Abstract
Triplex-forming oligonucleotides constitute an interesting DNA sequence-specific tool that can be used to target cleaving or cross-linking agents, transcription factors or nucleases to a chosen site on the DNA. They are not only used as biotechnological tools but also to induce modifications on DNA with the aim to control gene expression, such as by site-directed mutagenesis or DNA recombination. Here, we report the state of art of the triplex-based anti-gene strategy 50 years after the discovery of such a structure, and we show the importance of the actual applications and the main challenges that we still have ahead of us.
INTRODUCTION
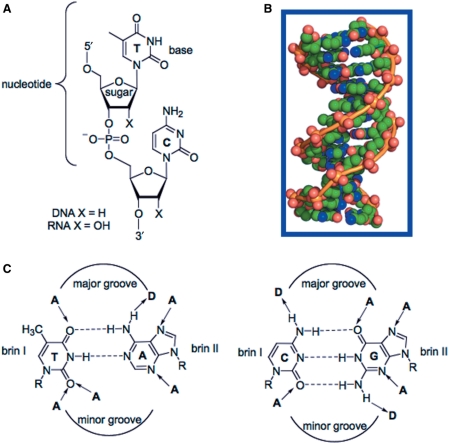
The beauty and the elegant simplicity of the structure of the DNA double helix dictate life. The discovery of the double-helical structure of the deoxyribonucleic acids by Watson, Crick, Wilkins and Franklin in 1953 (1–3) was an important milestone of modern biology. DNA is formed by two complementary strands where, through hydrogen bonds, an adenine pairs with thymine and guanine with cytosine-forming A•T and G•C base pairs, the stairs of the DNA ladder (Figure 1). The succession of base pairs defines the genetic information and gives the information to the cell to accomplish its vital functions. Four years later, Felsenfeld et al. (4) found that a chain of polyriboadenylic acid (polyrA) and two chains of polyribouridylic acid (polyrU) could form in the presence of Mg(II) a three-stranded structure. In fact, in the DNA major groove there are acceptor and donor groups that can form hydrogen bond interactions with a third strand (Figure 1C). This was the starting point for a number of studies, showing that double helices containing only purines in one chain could bind a third polynucleotide containing either pyrimidines [e.g. poly(rUC) binds poly(TC) • poly (dGA)] (5) or purines (e.g. polyG binds polyC • polyG) (6). The hydrogen bond interactions involved in triple-helix formation are different from the hydrogen-bonding pattern that holds together the Watson–Crick base pairs and they are referred to as Hoogsteen hydrogen bonds (7). But it was not till 30 years later, with the discovery that short oligonucleotides can bind in the major groove of the DNA duplex to form a triple-helical structure, that the implications and the potential of this structure were fully understood. Simultaneously, Dervan and co-workers (8) and Hélène and co-workers (9) showed that short oligonucleotides could be used to induce a DNA cleavage at a specific site on DNA through triplex formation. At the same time, Fresco and co-workers (10) and Wells and co-workers (11) contributed to this discovery by studying different triple-helical structures, suggesting a role in the control of gene expression.
Figure 1.
Chemical structure of DNA: (A) general structure of sugar-phosphate backbone; (B) space-filling model of cristal structure of a B-DNA dodecamer 5′-CGCGAATTCGCG-3′ (PDB 1BNA). The phosphoester backbones are in orange, whereas oxygen, nitrogen and carbon atoms are respectively in red, blue and green; (C) DNA base pairs bearing hydrogen bond donors (D) and acceptors (A).
Triplex-forming oligonucleotides (TFOs) are major groove ligands that target unique DNA sequences by forming DNA triple helices thanks to specific hydrogen bonding interactions between the TFO and the oligopurine strand of the duplex (Figure 2A). The use of TFO is limited to the presence of oligopyrimidine • oligopurine sequences in the DNA target and by the stability of the triple-helical structure. However, triple-helix target sites (TTS) are over-represented in the human genome and especially at promoter regions (12,13). According to Orozco and coworkers, even if TTS are not directly targeted by transcription factors, they may be important for gene functionality by acting as spacing fragment to help the correct positioning of transcription factors. Recently, it has been reported the example of an H-DNA structure (an intramolecular triplex) that modulates transcription in the human c-MYC promoter (14). Nevertheless, TFOs constitute an interesting DNA sequence-specific tool for many applications, since they are very specific DNA binders and are easy to synthesize. They have been used, for example, to target cleaving or cross-linking agents, transcription factors or nucleases to a chosen site on the DNA. Moreover, they have been used as tools for site-directed mutagenesis in order to induce modifications on DNA and to control gene expression, for example in mice (15). Finally, TFOs have found great application as biotechnological tools in various assays, for example to test translocation of proteins on DNA (16) or topoisomerase activity (17).
Figure 2.
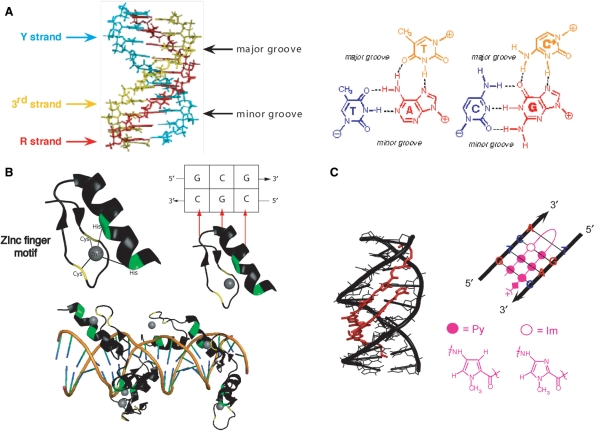
Specific recognition of DNA sequences by synthetic anti-gene molecules (TFOs, polyamides and zinc-finger proteins). (A) Triplex structure. The third strand is in yellow, whereas the oligopurine and the oligopyrimidine strand are respectively in red and blue. The triplet motifs of a pyrimidic triplex are represented, TA•T and CG•C+ respectively from left to right. (B) Crystal structure of a zinc-finger motif isolated from EGR1 (Early Growth Response protein 1, PDB 1P47). Zinc cation is coordinated to two histidine and cysteines aminoacids respectively represented in yellow and green. This zinc-finger protein binds to the DNA sequence 5′CGCGGGCGC-3′ (EGR-site). The interaction between one zinc finger and the 5′-CGC-3′ DNA sequence is represented by red arrows. Six zinc-finger motifs interact with the double strand EGR-DNA binding site. (C) Crystal structure of a polyamide dimer (HydroxyPyrrole-Imidazole-Pyrrole) on a B-DNA decamer 5’-CCAGATCTGG-3’ in red and black, respectively (PDB 1CVX). Formula of pyrrole and imidazole are represented in pink besides the schematic representation of an MGB hairpin on its double strand DNA target.
