed duplexes with 14 base-pairs because ion mobility/molecular dynamics studies have reported that DNA duplexes greater than 12 base pairs maintain the helical conformation in the gas-phase better than shorter duplexes.52,53 It may, however, be desirable to examine the KMnO4 oxidation of smaller duplexes for high throughput applications and targeted screening strategies. Since the oxidation reactions are undertaken in solution and mass spectrometry is simply used to detect the products in the present methodology, concerns about gas-phase conformations are not a primary issue. Experiments involving a seven base pair duplex, d(GCAGTGA/TCACTGC) (ds3) were undertaken to assess the lower size limit of the duplexes that can be examined by the ESI-MS/oxidation technique. For this shorter duplex, the dominant DNA ions are detected in the 3-charge state. The percent oxidation value for the [ds]3− ion after KMnO4 reaction in the absence of any DNA-interactive ligand was determined to be 66% as demonstrated by Figure S-1a shown in the Supporting Information section. This value is greater than what was observed for the 14 base pair duplexes in the absence of ligand, and suggests more extensive distortion of the shorter duplex in solution. This result is consistent with past studies that found that the B-form of duplex DNA is more easily distorted in shorter duplexes due to the reduced base stacking and hydrophobic interactions.54 For the present study, when the oxidation reaction is carried out on a solution containing echinomycin and d(GCAGTGA/TCACTGC), up to two oxidation adducts are formed for each 1:1 and 2:1 echinomycin/duplex complex. The percent oxidation for these complexes is 100% as no unoxidized form of the duplex remains in the ESI mass spectrum (Figure S-1b). These results reflect the greater distortion of smaller duplexes upon ligand binding compared to longer ones due to the smaller number of hydrogen bonds and stacking interactions, as well as the substantial degree of distortion caused by the intercalator ligand.
Experiments with a longer 21 base pair duplex were also undertaken. The oxidation of the duplex, d(GGACAGTGAGGGCAGTGAGGG/CCCTCACTGCCCTCACTGTCC) (ds4) was first evaluated in the absence of echinomycin (Figure S-2a), and the percent oxidation was determined to be 39%. After echinomycin binding, the percent oxidation value of the 1:1 echinomycin:duplex complexes was determined to be 60% and the value for the 2:1 complexes was 74% (Figure S-2b). While the increase in the degree of oxidation for ds4 compared to the echinomycin/ds4 complex is not as dramatic as was observed for the complexes containing the shorter duplexes (7-mers or 14-mers), it is reproducible and consistent with the expected duplex distortion caused by intercalation.
One of the main challenges of using longer DNA duplexes in an ESI-MS/KMnO4 experiment is that at higher charge states, it is more difficult to distinguish the formation of an oxidized thymine from a sodium adduct which are always present in ESI mass spectra of nucleic acids. The dominant charge state for drug/DNA complexes containing 14 base pair duplexes is 6-. The observed mass shift upon formation of an oxidized thymine in the 6- charge state is 5.7 Da, while the shift for a sodium adduct is 3.8 Da. The m/z values of the sodium and oxidation adducts in this charge state can be easily distinguished using a quadrupole ion trap mass spectrometer. However, complexes containing the 21 base pair duplex are observed in the 9- and 8- charge states, and at these high charge states, the m/z differences between the sodium and oxidation adducts are decreased. In the 8- charge state, the ions corresponding to sodium adducts are shifted by 2.9 Da, while those of the oxidation adducts are shifted by 4.3 Da. These peaks can generally distinguished in the mass spectrum; however, for the 9- charge state, the peaks for the sodium and oxidation adducts overlap since the mass shift for a sodium adduct is 2.5 Da, while the shift for an oxidation adduct is only 3.7 Da. Analyses of the KMnO4 reaction with DNA present in the 9- charge state or greater require a mass analyzer with greater mass accuracy and resolving power.
KMnO4 oxidation of complexes containing other drugs
KMnO4 reactions with complexes containing other types of DNA interactive ligands, including three other intercalators and a minor groove binding agent, were also examined by ESI-MS. Actinomycin-D (Figure 1) binds to DNA duplexes via the intercalation of the phenoxazone chromophore, preferably at 5'-GC-3' sites, while the two cyclic peptides bind to the minor groove.55,56 As a monointercalator, the unwinding angle and elongation of the duplex upon actinomycin-D binding is less substantial compared to the bis-intercalator echinomycin,51,57,58 therefore the KMnO4 reactivity of an actinomycin-D/duplex complex is expected to be reduced compared to complexes containing echinomycin. To enhance the duplex binding by actinomycin-D, the duplex d(GCGGGGATGGGGCG/CGCCCCATCCCCGC) (ds5) was used due to its G/C rich sequence. The ESI mass spectrum of the solution containing the duplex with actinomycin-D prior to the oxidation reaction reveals abundant complexes with binding stoichiometries of 1:1 in the 6- and 5- charge state. As summarized in Table 1, the percent oxidation value for this duplex in the absence of ligand was determined to be 20% (Figure S-3a). After 20 min. reaction with KMnO4, one low abundance oxidation adduct was formed for the actinomycin-D/duplex complexes (Figure S-3b). The percent oxidation for the 1:1 complexes was calculated to be 21%, which is not significantly different than the percent oxidation of the duplex in the absence of the ligand (Table 1).
