Abstract
The nucleotide sequence of gene 18 of bacteriophage T4 was determined by the Maxam-Gilbert method, partially aided by the dideoxy method. To confirm the deduced amino acid sequence of the tail sheath protein (gp18) that is encoded by gene 18, gp18 was extensively digested by trypsin or lysyl endopeptidase and subjected to reverse-phase high-performance liquid chromatography. Approximately 40 peptides, which cover 88% of the primary structure, were fractionated, the amino acid compositions were determined, and the corresponding sequences in DNA were identified. Furthermore, the amino acid sequences of 10 of the 40 peptides were determined by a gas phase protein sequencer, including N- and C-terminal sequences. Thus, the complete amino acid sequence of gp18, which consists of 658 amino acids with a molecular weight of 71,160, was determined.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amos L. A., Klug A. Three-dimensional image reconstructions of the contractile tail of T4 bacteriophage. J Mol Biol. 1975 Nov 25;99(1):51–64. doi: 10.1016/s0022-2836(75)80158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisaka F., Ishimoto L., Kassavetis G., Kumazaki T., Ishii S. Nucleotide sequence of the tail tube structural gene of bacteriophage T4. J Virol. 1988 Mar;62(3):882–886. doi: 10.1128/jvi.62.3.882-886.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arisaka F., Tschopp J., Van Driel R., Engel J. Reassembly of the bacteriophage T4 tail from the core-baseplate and the monomeric sheath protein P18: a co-operative association process. J Mol Biol. 1979 Aug 15;132(3):369–386. doi: 10.1016/0022-2836(79)90266-3. [DOI] [PubMed] [Google Scholar]
- Caspar D. L. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys J. 1980 Oct;32(1):103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Christensen A. C., Young E. T. T4 late transcripts are initiated near a conserved DNA sequence. Nature. 1982 Sep 23;299(5881):369–371. doi: 10.1038/299369a0. [DOI] [PubMed] [Google Scholar]
- Duda R. L., Eiserling F. A. Evidence for an internal component of the bacteriophage T4D tail core: a possible length-determining template. J Virol. 1982 Aug;43(2):714–720. doi: 10.1128/jvi.43.2.714-720.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giphart-Gassler M., Wijffelman C., Reeve J. Structural polypeptides and products of late genes of bacteriophage Mu: characterization and functional aspects. J Mol Biol. 1981 Jan 5;145(1):139–163. doi: 10.1016/0022-2836(81)90338-7. [DOI] [PubMed] [Google Scholar]