Today in the genomic era, sequence-specific DNA ligands, such as TFOs, have acquired all their importance (18). The identification of genes that play a key role in the progression and maintenance of specific diseases, such as oncogenes or tumour suppressors in cancers, calls for the use of agents able to act on specific DNA sequences. In addition to triplex-forming oligonucleotides, two other molecules are known to recognize DNA in a sequence-specific manner: zinc fingers and minor groove binders (MGBs) (Figure 2B and C).
Zinc fingers are derived from natural ligands that bind in the DNA major groove: the zinc-finger proteins (19) (Figure 2B). Zinc fingers can be synthesized by the transcription/translation machinery of the cell from introduced vectors to produce a protein able to target a DNA sequence. The binding activity of the zinc finger can be covalently linked to a transeffector (20), such as a transcription factor (21) or a catalytic domain of a protein [nucleases (18–24) or methylases (25–28)]. This strategy suffers from the difficulties in selecting the best zinc fingers with high specificity (not all three base pairs are recognized) and the cellular toxicity (29).
A number of small molecules bind specifically to the minor groove of the DNA helix. MGBs include carboxyamide ligands of N-methyl pyrrole (Py) and N-methylimidazole (Im) that form hydrogen bonds and adapt to the curvature of the DNA helix (Figure 2C) (30). A major limitation of the MGBs is their chemical stability and the length of the target DNA sequence (8–10 bp). Many efforts are carried out to increase the length of the target DNA to at least 16 bp.
In this review, we focused on the anti-gene approach based on the triplex strategy. Even if it has not been applied in therapy yet, its great potential has been widely demonstrated, both for therapeutical and biotechnological aspects. Many recent reviews have covered specific applications of triple helices (31,32). Here, we aim to report the state of the art of the triplex-based anti-gene strategy 50 years after the discovery of such structure and we intend to show the importance of actual applications and the main challenges that we still have ahead of us. We start by reviewing the general features of triplexes. Next, we review the most revealing in vitro applications, to conclude with the experiments in cells and in vivo that mostly underline how a triple helix can be used as a tool to study biological processes or interfere with them.
TRIPLEX MOTIFS
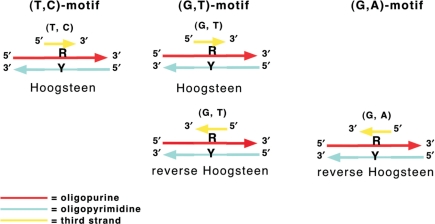
As illustrated in Figure 2A, the bases of the third strand form hydrogen bonds (Hoogsteen or reverse Hoogsteen) with the purine bases already involved in Watson–Crick base pairs forming base triplets. The rules of triplex formation are well described (33). Three classes of triple helices exist that differ in sequence composition and relative orientation of the backbone of the third strand to that of the oligopurine strand of the duplex (34) (Figure 3). In the TC triplex, the third strand is parallel (i.e. in the same 5′ to 3′ orientation) to the oligopurine strand of the oligopurine • oligopyrimidine duplex, forming T•A*T and C•G*C+ triplets in the Hoogsteen configuration. The pKa of the imino group of cytosine, which must be protonated, is well <7 making TC triplex formation pH dependent (35). In the GT triplex, the third strand can be either anti-parallel to the purine strand of the duplex by forming reverse Hoogsteen C•G*G and T•A*T triplets or parallel by forming Hoogsteen C•G*G and T•A*T triplets. In the GA triplex, the third strand is oriented anti-parallel to the purine strand of the duplex and forms reverse Hoogsteen C•G*G and T•A*A triplets.
Figure 3.
Orientation of the three triplex motifs.
The understanding of the structural features of triplexes is essential for the study of their biological functions and applications. In this context, a number of triplex structures have been investigated by NMR spectroscopy combined with molecular modelling (36–44).
The binding of the TFO to the target duplex generally results in a thermodynamically weaker interaction than the one observed between the two strands of the duplex itself (45). Moreover, the low stability of triplexes under physiological conditions is partly due to unfavourable charge repulsion between the three negatively charged DNA strands. As a consequence, high, non-physiological levels of multivalent cations, such as Mg2+ (46,47) or polyamines play a role in triplex stabilization (48,49). In the case of the purine-rich third strands, competing structures such as G-quadruplexes structures or GA homoduplexes can interfere with triplex formation. The former is stabilized by monovalent cations, such as K(I) (50), while the latter by divalent Mg(II) (51).
While triplex formation is straightforward under controlled conditions in vitro, the nuclear environment of living cells presents substantial obstacles. The third strand oligonucleotide must be nuclease resistant, overcome the charge repulsion between the third strand phosphates and those of the duplex target, not be blocked in a stable secondary structure, form a triplex in physiological pH, surmount entropic barriers to formation of a structure imposing constraints on both members of the complex and form a complex stable enough to interfere with the biological processes that act on DNA.
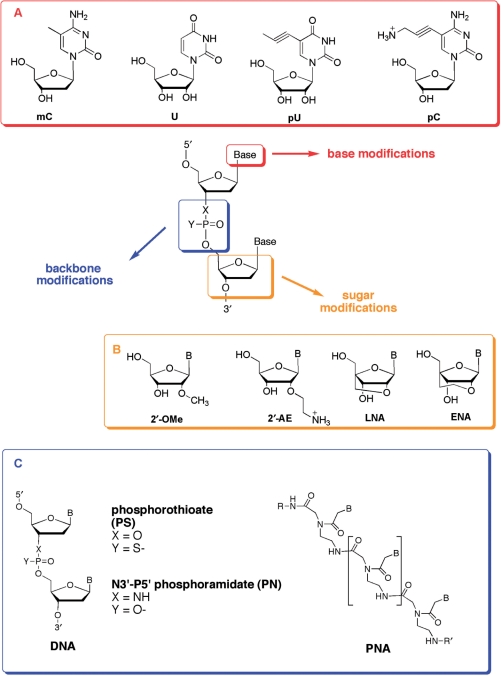
TFOs need to overcome these limitations in order to be effective. To address the issues mentioned above, chemical modifications have been incorporated in the bases (mainly C and T) (34,52), in the backbone [phosphoramidates (PN, 53), phosphothioates (PS, 54) and peptide nucleic acids (PNA, 55), or in the sugar (e.g. RNA, 56), morpholino (57), LNA (58)] (Figure 4). We list here some of the mainly used modifications.
Figure 4.
Chemical modifications introduced in TFOs. (A) Base modifications; (B) sugar modifications and (C) backbone modifications.