For comparison, the KMnO4 reaction with complexes containing echinomycin with ds5 were also examined. Echinomycin/duplex binding stoichiometries of 1:1, 2:1, and 3:1 were observed in the mass spectrum, and the corresponding percent oxidation values calculated for these complexes were 59%, 73%, and 76%, respectively. The smaller duplex distortion imparted by actinomycin-D compared to that of echinomycin is reflected in the lower percent oxidation values for the actinomycin-D/duplex complexes and is likely the result of the monointercalative binding mode of actinomycin-D. It is also possible that the cyclic peptides of the molecule hinder the access of the permanganate ions to the thymines.
Ethidium bromide is another monointercalator that has been shown to induce KMnO4 reactivity in previous studies.30 Like actinomycin-D, the DNA unwinding caused by ethidium bromide is not as significant as that of echinomycin,51 resulting in decreased thymine hyperactivity. The conformational changes induced by ethidium bromide binding were thus examined by our oxidation/ESI-MS protocol. Prior to the KMnO4 reaction, a solution containing ethidium bromide with ds2 was incubated, followed by C18 spin column purification. In the mass spectrum of the resulting solutions, abundant ethidium/duplexes complexes with binding stoichiometries of 1:1 and 2:1 were present in the mass spectra (Figure S-4a). The spectrum of the solution containing ethidium bromide with ds2 after the 20 min. reaction with KMnO4 showed no surviving ethidium bromide/ds2 complexes (Figure S-4b). However, the abundances of the oxidation adducts of the free duplex in the 5- charge state were slightly greater than those of the oxidation adducts of ds2 in the absence of ethidium bromide. The percent oxidation of ds2 in the absence of ethidium bromide was 55%, while after the addition of ethidium bromide, the percent oxidation increased to 59% despite the apparent lack of retention of the ligand by the duplexes after KMnO4 oxidation. The results were similar for ethidium bromide with ds1, however a much greater increase in the degree of thymine oxidation is observed. In the absence of ligand, the percent oxidation of ds1 was 15%, but after the addition of ethidium bromide, the percent oxidation was 46%.
These results suggest that ethidium bromide reacts with KMnO4, causing it to dislodge from the duplex, likely during the oxidation reaction. In the absence of KMnO4, ethidium bromide remains bound to the duplex after the spin column clean-up step, thus specifically pointing to KMnO4 as causing the disruption of the binding of ethidium bromide. The increase in the degree of oxidation of the drug-free [ds]5− ion in the spectrum obtained for the solution that contained both the duplex and ethidium bromide indicates that the ethidium bromide induced the thymine reactivity of the duplex prior to its dislocation. Thus, the KMnO4 reactivity induced by ethidium can be assessed by examining the relative increase in the percent oxidation of the free duplex. The percent oxidation results summarized in Table 2 for two duplexes, ds1 and ds2, indicate that the duplex distortion induced by ethidium bromide was greater than that caused by actinomycin-D, but not as dramatic as that caused by echinomycin.
Table 2.
Site of thymine oxidation for DNA/ligand complexes.
| Ligand | Duplex | Sequence and Thymine Oxidation Site(s)a |
|---|---|---|
| echinomycin | ds1 | d(GCGGATATATGGCG/CGCCATATATCCGC) |
| echinomycin | ds2 | d(GTAGAGTCGACCTG/CAGGTCGACTCTAC) |
| echinomycin | ds5 | d(GCGGGGATGGGGCG/CGCCCCATCCCCGC) |
| echinomycin | ds3 | d(GCAGTGA/TCACTGC) |
| echinomycin | ds4 | d(GGACAGTGAGGGCAGTGAGGG/CCCTCACTGCCCTCACTGTCC) |
| ethidium bromide | ds1 | d(GCGGATATATGGCG/CGCCATATATCCGC) |
| cis-C1 | ds5 | d(GCGGGGATGGGGCG/CGCCCCATCCCCGC) |
The site(s) of thymine oxidation are shown in bold and underlined.
In a recent study, we examined the binding affinities of a series of new threading bisintercalators47 for different DNA duplexes by ESI-MS/MS.11 One compound, cis-C1, demonstrated a high degree of binding specificity based on our ESI-MS screening results, and the bis-intercalative binding mode was confirmed by subsequent NMR studies.59 In the present study, we have examined the KMnO4 reactivity of DNA duplex complexes containing cis-C1 to determine the degree of oxidation induced by the new bis-intercalator compared to that of echinomycin. An abundant complex with 1:1 binding stoichiometry in the 6- charge state is present in the ESI mass spectrum of a solution containing cis-C1 with ds5 prior to the KMnO4 reaction. After the permanganate reaction, one oxidation adduct was formed for the cis-C1/ds5 complex, and the percent oxidation was calculated to be 42% as summarized in Table 1. In the absence of the bis-intercalator ligand, the percent oxidation of the duplex is 20%. The duplex distortion induced by cis-C1 is considerably greater than that caused by actinomycin with the same duplex (only 21%) which is expected since actinomycin is a monointercalator whereas cis-C1 is a bis-intercalator. However, the percent oxidation suggests that the distortion of the duplex by cis-C1 is not as great as that caused by echinomycin, as evidenced by the moderately greater percent oxidation value for 1:1 complexes containing echinomycin with ds5 (59%).