- Hunkapiller M. W., Lujan E., Ostrander F., Hood L. E. Isolation of microgram quantities of proteins from polyacrylamide gels for amino acid sequence analysis. Methods Enzymol. 1983;91:227–236. doi: 10.1016/s0076-6879(83)91019-4. [DOI] [PubMed] [Google Scholar]
- Ishii S., Yokosawa H., Kumazaki T., Nakamura I. Immobilized anhydrotrypsin as a specific affinity adsorbent for tryptic peptides. Methods Enzymol. 1983;91:378–383. doi: 10.1016/s0076-6879(83)91035-2. [DOI] [PubMed] [Google Scholar]
- KELLENBERGER E., DELATOUR E. B. ON THE FINE STRUCTURE OF NORMAL AND "POLYMERIZED" TAIL SHEATH OF PHAGE T4. J Ultrastruct Res. 1964 Dec;11:545–563. doi: 10.1016/s0022-5320(64)80081-2. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., King J. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J Mol Biol. 1975 Dec 25;99(4):695–716. doi: 10.1016/s0022-2836(75)80180-x. [DOI] [PubMed] [Google Scholar]
- King J., Mykolajewycz N. Bacteriophage T4 tail assembly: proteins of the sheath, core and baseplate. J Mol Biol. 1973 Apr 5;75(2):339–358. doi: 10.1016/0022-2836(73)90025-9. [DOI] [PubMed] [Google Scholar]
- Kretsinger R. H., Nockolds C. E. Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem. 1973 May 10;248(9):3313–3326. [PubMed] [Google Scholar]
- Kumazaki T., Nakako T., Arisaka F., Ishii S. A novel method for selective isolation of C-terminal peptides from tryptic digests of proteins by immobilized anhydrotrypsin: application to structural analyses of the tail sheath and tube proteins from bacteriophage T4. Proteins. 1986 Sep;1(1):100–107. doi: 10.1002/prot.340010115. [DOI] [PubMed] [Google Scholar]
- Leberman R., Egner U. Homologies in the primary structure of GTP-binding proteins: the nucleotide-binding site of EF-Tu and p21. EMBO J. 1984 Feb;3(2):339–341. doi: 10.1002/j.1460-2075.1984.tb01808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel J. A., Goldstein R. N., Marsh M., Calendar R. Structure of the bacteriophage P2 tail. Virology. 1974 Nov;62(1):161–174. doi: 10.1016/0042-6822(74)90312-2. [DOI] [PubMed] [Google Scholar]
- Lepault J., Leonard K. Three-dimensional structure of unstained, frozen-hydrated extended tails of bacteriophage T4. J Mol Biol. 1985 Apr 5;182(3):431–441. doi: 10.1016/0022-2836(85)90202-5. [DOI] [PubMed] [Google Scholar]
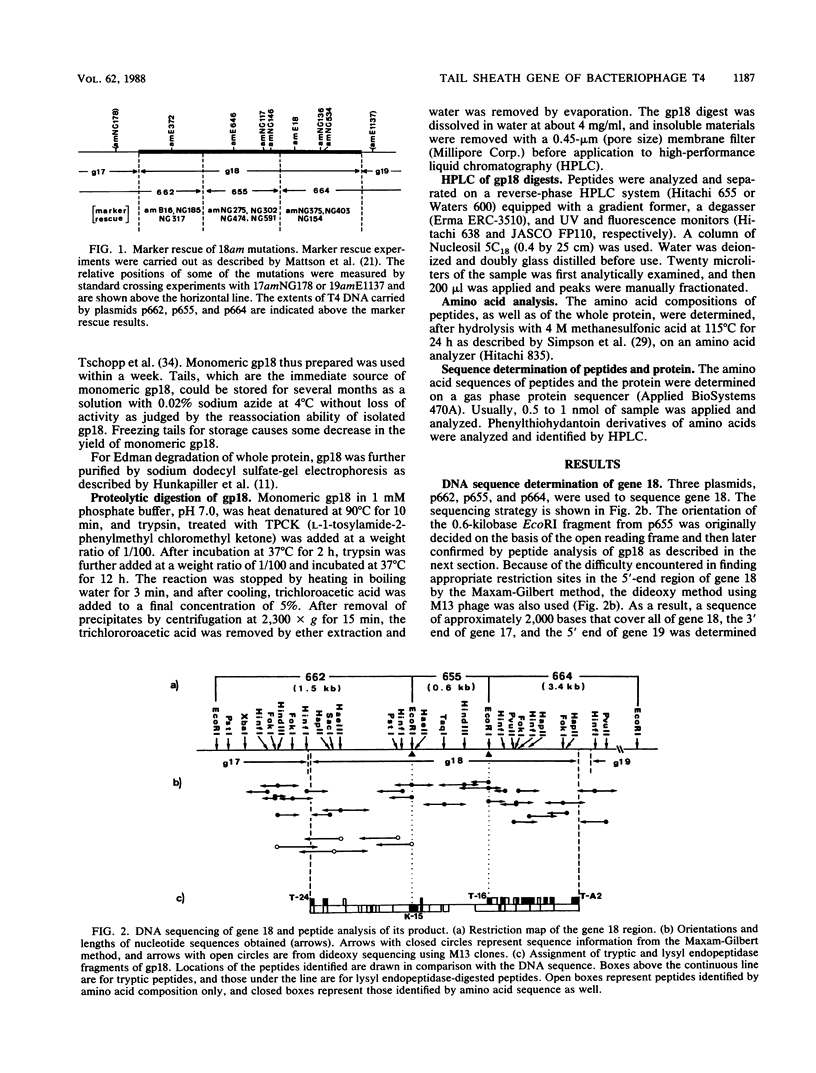