Nucleobase modifications
Base modifications (Figure 4A) have been studied in an attempt to stabilize triple-stranded structures. Among well-known base modifications, 5-methylcytosine (mC) was shown many years ago to ameliorate the pH restrictions on TFOs in the pyrimidine motif (59). Thymidine has also been replaced by deoxyuridine (U) (60) or 5-propynyl-deoxyuridine (pU) (61). More recently, the stabilizing properties of the positively charged C5-propargylamino-2′-deoxyuridine (UP) have been described (62–64). Such stabilization, as well as for pU, is largely independent of salt concentration but does show a pH dependence. Other similar modifications have been studied and in all cases the stabilizing effects of the propynyl or propargylamino modifications are likely to arise principally from favourable base stacking interactions (64–66).
We will not list here all purine bases that have been modified, in particular to decrease the competitive self-structures (34,52,67) [some examples are 8-aminopurines and 2′-deoxy-6-thioguanosines (68,69)].
The recognition of mixed purine/pyrimidine sequences by TFOs remains a challenge. In the last years, two main approaches, which involve a number of chemical modifications have been proposed to overcome this sequence limitation. One of these strategies is the universal base approach, which consists into conjugating intercalating agents to the 5′- or 3′-end or to internal positions of the TFO in order to stabilize the triplex containing base-pair interruptions in the purine motif (67). An alternative approach is the specific base strategy, which calls for the synthesis of new modified bases able to form hydrogen bonds with one or both partners of the A–T or the G–C Watson–Crick inverted base pairs in the major groove (70–73). When two short oligopurine sequences are present alternatively on the two strands of double-stranded DNA, two covalently linked TFOs have been used (74,75).
Sugar modifications
Modifications in the sugar moiety have also been developed. RNA is the most obvious example of substitution on the deoxyribose sugar moiety. Let us not forget that the first example of triple helix was observed with ribonucleotides polymers (4). The observation that RNA third strands formed more stable triplexes than their deoxy counterparts (56) prompted the synthesis and characterization of TFOs containing a number of ribose analogues. 2′-methoxylation (2′-OMe) (76,77) stabilizes the C3′-endo conformation of the sugar, which favours triplex formation because of least distortion of the duplex target (78). This modification allows also to avoid degradation by RNA nucleases. The 2′-aminoethylribose analogue (2′-AE) combines the C3′-endo character with a positive charge, as the amine is protonated at physiological pH (79).
It is well established that conformational restriction may lead to favorable complex formation because of an entropic advantage. Another successful approach has been to conformationally restrict the sugar part of one or more of the nucleotides of the TFO in the C3′-endo conformation, by covalently blocking the sugar using locked nucleic acid (LNA) monomers (O2′, O4′-methylene-linked nucleic acid) (80). LNA monomers have previously been shown to significantly enhance triplex stabilities (53,81), although TFOs composed entirely of LNA monomers do not form triplexes (82). O2′,O4′-ethylene-linked nucleic acid (ENA) induces slightly lower thermal stabilities of triplexes than LNA, even though fully modified ENA–TFOs are able to form stable triplexes even at pH 7.2. This kind of modifications has been differently combined and further modified in order to evaluate triplex stability (55,65). Morpholino groups confer also interesting stabilizing properties to TC triplexes in the absence of Mg(II) (57).
Most recently, a new LNA-type TFOs were described. These six-membered bridged nucleic acids analogues (called 2′-4′-BNANC) contain an N–O linkage, where the amino nitrogen can be easily substituted (83). These modified TFO in the TC motif have greater affinity for the duplex target than the corresponding LNA and ENA and improved resistance to nuclease degradation. In addition, a fully modified 2′-4′-BNANC[NH] TFO still forms a stable triplex at neutral pH.
Backbone modifications
Several backbone modifications, such as PS and PN, have been conceived in order to change the electrostatic properties of the negative phosphodiester backbone of natural DNA molecules and achieve greater degrees of nuclease resistance and cell membrane permeability (34). These cationic backbone modifications can produce both zwitterionic and fully modified cationic TFOs, thus resulting in favourable electrostatic interactions (84,85).
As described in this section, the large set of modifications available allows for a great improvement in triplex formation and stability, thus rendering the anti-gene strategy useful for various applications. The following sections of this review will try to give an overall view of the applications that have been performed and that can be envisaged.
IN VITRO APPLICATIONS OF TFOS AS GENE MODULATORS
Site-specific recognition of duplex DNA by TFOs offers a useful approach for the modification of gene structure and functions both in vitro and in vivo. In fact, triplex formation can lead to site-specific modulation of gene expression, modulation of protein binding, targeting of DNA damage, mutagenesis and enhancement of homologous recombination, thus providing a tool for gene-specific manipulation of DNA (31,86–89). At the same time, triplex technology has wide applications in molecular biology and biochemistry as a tool for delivering DNA-damaging drugs to a specific site, for sequence-specific labelling of DNA duplexes, for recognition and purification of DNA and for the study of complex DNA–protein interactions.
The efficient and specific modulation of gene expression relies on one hand on the ability of the oligonucleotide to bind with high affinity to the duplex target and to remain bound long enough in order to efficiently interfere with one of the DNA metabolic processes, on the other hand, on the specificity of recognition. This section will present an overview of the in vitro applications of triplex approach as gene modulators. Some examples will be reported with particular attention to more recent discoveries.
Modulation of transcription
The presence of a TFO in the major groove of a DNA duplex leads to major modifications in the capacity of the target duplex to be recognized by specific proteins and produces major changes in the functionality of the DNA (90–93). TFOs have been shown in vitro to alter gene expression during the transcription process (Figure 5A and B), by interfering either with the binding of transcription factors (94,95) or with the formation of the initiation complex (96). It can also arrest transcription elongation by binding to the transcribed position of the targeted gene (97). It has been shown that the affinity of the TFO for the target leads to a complex which has comparable, or even greater, stability than the complex formed by DNA and the transcription factors.
Figure 5.
Examples of use of triplexes to modulate gene expression: (A) physical block of transcription or replication elongation; (B) blockage of transcription and replication initiation and (C) targeting of DNA modifying agents such as cross-linking and cleaving agents.
A great number of examples describing the efficient transcription elongation inhibition have been reported. TFOs are used alone or conjugated to psoralen and other DNA damaging agents (98,99).
Transcription inhibition has been described for plasmid-harbored genes (87,100,101), for foreign sequences in the cellular genome (102), and in several endogenous genes including c-myc (103,104), ets2 (105), tie1 (94), HER2/neu (98), bcr/abl (107), the inflammatory mediators TNF-α (108), MCP-1 (109), or GMF/CSF (110) and the cell adhesion molecule ICAM-1 (111). A recent example of inhibition of transcription by triplex strategy has been reported on tie-1 promoter, where the formation of a stable triplex prevents the binding of Ets transcription factors that are essential for the promoter activity (94). In this case, PS TFOs inhibit promoter activity and tie1 expression in vitro and in endothelial cells.