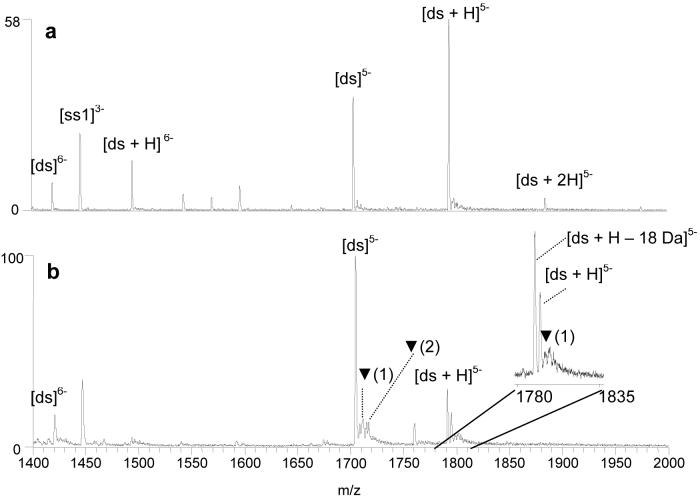
Hoechst 33342 (structure shown in Figure 1) is a well-studied minor-groove binder known to target A/T rich regions of a duplex. Compounds that bind to the minor groove of DNA typically do not distort the duplex as significantly as do intercalators53, 54 and as such, do not induce KMnO4 reactivity. The ESI mass spectrum of a solution containing Hoechst 33342 (abbreviated as “H”) and d(GCGGATATATGGCG/CGCCATATATCCGC) (ds1) prior to the permanganate reaction is shown in Figure 5a. An abundant 1:1 complex is present in the 5-charge state at m/z 1795 and a much less abundant 2:1 complex is present at m/z 1885. The corresponding mass spectrum of the solution after the KMnO4 reaction is shown in Figure 5b. The relative abundance of the Hoechst 33342/duplex complex at m/z 1795 has decreased, and a new peak is present at m/z 1791 that is consistent with a dehydrated Hoescht 33342/duplex species, not a typical oxidation adduct. To identify the new species at m/z 1791, CAD experiments were undertaken on the ion. The ion dissociated to form an ion at m/z 1705, which corresponds to the m/z of the original duplex (ds5−). Upon a second stage of collisional activation, the resulting ion of m/z 1705 dissociated in a manner consistent with the fragmentation of native duplex ions in the 5- charge state (spectra not shown). These results suggest the oxidation of the bound Hoechst 33342 by KMnO4 but not the oxidation of the duplex. The Hoechst ligand apparently reacts with KMnO4, and the oxidized ligand remains bound to the duplex to form the observed product of m/z 1791. This reaction also caused the decrease in the relative abundance of the original 1:1 Hoechst 33342/duplex complex. We speculate that the permanganate ion oxidizes the aniline nitrogen of the piperazine ring of the Hoechst 33342 molecule which then undergoes further reaction to lose 18 Da.
Figure 5.
ESI-mass spectra showing solutions containing the duplex d(GCGGATATATGGCG/CGCCATATATCCGC) and Hoechst 33342 (H) (a) before reaction with KMnO4 and (b) after 20 min. reaction with KMnO4. Ions containing oxidized thymines are labeled with ▼, with the number in parenthesis indicating the number of adducts each peak contains.
Low abundance oxidation adducts associated with the complex at m/z 1791 and 1795 are present as shown in the spectral enlargement in the inset of Figure 5b; however, the percent oxidation for these complexes does not exceed 22% and in fact is not much greater than the oxidation value of the drug-free duplex. The significantly lower percent oxidation value of the Hoechst 33342/duplex complexes compared to the echinomycin/duplex complexes is consistent with the low degree of distortion of the duplex by minor groove binder ligands.
CAD of oxidized DNA
In addition to assessing the degree of oxidation induced by ligand binding to duplex DNA, another important aspect of the KMnO4 experiment is determination of the site of oxidation. In traditional gel-based experiments, oxidation sites are mapped via a multi-step procedure in which the oxidized DNA is cleaved adjacent to the modification sites by a piperidine heat treatment, followed by DNA precipitation, and then subsequent gel-analysis of the products.23, 24 Rather than employing the piperidine treatment, our strategy entails direct analysis of the oxidized DNA adducts based on CAD to determine the site of oxidation, thus simplifying the analytical scheme.