- Mattson T., Van Houwe G., Bolle A., Selzer G., Epstein R. Genetic identification of cloned fragments of bacteriophage T4 DNA and complementation by some clones containing early T4 genes. Mol Gen Genet. 1977 Sep 9;154(3):319–326. doi: 10.1007/BF00571289. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Moody M. F. Sheath of bacteriophage T4. 3. Contraction mechanism deduced from partially contracted sheaths. J Mol Biol. 1973 Nov 15;80(4):613–635. doi: 10.1016/0022-2836(73)90200-3. [DOI] [PubMed] [Google Scholar]
- Ooi T., Takanami M. A computer method for construction of secondary structure from polynucleotide sequence. Possible structure of the bacterial replication origin. Biochim Biophys Acta. 1981 Sep 28;655(2):221–229. doi: 10.1016/0005-2787(81)90012-5. [DOI] [PubMed] [Google Scholar]
- Parker M. L., Eiserling F. A. Bacteriophage SPO1 structure and morphogenesis. I. Tail structure and length regulation. J Virol. 1983 Apr;46(1):239–249. doi: 10.1128/jvi.46.1.239-249.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serysheva I. I., Tourkin A. I., Venyaminov SYu, Poglazov B. F. On the presence of guanosine phosphate in the tail of bacteriophage T4. J Mol Biol. 1984 Nov 5;179(3):565–569. doi: 10.1016/0022-2836(84)90081-0. [DOI] [PubMed] [Google Scholar]
- Shinomiya T., Shiga S. Bactericidal activity of the tail of Pseudomonas aeruginosa bacteriophage PS17. J Virol. 1979 Dec;32(3):958–967. doi: 10.1128/jvi.32.3.958-967.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
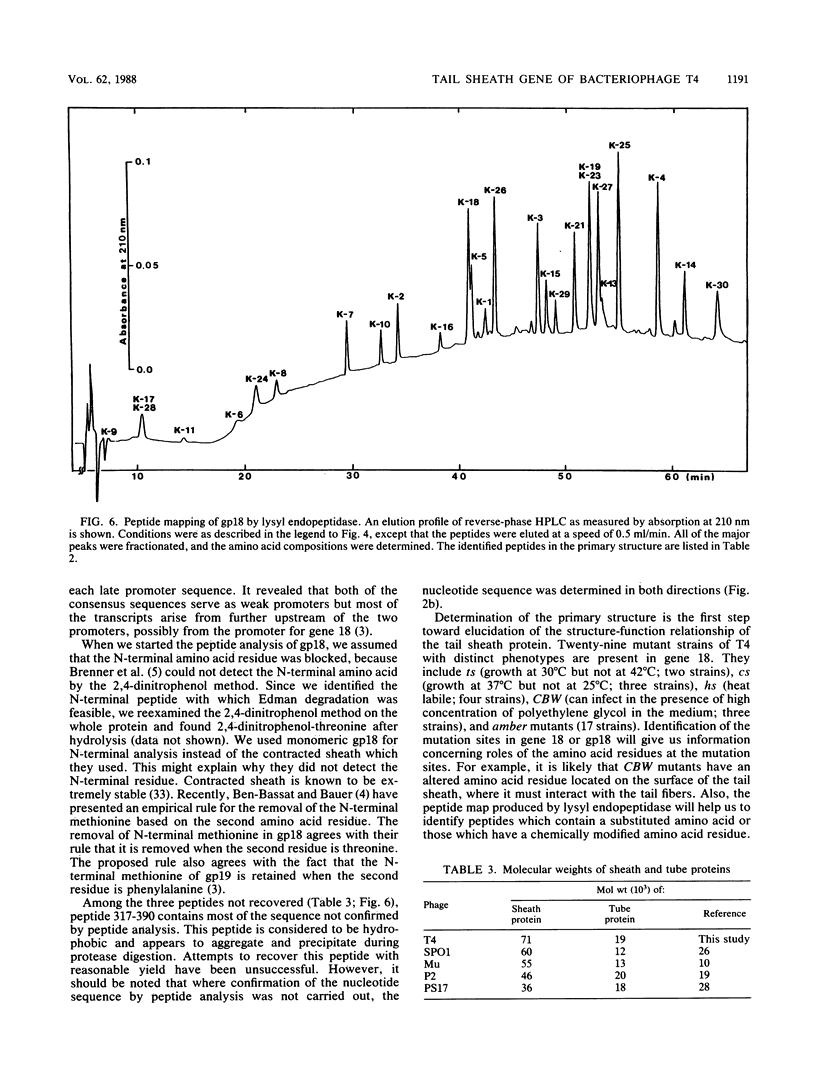
- Simpson R. J., Neuberger M. R., Liu T. Y. Complete amino acid analysis of proteins from a single hydrolysate. J Biol Chem. 1976 Apr 10;251(7):1936–1940. [PubMed] [Google Scholar]
- Smith P. R., Aebi U. Studies of the structure of the T4 bacteriophage tail sheath. I. The recovery of three-dimensional structural information from the extended sheath. J Mol Biol. 1976 Sep 15;106(2):243–271. doi: 10.1016/0022-2836(76)90083-8. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Crasemann J. M., Yegian C., Stahl M. M., Nakata A. Co-transcribed cistrons in bacteriophage T4. Genetics. 1970 Feb;64(2):157–170. doi: 10.1093/genetics/64.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg M. J., Taylor W. R. Modelling the ATP-binding site of oncogene products, the epidermal growth factor receptor and related proteins. FEBS Lett. 1984 Oct 1;175(2):387–392. doi: 10.1016/0014-5793(84)80774-7. [DOI] [PubMed] [Google Scholar]
- To C. M., Kellenberger E., Eisenstark A. Disassembly of T-even bacteriophage into structural parts and subunits. J Mol Biol. 1969 Dec 28;46(3):493–511. doi: 10.1016/0022-2836(69)90192-2. [DOI] [PubMed] [Google Scholar]