Increasing expression of genes that are transcribed at low levels can be used as a practical treatment for some genetic diseases, such as β-globin disorder or sickle cell anemia. Song et al. (112) demonstrated that psoralen can be used to activate gene expression up to 4-fold when targeted to sites upstream the promoter. These authors designed a psoralen–TFO adapter to deliver an artificial enhancer to a disabled gene in order to recruit a transcription factor and activate transcription. However, they showed that the majority of the activation effect observed is due instead to the psoralen cross-link itself. In analogy, Xu et al. (113) reported that a TFO conjugated to psoralen activated the transcription of the γ-globin gene and a 4-fold increase in gene expression has been demonstrated in cells. In this case, the mutation introduced in the transcription factor binding site is directly responsible for gene activation. It is also possible to activate the transcription process when the triplex structure is formed in a repressor site, thus leading to a stimulation of the expression of the gene of interest (102,112). The activation of transcription has also been demonstrated in vivo with hairpin TFO in Saccharomyces cerevisiae (114). Furthermore, it has been demonstrated that hybrid molecules containing a triplex-forming sequence, linked through a phosphoroamidate bond to several minimal transcriptional activation domains derived from Herpes simplex virus protein 16 (VP16) can specifically recognize its DNA target at physiological salt and pH conditions. Bound to the double-stranded target DNA in a promoter region, the TFO–VP16 is able to activate gene expression (115).
Inhibition of replication
Some examples of inhibition of the replication process (Figure 5C) have also been reported (116,117). DNA polymerase elongation can be inhibited by in vitro triplex-formation upstream (118) or downstream (119,120) the initiation site. It has also been shown that a conjugate oligothymidine–acridine targeted toward the initiation site of SV40 can inhibit viral replication in cells (121). Even if the mechanism of this inhibition has not yet been completely elucidated, it is possible that the triple helix acts by inhibiting the interaction with helicases associated with the replication complex (122).
The interaction of triplex structures with proteins has also been studied. Several proteins, other than transcription factors and helicases have been recognized to interact specifically with triplex structures (123–127).
Site-specific mutagenesis
TFOs have been used to induce site-specific DNA damage (15,128–130), thereby enhancing the frequencies of mutation (131–133) and recombination (134–136) in vitro and in vivo. Regarding site-directed mutagenesis and recombination by triplex-forming oligonucleotides, two recent reviews described the latest discoveries in detail (137,138). Here, we just briefly summarize the essential findings. In order to trigger direct DNA damage on the target duplex, oligonucleotides have been coupled to DNA damaging agents. DNA damage from UV light, alkylating agents and photoreactive molecules such as psoralen has been shown to be recombinagenic but in a non-site-specific way. However, once the DNA damage agents are conjugated to TFO, these are able to induce site-specific mutagenesis and recombination (31). For example, the conjugation of psoralen to TFOs can mediate the introduction of base pair-specific psoralen adducts (and consequently mutations) in the target DNA (139). It has also been demonstrated that a TFO itself, without conjugation, is able to induce recombination if the third strand is capable of high-affinity binding to the target DNA and is able to stimulate recombination in a pathway dependent on nucleotide excision repair (NER) (135). These results indicate that, with efficient delivery, TFOs may be useful in promoting site-specific recombination.
Diverse damaging agents have been coupled to TFOs: (i) photoactivatable agents (140), (ii) metal complexes, such as Fe–EDTA (141), orthophenantroline (142,143) or metalloporphirines (144) and (iii) enzymes such as nucleases (145,146). The most commonly used is psoralen. It has however some serious drawbacks, such as a limited reactivity (restricted to 5′-TA or AT sites) (147,148), which greatly reduces the target options for gene mutagenesis. Nagatsugi et al. (120) recently demonstrated that a new nucleoside derivative (2-amino-6-vinylpurine derivative) exhibited triplex-mediated reactivity with high selectivity toward cytosines at a GC target site. This derivative, conjugated to a TFO, has been used to achieve site-specific modification in the supF reporter gene. TFO–chlorambucil conjugates have also been used with success thanks to the formation of a site-specific covalent guanine adducts and applied to the HER-2-neu promoter sequence (149). On the TFO side, PNAs (Figure 4) have been used with success to trigger in vitro mutation and recombination once conjugated to psoralen (130,150–152), benzophenone or anthraquinone molecules (153–155). More recently, Kim et al. (23) conceived dimeric bis-PNAs conjugated to psoralen designed to form a triplex invasion complex within the supF reporter gene and to direct site-specific photoadduct formation by the conjugated psoralen. After in vitro binding, these compounds have been shown to induce mutations at frequencies in the range of 6.5-fold and in cells at frequencies of 3.5-fold higher than the background.
Recently, it has been shown that site-specific DNA cleavage can be obtained by attaching a restriction enzyme to a TFO, as PvuII (156). Other approaches, based on the use of nucleases or restriction enzymes, have been used in the 1990s (146,157). Furthermore, it is important to underline that the presence of the triplex structure on the DNA can trigger DNA repair systems, such as the NER pathway, and be processed (158).
Improving the intracellular efficacy of the anti-gene strategy
Despite the fact that triplexes are now well characterized and validated for in vitro applications, the success of the strategy in cells still encounters some limitations, such as limited cellular penetration, target sequence accessibility in the nucleus and intracellular instability of the oligonucleotides. Fortunately, recent studies demonstrated that it is possible to overcome these difficulties and to envisage the use of the anti-gene strategy in vivo.
Because oligonucleotides are polyanions and thus do not readily penetrate biological membranes, various strategies have been developed to increase their cellular uptake. Viral delivery systems have been used in many applications and clinical trials; however, the immune response to viral proteins continues to be a daunting problem (159,160). Complex formation with cationic lipids or polycations (such as polyethylenimine) is presently commonly applied to facilitate their uptake by cells in culture, but it is not very suited for use in vivo (161–163). Another way to deliver TFOs into the cell relies on the use of dendrimers, highly branched 3D molecules (164,165). Dendrimers showed to enhance the uptake of oligonucleotides in cells (166,167). An approach that was found to be effective, both in vitro and in vivo, is conjugation of oligonucleotides to hydrophobic compounds such as cholesterol, alkyl chain or lipid, which have shown to have higher cellular uptake than their unconjugated counterparts. Cheng et al. (168) recently demonstrated that TFOs conjugated to cholesterol are not only able to penetrate efficiently the cellular and the nuclear membranes but also to inhibit transcription. Furthermore, these cholesterol-conjugated TFOs have been administrated to rats showing the desired biodistribution with enhanced uptake by liver cells, in particular hepatic stellate cells (HSCs) and hepatocytes.