The CAD spectra of intercalator/DNA complexes have been previously evaluated.8,62-63 These complexes typically dissociate via the ejection of the ligand, leaving the drug-free duplex ion, or by separation of the single strands of the duplex with the ligand remaining bound to one of the strands. The MS/MS dissociation pathways of oligonucleotides have also been extensively examined,44,64 and systematic interpretation of the fragmentation patterns has been used to determine the site of covalent modifications of nucleic acids based on diagnostic a – Band w sequence ions that result from the backbone fragmentation of single strand oligonucleotides.24-27,65
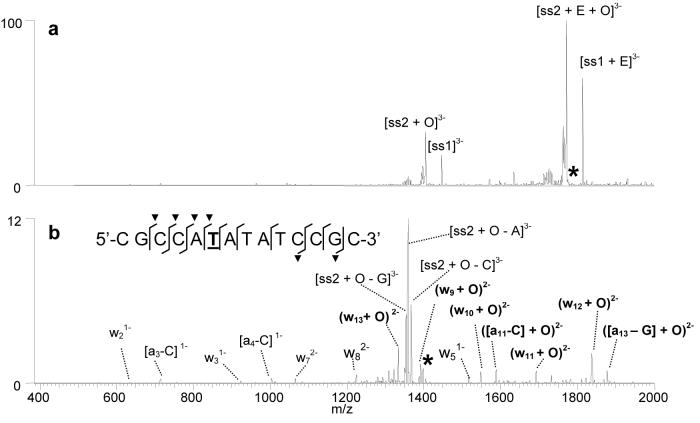
In the present study, the most abundant oxidation adducts were isolated in the trap and subjected to CAD. For example, Figure 6a shows the CAD spectrum of the [ds + 2 x E + O]6− ion containing two molecules of echinomycin and duplex ds1 that was originally observed in the ESI mass spectrum shown in Figure 4c at m/z 1789. After the first stage of CAD, the complex dissociates by strand separation with one echinomycin molecule remaining bound to each single strand. The mass of the ions containing the d(CGCCATATATCCGC) single strand (labeled ss2 in Figure 6a) is shifted by 34 Da, indicating that the oxidation occurred exclusively on ss2 rather than ss1. The [ss2 + O]3− product ion was subjected to a second stage of collisional activation (MS3) to gain information about the specific position of the oxidized base. As shown in Figure 6b, the MS3 spectrum contains many informative a - B and w ions, some of which contain the mass shift characteristic of the oxidized base. The sequence overlaid on Figure 6b summarizes the diagnostic cleavages (labeled with slash marks) in the spectrum, with those containing the mass shift labeled with a “▼”. Based on the cleavage pattern, T6 can be identified as the sole oxidized residue. The key fragment ions for making this assignment are the w8 ion that does not contain the mass shift and the (w9 + O) ion that does contain the oxidation mass shift. Because the echinomycin molecule binds to the G/C rich regions of the duplex, it follows that the thymines closest to this region, like T6, will be most easily oxidized. The clarity of the fragmentation pattern is remarkable, as well as the fact that the oxidation occurs exclusively at one specific thymine site. The abundant oxidation of T6 over T10, the other thymine in the sequence that is close to the G/C rich region of the sequence, may occur because this thymine is flanked by two adenines, and previous studies have found thymines adjacent to short A-tracts are more susceptible to the permanganate reaction.66 The preference for thymine reaction on the C-rich single strand is not full understood at this time and merits further study. The same thymine was identified as containing the oxidation adduct in the ESI-MS3 spectra for [ds + E + O]6−as shown in Figure S-5.
Figure 6.
ESI-MS3 experiments for [ds + 2E + O]6− containing d(GCGGATATATGGCG/CGCCATATATCCGC) and echinomycin (E): (a) CAD spectrum of the initial [ds + 2E + O]6− complex and (b) MS3 spectrum of the [ss2 + O]3− product ion. “O” is indicative of an oxidation adduct. The sequence structure in Figure 5b summarizes the sequence coverage. The fragments containing an oxidation adduct are labeled with a “▼” symbol. The thymine that was determined to be oxidized is underlined in the sequence shown in Figure 2b. The d(GCGGATATATGGCG) single strand is abbreviated as ss1, while d(CGCCATATATCCGC) is ss2. The selected precursor ion is labelled with an asterisk.
The sites of thymine oxidation determined by CAD experiments for the other drug/DNA complexes examined in this study are summarized in Table 2. In most cases, the CAD results confirm a single thymine oxidation, not a distribution or ensemble of oxidation sites. Thus, the oxidation process displays striking selectivity that makes mass spectrometric analysis a natural fit for pinpointing the oxidation sites. The thymines identified in Table 2 are all within four or five bases of the proposed 5'-CG-3' intercalator binding sites or adjacent to G/C rich regions of the sequence that are likely intercalator binding sites. Future studies will be aimed at examining trends in the sites of oxidation in greater detail. One result of note involves the complexes containing echinomycin and d(GTAGAGTCGACCTG/CAGGTCGACTCTAC) (ds2). As discussed above, the oxidation of complexes containing this duplex were examined to maintain consistency with a previous gel electrophoresis study of the binding of echinomycin to duplex DNA assessed by various chemical probes, including KMnO4.31 The high affinity binding site of echinomycin on the duplex was determined to be the central 5'-TCGA-3' site.31 Based on the strand separation products present in the CAD spectrum of the [ds + E + O]6− complex in our present study, the site of oxidation was determined to be located on the first single strand, d(GTAGAGTCGACCTG). However, the MS3 spectrum of the [ss + O]3− ion resulting from the first stage of CAD was inconclusive since there were product ions suggesting that the oxidized thymine could be the T2 or T13 (Table 2). It is likely that the [ss + O]3− species consisted of two population of ions in which the thymine at position 2 was oxidized in one population, while the thymine at position 13 was oxidized in the other, suggesting that the duplex can unravel at both ends. Unlike the other DNA sequences used in this study, d(GTAGAGTCGACCTG) contains two thymines (T2 and T13) that are located at equivalent positions from the ends of the duplex, and thus may have roughly equal susceptibility to thymine oxidation upon echinomycin binding.
To further ascertain the regions of DNA unwinding for this duplex, CAD studies were performed on the [ds2 + E + 2 × O]6− complex, the complex containing two sites of oxidation. Upon the first stage of CAD, the resulting strand separation products revealed that the second site of thymine oxidation was located on the second strand, d(CAGGTCGACTCTAC). Subsequent MS3 experiments indicated that T12 is the position of the second oxidized thymine (spectra not shown). This is the same thymine that was found to be oxidized in the gel-based study by Bailly et al. using a longer duplex.31 When the structure of the duplex is considered, the thymine at the twelfth position on the second single strand d(CAGGTCGACTCTAC) is adjacent to the second thymine on the complementary first strand, d(GTAGAGTCGACCTG), which confirms that this region of the DNA is especially distorted upon binding echinomycin. The sites of thymine oxidation correlate with the results of the past gel experiments24 and support that echinomycin is bound to the 5'-TCGA-3' region, thus promoting oxidation around this site.
IRMPD of oxidized DNA
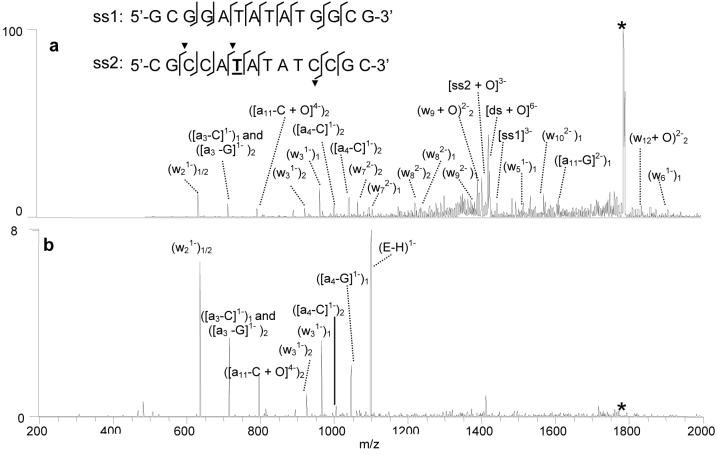
IRMPD has also proven useful for gaining sequence information of DNA molecules and sites of modification.45,46 During IRMPD, ion activation is independent of the RF voltage applied to the trap which eliminates the low mass cut-off problem characteristic of CAD experiments in a quadrupole ion trap.45 Ion activation by IR photoabsorption is a non-resonant process, resulting in increased secondary fragmentation of DNA and subsequently more a - B and w ions without the need for sequential stages of ion isolation, activation and dissociation that are common for CAD strategies.45 To explore the potential merits of IRMPD for the characterization of DNA oxidation adducts and determination of oxidation sites, several pilot IRMPD experiments were undertaken in comparison to the CAD results described above. Figure 7a shows the IRMPD spectrum of the [ds + 2 × E +O]6− complex containing echinomycin and duplex ds5 using 50 W laser power and an activation time of only 1.7 ms. The types and relative abundances of the fragment ions present in the IRMPD mass spectrum are quite different from those in the CAD spectrum of the same complex shown in Figure 6a. While strand separation ions dominated the CAD spectrum, they are essentially absent in the IRMPD spectrum because these ions have undergone efficient IR absorption and been converted into informative sequence a - B and w ions. The site of the oxidation can be determined readily from these sequence ions, as shown by the cleavages marked on the duplex sequence structure overlaid in Figure 6a, thus eliminating the need for a second stage of activation which is typically necessary for more complete sequence coverage in the analogous CAD experiments.
Figure 7.
IRMPD spectra of [ds + 2 × E + O]6− using (a) 99% laser power and 1.7 ms irradiation time and (b) 99% laser power and 2.5 ms irradiation time. The d(GCGGATATATGGCG) single strand is abbreviated as ss1, while d(CGCCATATATCCGC) is ss2. The subscripts on the ion labels denote whether the fragment ion arises from the first or second strand of the duplex. The selected precursor ion is labelled with an asterisk.