Since DNA in cells is typically bound to histones and tightly packed into chromatin, the binding and activity of TFOs must be determined in this context. Chromatin structure is one of the major barriers since it may preclude TFO access to target sequences. Many efforts to demonstrate triplex formation in chromosomal environment have been reported (106,131,169–175). It has been shown that the frequency of mutagenesis induced by TFOs conjugated to psoralen differs in quiescent and S phase cells. This difference probably reflects the accessibility of the target sequence and thus the levels of targeted cross-linking in the two-cell populations (176). Many studies are still carried out in order to show triplex formation inside living cells, since other mechanisms, beside triplex formation, could be involved in oligonculeotide-induced gene inhibition. An additional barrier to the detection of triplex formation in cells is the extremely low concentration of TFO in the nuclei because of its low-penetration efficacy. Recently, Ye et al. (175) employed a TFO conjugated to psoralen into HSCs of fibrotic rats in order to target the α1 collagen gene. This gene is responsible for the synthesis of collagen, which is involved in fibrosis. The inhibition of collagen synthesis should therefore prevent fibrosis. The extent of psoralen photoadducts was evaluated using real-time PCR. The inhibition of gene transcription demonstrated that there is a strong correlation between triplex formation and transcription inhibition of TFOs.
TFOs can be also conjugated to DNA anti-cancer drugs in order to target these latter to specific sequences on DNA and increase their specificity. Carbone et al. demonstrated the efficacy of TFO–daunomycin (DNM) conjugates in inhibiting transcription of a gene involved in tumour growth: c-myc gene (104,177). DNM belongs to the family of the anthracyclins, which are among the most commonly used and effective anti-cancer drugs. Attachment of DNM to TFOs resulted in increased triplex stability thanks to the intercalating activity of DNM. Furthermore, DNM–TFO conjugates showed sequence-specific inhibition of the targeted gene, without however an effect of the anti-cancer activity of DNM. We recently demonstrated that conjugation of DNM to TFOs allowed to inhibit transcription of the target gene (MDR1) by a specific role of the DNM moiety (178). Furthermore, if these conjugates are employed in DNM-resistant cell lines, the uptake of DNM itself is permitted by the presence of the oligonucleotide, thus showing a mutual action of the partners of these conjugates, the DNM and the TFO. Recently, we have also shown that TFO conjugates of the anti-tumour camptothecin, a potent inhibitor of human topoisomerase IB, are able to induce topoisomerase I-mediated DNA cleavage in cells and to inhibit specifically the expression of a transient reporter gene in cells (100).
Interestingly, Christensen et al. (179) showed that it is possible to increase the action of anti-cancer agents when these latter are used in combination with TFOs without conjugation. When used in combination, TFOs, directed against the promoter sequence of c-myc, increased the incorporation of the anti-cancer nucleoside gemcitabine at the targeted site 4-folds due to induction of replication independent DNA synthesis. Finally, cells treated with these TFOs and gemcitabine showed a reduction in both cell survival and capacity of anchorage-independent growth.
Therapeutic potential of TFOs
Despite the great number of in vitro applications and an example of site-directed mutagenesis in mice (15), the anti-gene strategy did not lead to new therapeutical agents so far. However, the improvements in the stability of triplexes in physiological conditions and the examples reported so far clearly demonstrate that the anti-gene strategy can be very effective. Furthermore, bioinformatics is giving an essential support to the future applications of TFOs in therapy. In fact, a wide analysis along annotated regions of the genome allowed to demonstrate that the largest relative concentration of TTS is found in regulatory regions, especially in promoter regions (12,13). This suggests a great potentiality for triplex strategy in the control of gene expression. Softwares able to identify high-affinity TTS have also been developed (180). While a certain number of practical problems (stability, sequence restrictions, susceptibility to nucleases and delivery in the cellular nucleus) have to be solved before to apply the anti-gene strategy in therapy, some new biotechnological applications are emerging as an important way to exploit the potential of triplex formation. The following section is devoted to the description of the use of TFOs as biochemical tools.
TFOS AS TOOLS IN MOLECULAR BIOLOGY AND BIOCHEMISTRY
While the applications described so far regard the use of the triplex approach in the modulation of gene expression, the following section will examine how triplexes, mainly the same conjugates as above, can be employed as biotechnological tools. In this context, TFOs can be employed in targeting of drugs to study, for example, molecular mechanisms of protein and enzymes interacting with DNA, site-specific labelling of DNA and in the recognition and purification of DNA (Figure 6).
Figure 6.
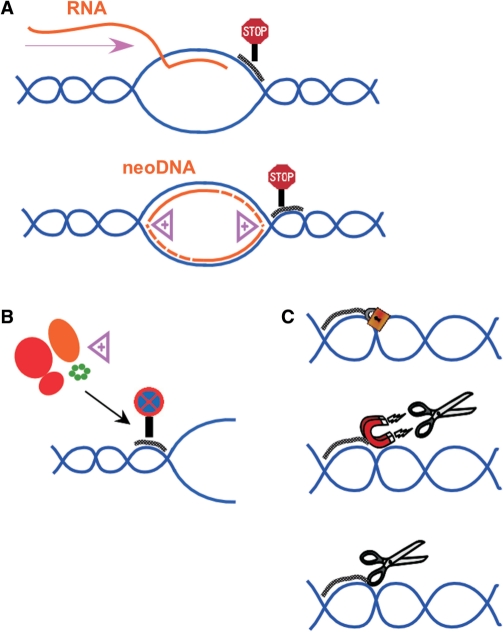
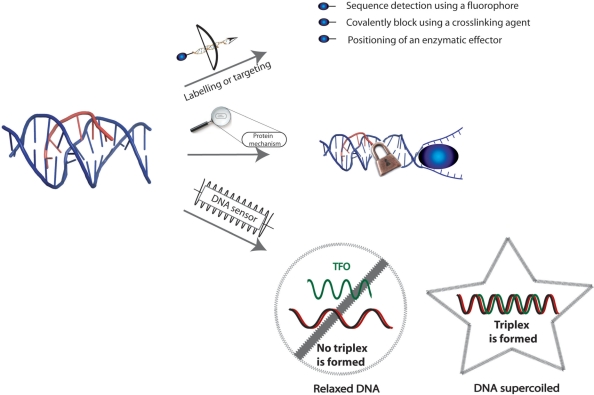
Triplexes as biotechnological tools: to target a molecule of interest on a specific DNA sequence, such as fluorophores, enzymatic effectors (transcription factors, restriction enzymes), cross-linking agents; to study the mechanisms of proteins that act on DNA, such as translocases; to sense DNA topology: using immobilized TFOs, it is possible to trap negatively supercoiled plasmids versus relaxed plasmids.
First of all, targeting of therapeutic agents to specific sites of DNA revealed to be a useful strategy to induce the effect of these drugs in a sequence-specific manner. The first example of this approach has been reported by Matteucci et al. (181), who described a system where a topoisomerase I inhibitor, camptothecin, has been coupled to a TFO in order to target the inhibition of the enzyme at proximity of the triplex. Topoisomerase I is a cellular enzyme whose function is to relax the superhelical twist in DNA by cleavage of one strand, rotation around the cleaved position and religation of the DNA (182). Inhibitors of this enzyme, such as camptothecin, trap the enzyme in the cleavable complex where the DNA is cleaved and covalently linked to the enzyme, thus leading to durable lesions of DNA. When camptothecin is conjugated to a TFO, subsequent triplex formation at the target sequence positions the drug selectively at the triplex site, thereby stimulating topoisomerase I-mediated cleavage at this site. The specificity and the efficacy of cleavage depend markedly on the length of both the triple-helical structure and the linker between the oligonucleotide and the poison (183,184). Diverse analogues of camptothecin were studied by this approach and it has been demonstrated that even poorly active or inactive camptothecin derivatives were able to stabilize the ternary complex (185). Other topoisomerase I inhibitors have been used for this approach, such as for example rebeccamycin (186) and indolocarbazole (187). Most recently, we have used these conjugates to position camptothecins at one specific site on a ∼19.2 kb DNA to study, in a single-molecule set-up, the physical parameters of the inhibition of topoisomerase I by the drug (188).
The same principle has been applied to topoisomerase II inhibitors, such as anthracyclines, widely used and very effective anti-cancer agents (189,190). The conjugation of DNM to a TFO allows the delivery of the anthracycline to a specific gene and has been applied to the PPT sequence of HIV-1 in vitro, as well as to c-myc gene in cells (104,177) (see Site-specific mutagenesis section). Recently, we reported that new derivatives of etoposide (a topoisomerase II inhibitor widely used in therapy) have been coupled to TFOs in order to increase the sequence specificity of these drugs (191,192). The active topoisomerase II poisons, once linked, induced cleavage at 13–14 bp from the triplex end where the drug was attached. Thus, also in this case, the use of triple-helical DNA structures offered an efficient strategy for targeting topoisomerase II-mediated cleavage to DNA-specific sequences. The use of an etoposide analogue able to photocrosslink the DNA target allowed to draw a model for the interaction of these conjugated analogues in the ternary complex with topoisomerase II, where the inhibitor is not positioned as expected in the catalytic site of the enzyme, but in an outer part of topoisomerase II, most likely in a key region that is important for the conformational changes of the enzyme through the catalytic cycle. Thus, the triplex approach revealed its usefulness as a tool to unravel mechanistic details of the complex process of topoisomerase poisoning.
Concerning the study of protein–DNA interactions, the triplex approach can help understand the detailed molecular mechanism, as described for topoisomerase inhibitors and their interaction with DNA and the enzyme. Triplex directed site-specific psoralen interstrand cross-link (ICL) has been used as a model substrate to explore the molecular mechanism of psoralen ICL repair in human cells. It is known that NER system is involved in a mutagenic repair of psoralen ICLs in mammalian cells (132,134,193,194). NER is a multistep process involving at least 25 proteins, including the damaged DNA recognition factors XPA and RPA. Recently, it has been demonstrated that other proteins are also crucial for efficient processing and error-free repair of psoralen ICLs, showing how the triplex approach can be applied in the study of repair pathways (88,195,196).
Reddy et al. (195) studied the recognition of psoralen-cross-linked triplex structure by the HMGB1 and HMGB2 proteins and their interactions with the NER damage recognition proteins, XPA and RPA, on these lesions. Authors reported that the human HMGB1 protein recognizes and binds to psoralen-cross-linked triplex DNA with high affinity and specificity even in the presence of the XPA–RPA complex. This binding is supposed to modulate the repair of these mutagenic structures.
Another possible application is the recognition and purification of nucleic acids. A big advantage is the fact that pyrimidine triplex formation is pH-dependent and thus tunable. Triplexes can be used in order to select a sequence of DNA containing oligopurine•oligopyrimidine sequences in a mixture of duplex DNAs (197). This has been realized by attaching a TFO to an affinity column or magnetic beads. In the presence of a mixture of duplexes, for example plasmids containing a sequence able to form a triple helix can be selectively recognized inside a bacterial lysate (198–201).
This approach has been also used in order to recognize the interaction sites of proteins on DNA, since the third strand can act as a competitor for the protein (125,202,203). An important example is the identification of topoisomerase II binding and cleavage site (204). Triplexes have been used as a basis for assays for DNA translocation (205,206). The principle of these assays is the displacement of a fluorescent-labelled TFO by the translocating enzyme. Maxwell et al. (17) exploited the observation that triplex formation is favoured by negative supercoiling in order to develop methods for assaying topoisomerases, based on the differential capture of negatively supercoiled versus relaxed plasmids by immobilized TFOs.
TFOs can also be used in order to label a target DNA. Biotin has been employed individually in studies based on triplex structures (198,207,208). Biotin has also been used in combination with psoralen each attached to one extremity of a TFO (209). These probes were directed to different target sites on plasmids and were photocrosslinked to the target DNAs via the psoralen moiety. The yield of triple helix can be evaluated by chemiluminescence using avidin or streptavidin as protein tags and the covalent adduct can be visualized by scanning force microscopy. Grimm et al. (210) developed a simple synthetic method for conjugation of an intercalating ruthenium diphenanthroline dipyridophenazine complex with a TFO. Once the triplex is formed with the target duplex (the PPT sequence of HIV1), the ruthenium complex intercalates at the triplex–duplex junction leading to an important increasing of fluorescence that allows to follow the kinetics of duplex and triplex formation by fluorescence spectrometry.
Escudé et al. (211) have shown that a TFO could be circularized around its target, yielding the so-called ‘padlock’ oligonucleotide. After sequence-specific recognition of a double-stranded DNA target through triple-helix formation, the ends of the TFO were joined through the action of T4 DNA ligase, thus creating a circular DNA molecule catenated to the plasmid containing the target sequence. Padlock oligonucleotides have been used for fluorescent site-specific labelling (211–213). Géron-Landre et al. (214) reported the ligation of a fluorescent DNA fragment to a stem-loop TFO to visualize DNA. This is the first report where a non-repeated sequence as short as 15 bp has been visualized by optical microscopy. Thus, this method could be used for single molecule studies of DNA. The same group reported the use of radioactive labelling applied to an analogous approach (215).