The relative abundance of the sequence ions produced upon IRMPD can be controlled by varying the irradiation time. This is demonstrated in Figure 7b which shows the IRMPD spectrum of the same complex that was dissociated in Figure 7a, [ds + 2 × E +O]6−, with an irradiation time of 2.5 ms. The precursor ion has been totally converted to fragment ions, and the relative abundances of the lower mass a - B and w ions are significantly increased compared to those in Figure 7a. The ability to effectively “tune” the relative abundances of the sequence ions using different irradiation times during an IRMPD experiment is particularly beneficial for identifying the site of thymine oxidation without requiring more elaborate MS3 experiments.
Conclusions
An ESI-MS technique employing KMnO4 oxidation reactions has been developed to assess the distortion of DNA duplexes upon ligand binding. Adducts consistent with the oxidation of thymine nucleobases are detected in the ESI mass spectrum, while CAD and IRMPD can be used to identify the specific sites of oxidation. Ligand binding sites can be determined based on the thymine oxidation pattern. The technique presented here represents an alternative to traditional gel-based KMnO4 experiments and is attractive due to the elimination of the need to use both radiolabeled DNA and the piperidine heat treatment required to identify the site of oxidation, while also offering excellent sensitivity and facile adaptation to high throughput screening applications.
Significant differences in the extent of oxidation of thymines in single strand DNA compared to duplex DNA were observed, and the susceptibility of thymines to oxidation in drug/duplex complexes was significantly enhanced, presumably due to conformational changes of the duplex upon drug binding. The bis-intercalator echinomycin caused the most extensive thymine oxidation, followed by the threading bis-intercalator cis-C1, and the monointercalator ethidium bromide. Actinomycin-D, another monointercalator, and the minor groove binder Hoechst 33342 did not substantially increase the reactivity of thymines with KMnO4. In addition to the type of drug bound to the duplex, other factors that influenced the extent of thymine oxidation were the size of the duplex, the number of bound ligands, and the proximity of the thymines to the termini of the duplex. The ability to assess the KMnO4 reactivity induced by ligands that become unbound from the duplex, either by reacting with the permanganate ion or as a result of the post-oxidation C18 clean-up procedure, is also demonstrated. Multi-stage CAD experiments allowed confident determination of the positions of the oxidized thymines. As an alternative to CAD and MS3 approaches, IRMPD offers a promising option that results in more abundant sequence ions due to enhanced secondary fragmentation.
Supplementary Material
Acknowledgements
Funding from the Robert A. Welch Foundation (F-1155) and the National Institutes of Health (RO1 GM65956) is gratefully acknowledged.
Footnotes
Supplementary Material Available: ESI-mass spectra of d(GCAGTGA/TCACTGC) before the KMnO4 reaction and with echinomycin after the KMnO4 reaction, ESI-mass spectra of d(GGACAGTGAGGGCAGTGAGGG/CCTGTCACTCCCGTCACTCCC) before the KMnO4 reaction and with echinomycin after the KMnO4 reaction, ESI-mass spectra of d(GCGGGGATGGGGCG/CGCCCCATCCCCGC) before the KMnO4 reaction and with actinomycin-D after the KMnO4 reaction, ESI-mass spectra of d(GTAGAGTCGACCTG/CAGGTCGACTCTAC) with ethidium bromide before and after the KMnO4 reaction, ESI-MS3 spectra for [ds + E + O]6− containing d(GCGGATATATGGCG/CGCCATATATCCGC) and echinomycin is available as Supporting Information. Current ordering information is found on the masthead page.
References
- 1.Hofstadler SA, Sannes-Lowery KA. Nat. Rev. Drug Discov. 2006;5:585–595. doi: 10.1038/nrd2083. [DOI] [PubMed] [Google Scholar]
- 2.Hofstadler SA, Griffey RH. Chem. Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 3.Beck JL, Colgrave ML, Ralph SF, Sheil MM. Mass Spectrom. Rev. 2001;20:61–87. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- 4.Gale DC, Smith RD. J. Am. Soc. Mass Spectrom. 1995;6:1154–1164. doi: 10.1016/1044-0305(95)00530-7. [DOI] [PubMed] [Google Scholar]