In 2003, Hausmann et al. (216) introduced combinatorial oligonucleotide fluorescence in situ hybridization (COMBO-FISH), which uses TFOs to label chromosomes in a cell nucleus under non-denaturing conditions. More recently, it has been demonstrated that pyrene-labelled oligonucleotides are great probes that are able to discriminate single mutation in purine stretches. Such probes have potential for use in fluorescence in vivo hybridization under vital conditions and in living cells (217).
CONCLUSIONS
Fifty years later, we know now how to design short oligonucleotides able to form stable triplexes on the oligopyrimidine•oligopurine sequences present in the genome. We have also learned to deliver them and improve their cellular stability, i.e. controlling the affinity for the duplex target, salt concentration and pH dependence or overcome intracellular barriers. Triplexes have become useful tools that are widely used to target DNA modifying agents, with the aim either to study biological processes or to modulate them (it is the case of gene expression, mutagenesis and recombination); to label DNA, study enzymes that act on DNA, purify DNA etc. An important step forward in the anti-gene strategy will be the use of triplexes in vivo as therapeutic agents for a wide variety of diseases including cancer or viral infections, in which a sequence can be identified as responsible of the disease. Their high specificity leads also to a theoretical lack of toxic effects.
On the other hand, other oligonucleotides-based therapies are in use, such as anti-sense strategy (218); siRNAs have also emerged as a very effective way to modulate gene expression (219). However, both target the second step of the genetic information flux without affecting the real source, the DNA.
Based on the great progresses made these recent years and summarized in this review, it is definitely possible to overcome the practical problems that are still limiting the anti-gene approach and to finally apply this approach in therapy.
Finally, the importance of the triplex strategy resides also in its wide applications in biotechnology, where TFOs have shown to be versatile tools to deliver drugs to a specific site in the genome, to study the molecular mechanism of enzymes, to label DNA site-specifically and to recognize and purify DNA.
FUNDING
Funding to pay the Open Access publication charges for the article was provided by INSERM U565.
Conflict of interest statement. None declared.
REFERENCES
- 1.Franklin RE, Gosling RG. Evidence for 2-chain helix in crystalline structure of sodium deoxyribonucleate. Nature. 1953;172:156–157. doi: 10.1038/172156a0. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Wilkins MH, Stokes AR, Wilson HR. Molecular structure of deoxypentose nucleic acids. Nature. 1953;171:738–740. doi: 10.1038/171738a0. [DOI] [PubMed] [Google Scholar]
- 4.Felsenfeld G, Davies DR, Rich A. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023. [Google Scholar]
- 5.Morgan AR, Wells RD. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J. Mol. Biol. 1968;37:63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- 6.Beal PA, Dervan PB. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 7.Hoogsteen K. The structures of crystals containing a hydrogen complex bonding of 1-methylthymine and 9-methyladenine. Acta Cryst. 1959;12:822–823. [Google Scholar]
- 8.Moser HE, Dervan PB. Sequence specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 9.Le Doan T, Perrouault L, Praseuth D, Habhoub N, Decout JL, Thuong NT, Lhomme J, Helene C. Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-[alpha]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broitman SL, Im DD, Fresco JR. Formation of the triple-stranded polynucleotide helix, poly(A.A.U) Proc. Natl Acad. Sci. USA. 1987;84:5120–5124. doi: 10.1073/pnas.84.15.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells RD, Collier DA, Hanvey JC, Shimizu M, Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988;2:2939–2949. [PubMed] [Google Scholar]
- 12.Goni JR, de la Cruz X, Orozco M. Triplex-forming oligonucleotide target sequences in the human genome. Nucleic Acids Res. 2004;32:354–360. doi: 10.1093/nar/gkh188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goni JR, Vaquerizas JM, Dopazo J, Orozco M. Exploring the reasons for the large density of triplex-forming oligonucleotide target sequences in the human regulatory regions. BMC Genomics. 2006;7:63. doi: 10.1186/1471-2164-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007;282:32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 15.Vasquez KM, Narayanan L, Glazer PM. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 16.Levy O, Ptacin JL, Pease PJ, Gore J, Eisen MB, Bustamante C, Cozzarelli NR. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc. Natl Acad. Sci. USA. 2005;102:17618–17623. doi: 10.1073/pnas.0508932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maxwell A, Burton NP, O'Hagan N. High-throughput assays for DNA gyrase and other topoisomerases. Nucleic Acids Res. 2006;34:e104. doi: 10.1093/nar/gkl504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pingoud A, Silva GH. Precision genome surgery. Nat. Biotechnol. 2007;25:743–744. doi: 10.1038/nbt0707-743. [DOI] [PubMed] [Google Scholar]
- 19.Desjarlais JR, Berg JM. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proc. Natl Acad. Sci. USA. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandell JG, Barbas C.F., III Zinc finger tools: custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006;34:W516–W523. doi: 10.1093/nar/gkl209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beerli RR, Barbas C.F., III Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 22.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 23.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 24.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 25.Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, Jeltsch A. Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic Acids Res. 2007;35:100–112. doi: 10.1093/nar/gkl1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minczuk M, Papworth MA, Kolasinska P, Murphy MP, Klug A. Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. Proc. Natl Acad. Sci. USA. 2006;103:19689–19694. doi: 10.1073/pnas.0609502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nomura W, Barbas C.F., III In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J. Am. Chem. Soc. 2007;129:8676–8677. doi: 10.1021/ja0705588. [DOI] [PubMed] [Google Scholar]