- 5.Gabelica V, De Pauw E, Rosu F. J. Mass Spectrom. 1999;34:1328–1337. doi: 10.1002/(SICI)1096-9888(199912)34:12<1328::AID-JMS889>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 6.Kapur A, Beck JL, Sheil MM. Rapid Commun. Mass Spectrom. 1999;13:2489–2497. doi: 10.1002/(SICI)1097-0231(19991230)13:24<2489::AID-RCM816>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 7.Gabelica V, Rosu F, Houssier C, De Pauw E. Rapid Commun. Mass Spectrom. 2000;14:464–467. doi: 10.1002/(SICI)1097-0231(20000331)14:6<464::AID-RCM895>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 8.Wan KX, Gross ML, Shibue T. J. Am. Soc. Mass Spectrom. 2000;11:450–457. doi: 10.1016/S1044-0305(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 9.Wan KX, Shibue T, Gross ML. J. Am. Chem. Soc. 2000;122:300–307. [Google Scholar]
- 10.Rosu F, Gabelica V, Houssier C, De Pauw E. Nucleic Acids Res. 2002;30 doi: 10.1093/nar/gnf081. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazzitelli CL, Chu Y, Reczek JJ, Iverson BL, Brodbelt JS. J. Am. Soc. Mass Spectrom. 2006 doi: 10.1016/j.jasms.2006.09.021. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyzer ML, Brodbelt JS, Kerwin SM, Kumar D. Nucleic Acids Res. 2001;29:e103. doi: 10.1093/nar/29.21.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oehlers L, Mazzitelli CL, Brodbelt JS, Rodriguez M, Kerwin S. J. Am. Soc Mass Spectrom. 2004;15:1593–1603. doi: 10.1016/j.jasms.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 14.Rosu F, Gabelica V, Houssier C, Colson P, De Pauw E. Rapid Commun. Mass Spectrom. 2002;16:1729–1736. doi: 10.1002/rcm.778. [DOI] [PubMed] [Google Scholar]
- 15.David WM, Brodbelt J, Kerwin SM, Thomas PW. Anal. Chem. 2002;74:2029–2033. doi: 10.1021/ac011283w. [DOI] [PubMed] [Google Scholar]
- 16.Guittat L, Alberti P, Rosu F, Van Miert S, Thetiot E, Pieters L, Gabelica V, De Pauw E, Ottaviani A, Riou JF, Mergny JL. Biochimie. 2003;85:535–547. doi: 10.1016/s0300-9084(03)00035-x. [DOI] [PubMed] [Google Scholar]
- 17.Rosu F, De Pauw E, Guittat L, Alberti P, Lacroix L, Mailliet P, Riou JF, Mergny JL. Biochemistry. 2003;42:10361–10371. doi: 10.1021/bi034531m. [DOI] [PubMed] [Google Scholar]
- 18.Rosu F, Gabelica V, Shin-ya K, De Pauw E. Chem. Commun. 2003:2702–2703. doi: 10.1039/b309394h. [DOI] [PubMed] [Google Scholar]
- 19.Guittat L, De Cian A, Rosu F, Gabelica V, De Pauw E, Delfourne E, Mergny JL. Biochim. Biophys. Acta. 2005;1724:375–384. doi: 10.1016/j.bbagen.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 20.Baker ES, Lee JT, Sessler JL, Bowers MT. J. Am. Chem. Soc. 2006;128:2641–2648. doi: 10.1021/ja0564968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazzitelli CL, Kern JT, Rodriguez M, Brodbelt JS, Kerwin SM. J. Am. Soc. Mass Spectrom. 2006;17:593–604. doi: 10.1016/j.jasms.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen PE. J. Mol. Recognit. 1990;3:1–25. doi: 10.1002/jmr.300030102. [DOI] [PubMed] [Google Scholar]
- 23.Bui CT, Rees K, Cotton RGH. Curr. Pharmacogenomics. 2004;2:325–332. [Google Scholar]
- 24.Kellersberger KA, Yu E, Kruppa GH, Young MM, Fabris D. Anal.Chem. 2004;76:2438–2445. doi: 10.1021/ac0355045. [DOI] [PubMed] [Google Scholar]
- 25.Yu ET, Zhang QG, Fabris D. J. Mol. Biol. 2005;345:69–80. doi: 10.1016/j.jmb.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 26.Yu E, Fabris D. Anal. Biochem. 2004;334:356–366. doi: 10.1016/j.ab.2004.07.034. [DOI] [PubMed] [Google Scholar]
- 27.Yu E, Fabris D. J. Mol. Biol. 2003;330:211–223. doi: 10.1016/s0022-2836(03)00589-8. [DOI] [PubMed] [Google Scholar]
- 28.Bui CT, Rees K, Cotton RGH. Nucleos. Nucleot. Nucl. 2003;22:1835–1855. doi: 10.1081/NCN-120023276. [DOI] [PubMed] [Google Scholar]
- 29.Sasse-Dwight S, Gralla JD. J. Biol. Chem. 1989;264:8074–8081. [PubMed] [Google Scholar]
- 30.Jeppesen C, Nielsen PE. Febs Lett. 1988;231:172–176. doi: 10.1016/0014-5793(88)80725-7. [DOI] [PubMed] [Google Scholar]
- 31.Bailly C, Gentle D, Hamy F, Purcell M, Waring MJ. Biochem. J. 1994;300:165–173. doi: 10.1042/bj3000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gille H, Messer W. Embo J. 1991;10:1579–1584. doi: 10.1002/j.1460-2075.1991.tb07678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh DJ, Camiolo SM, Yates JL. Embo J. 1993;12:4933–4944. doi: 10.1002/j.1460-2075.1993.tb06187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farah JA, Smith GR. J. Mol. Biol. 1997;272:699–715. doi: 10.1006/jmbi.1997.1259. [DOI] [PubMed] [Google Scholar]
- 35.Schaubach OL, Dombroski AJ. J. Biol. Chem. 1999;274:8757–8763. doi: 10.1074/jbc.274.13.8757. [DOI] [PubMed] [Google Scholar]
- 36.Rubin CM, Schmid CW. Nucleic Acids Res. 1980;8:4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones AS, Walker RT. Nature. 1964;202 [Google Scholar]
- 38.Jiang H, Zacharias W, Amirhaeri S. Nucleic Acids Res. 1991;19:6943–6948. doi: 10.1093/nar/19.24.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Desantis P, Palleschi A, Savino M, Scipioni A. Biochemistry. 1990;29:9269–9273. doi: 10.1021/bi00491a023. [DOI] [PubMed] [Google Scholar]
- 40.Mitas M, Yu A, Dill J, Kamp TJ, Chambers EJ, Haworth IS. Nucleic Acids Res. 1995;23:1050–1059. doi: 10.1093/nar/23.6.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klysik J, Rippe K, Jovin TM. Biochemistry. 1990;29:9831–9839. doi: 10.1021/bi00494a012. [DOI] [PubMed] [Google Scholar]
- 42.Bui CT, Lambrinakos A, Cotton RGH. Biopolymers. 2003;70:628–636. doi: 10.1002/bip.10543. [DOI] [PubMed] [Google Scholar]
- 43.Cotton RGH. Biochem. J. 1989;263:1–10. doi: 10.1042/bj2630001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLuckey SA, Van Berkel GJ, Glish GL. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 45.Keller KM, Brodbelt JS. Anal. Biochem. 2004;326:200–210. doi: 10.1016/j.ab.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 46.Wilson JJ, Brodbelt JS. Anal. Chem. 2006;78:6855–6862. doi: 10.1021/ac060760d. [DOI] [PubMed] [Google Scholar]
- 47.Chu YJ, Lynch V, Iverson BL. Tetrahedron. 2006;62:5536–5548. [Google Scholar]
- 48.Vandyke MM, Dervan PB. Science. 1984;225:1122–1127. doi: 10.1126/science.6089341. [DOI] [PubMed] [Google Scholar]
- 49.Low CML, Drew HR, Waring MJ. Nucleic Acids Res. 1984;12:4865–4879. doi: 10.1093/nar/12.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao X, Patel DJ. Q. Rev. Biophysics. 1989;22:93–138. doi: 10.1017/s0033583500003814. [DOI] [PubMed] [Google Scholar]
- 51.Waring MJ, Wakelin LPG. Nature. 1974;252:653–657. doi: 10.1038/252653a0. [DOI] [PubMed] [Google Scholar]
- 52.Rueda M, Luque FJ, Orozco M. J. Am. Chem. Soc. 2005;127:11690–11698. doi: 10.1021/ja0422110. [DOI] [PubMed] [Google Scholar]
- 53.Gidden J, Ferzoco A, Baker ES, Bowers MT. J. Am. Chem. Soc. 2004;126:15132–15140. doi: 10.1021/ja046433+. [DOI] [PubMed] [Google Scholar]
- 54.Sinden RR. DNA Structure and Function. Academic Press; San Diego: 1994. [Google Scholar]
- 55.Sobell HM. Proc. Natl. Acad. Sci. U.S.A. 1985;82:5328–5331. doi: 10.1073/pnas.82.16.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sobell HM, Jain SC, Sakore TD, Nordman CE. Nature new. Biol. 1972;231:200–205. doi: 10.1038/newbio231200a0. [DOI] [PubMed] [Google Scholar]
- 57.Wakelin LPG, Waring MJ. Biochem. J. 1976;157:721–740. doi: 10.1042/bj1570721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang JC. Biochim. Biophys. Acta - Nucleic Acids and Protein Synthesis. 1971;232:246–251. doi: 10.1016/0005-2787(71)90576-4. [DOI] [PubMed] [Google Scholar]
- 59.Chu Y, Sorrey S, Hoffman DW, Iverson BL. J. Am. Chem. Soc. 2006 in press. [Google Scholar]
- 60.Neidle S. Biopolymers. 1997;44:105–121. [Google Scholar]
- 61.Neidle S, Pearl LH, Skelly JV. Biochem. J. 1987;243:1. doi: 10.1042/bj2430001. &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keller KM, Zhang JM, Oehlers L, Brodbelt JS. J. Mass Spectrom. 2005;40:1362–1371. doi: 10.1002/jms.927. [DOI] [PubMed] [Google Scholar]
- 63.Gabelica V, De Pauw E. J. Am. Soc. Mass Spectrom. 2002;13:91–98. doi: 10.1016/s1044-0305(01)00335-x. [DOI] [PubMed] [Google Scholar]
- 64.McLuckey SA, Habibigoudarzi S. J. Am. Chem. Soc. 1993;115:12085–12095. [Google Scholar]
- 65.Wang YS, Zhang QC, Wang YS. J. Am. Soc. Mass Spectrom. 2004;15:1565–1571. doi: 10.1016/j.jasms.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 66.McCarthy JG, Williams LD, Rich A. Biochemistry. 1990;29:6071–6081. doi: 10.1021/bi00477a027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.