- 28.Smith AE, Ford KG. Specific targeting of cytosine methylation to DNA sequences in vivo. Nucleic Acids Res. 2007;35:740–754. doi: 10.1093/nar/gkl1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Kandavelou K, Chandrasegaran S. Custom-designed zinc finger nucleases: what is next? Cell. Mol. Life Sci. 2007;64:2933–2944. doi: 10.1007/s00018-007-7206-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dervan PB, Doss RM, Marques MA. Programmable DNA binding oligomers for control of transcription. Curr. Med. Chem. Anticancer Agents. 2005;5:373–387. doi: 10.2174/1568011054222346. [DOI] [PubMed] [Google Scholar]
- 31.Rogers FA, Lloyd JA, Glazer PM. Triplex-forming oligonucleotides as potential tools for modulation of gene expression. Curr. Med. Chem. Anticancer Agents. 2005;5:319–326. doi: 10.2174/1568011054222300. [DOI] [PubMed] [Google Scholar]
- 32.Jain A, Wang G, Vasquez KM. DNA triple helices: biological consequences and therapeutic potential. Biochimie. 2008 doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thuong NT, Hélène C. Sequence-specific recognition and modification of double-helical DNA by oligonucleotides. Angew. Chem. Int. Ed. 1993;32:666–690. [Google Scholar]
- 34.Buchini S, Leumann CJ. Recent improvements in antigene technology. Curr. Opin. Chem. Biol. 2003;7:717–726. doi: 10.1016/j.cbpa.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Manzini G, Xodo LE, Gasparotto D, Quadrifoglio F, van der Marel GA, van Boom JH. Triple helix formation by oligopurine-oligopyrimidine DNA fragments. Electrophoretic and thermodynamic behavior. J. Mol. Biol. 1990;213:833–843. doi: 10.1016/S0022-2836(05)80267-0. [DOI] [PubMed] [Google Scholar]
- 36.de los Santos C, Rosen M, Patel D. NMR studies of DNA (R +)n.(Y-)n.(Y+)n triple helices in solution: imino and amino proton markers of T.A.T and C.G.C+ base-triple formation. Biochemistry. 1989;28:7282–7289. doi: 10.1021/bi00444a021. [DOI] [PubMed] [Google Scholar]
- 37.Kan LS, Pasternack L, Wey MT, Tseng YY, Huang DH. The paperclip triplex: understanding the role of apex residues in tight turns. Biophys. J. 2006;91:2552–2563. doi: 10.1529/biophysj.106.084137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koshlap KM, Schultze P, Brunar H, Dervan PB, Feigon J. Solution structure of an intramolecular DNA triplex containing an N7-glycosylated guanine which mimics a protonated cytosine. Biochemistry. 1997;36:2659–2668. doi: 10.1021/bi962438a. [DOI] [PubMed] [Google Scholar]
- 39.Macaya R, Wang E, Schultze P, Sklenar V, Feigon J. Proton nuclear magnetic resonance assignments and structural characterization of an intramolecular DNA triplex. J. Mol. Biol. 1992;225:755–773. doi: 10.1016/0022-2836(92)90399-5. [DOI] [PubMed] [Google Scholar]
- 40.Pasternack LB, Lin SB, Chin TM, Lin WC, Huang DH, Kan LS. Proton NMR studies of 5′-d-(TC)(3) (CT)(3) (AG)(3)-3′—a paperclip triplex: the structural relevance of turns. Biophys. J. 2002;82:3170–3180. doi: 10.1016/S0006-3495(02)75659-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radhakrishnan I, Patel DJ. Solution structure of a pyrimidine.purine.pyrimidine DNA triplex containing T.AT, C+.GC and G.TA triples. Structure. 1994;2:17–32. doi: 10.1016/s0969-2126(00)00005-8. [DOI] [PubMed] [Google Scholar]
- 42.Radhakrishnan I, Patel DJ. Solution structure and hydration patterns of a pyrimidine.purine.pyrimidine DNA triplex containing a novel T.CG base-triple. J. Mol. Biol. 1994;241:600–619. doi: 10.1006/jmbi.1994.1534. [DOI] [PubMed] [Google Scholar]
- 43.Rajagopal P, Feigon J. NMR studies of triple-strand formation from the homopurine-homopyrimidine deoxyribonucleotides d(GA)4 and d(TC)4. Biochemistry. 1989;28:7859–7870. doi: 10.1021/bi00445a048. [DOI] [PubMed] [Google Scholar]
- 44.Warmlander S, Sandstrom K, Leijon M, Graslund A. Base-pair dynamics in an antiparallel DNA triplex measured by catalyzed imino proton exchange monitored via 1H NMR spectroscopy. Biochemistry. 2003;42:12589–12595. doi: 10.1021/bi034479u. [DOI] [PubMed] [Google Scholar]
- 45.Shafer RH. In: Progress in Nucleic Acid Research. Moldave K, editor. Vol. 59. Academic Press Inc: San Diego, CA; 1998. pp. 55–94. [DOI] [PubMed] [Google Scholar]
- 46.Floris R, Scaggiante B, Manzini G, Quadrifoglio F, Xodo LE. Effect of cations on purine.purine.pyrimidine triple helix formation in mixed-valence salt solutions. Eur. J. Biochem. 1999;260:801–809. doi: 10.1046/j.1432-1327.1999.00219.x. [DOI] [PubMed] [Google Scholar]
- 47.Blume SW, Lebowitz J, Zacharias W, Guarcello V, Mayfield CA, Ebbinghaus SW, Bates P, Jones D.E., Jr, Trent J, Vigneswaran N, et al. The integral divalent cation within the intermolecular purine*purine. pyrimidine structure: a variable determinant of the potential for and characteristics of the triple helical association. Nucleic Acids Res. 1999;27:695–702. doi: 10.1093/nar/27.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas T, Thomas TJ. Selectivity of polyamines in triplex DNA stabilization. Biochemistry. 1993;32:14068–14074. doi: 10.1021/bi00213a041. [DOI] [PubMed] [Google Scholar]
- 49.Thomas TJ, Kulkarni GD, Greenfield NJ, Shirahata A, Thomas T. Structural specificity effects of trivalent polyamine analogues on the stabilization and conformational plasticity of triplex DNA. Biochem. J. 1996;319:591–599. doi: 10.1042/bj3190591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 51.Arimondo PB, Barcelo F, Sun JS, Maurizot JC, Garestier T, Helene C. Triple helix formation by (G,A)-containing oligonucleotides: asymmetric sequence effect. Biochemistry. 1998;37:16627–16635. doi: 10.1021/bi9805588. [DOI] [PubMed] [Google Scholar]
- 52.Seidman MM, Glazer PM. The potential for gene repair via triple helix formation. J. Clin. Invest. 2003;112:487–494. doi: 10.1172/JCI19552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torigoe H, Hari Y, Sekiguchi M, Obika S, Imanishi T. 2′-O,4′-C-methylene bridged nucleic acid modification promotes pyrimidine motif triplex DNA formation at physiological pH: thermodynamic and kinetic studies. J. Biol. Chem. 2001;276:2354–2360. doi: 10.1074/jbc.M007783200. [DOI] [PubMed] [Google Scholar]
- 54.Lacoste J, Francois JC, Helene C. Triple helix formation with purine-rich phosphorothioate-containing oligonucleotides covalently linked to an acridine derivative. Nucleic Acids Res. 1997;25:1991–1998. doi: 10.1093/nar/25.10.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hojland T, Kumar S, Babu BR, Umemoto T, Albaek N, Sharma PK, Nielsen P, Wengel J. LNA (locked nucleic acid) and analogs as triplex-forming oligonucleotides. Org. Biomol. Chem. 2007;5:2375–2379. doi: 10.1039/b706101c. [DOI] [PubMed] [Google Scholar]
- 56.Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 57.Lacroix L, Arimondo PB, Takasugi M, Helene C, Mergny JL. Pyrimidine morpholino oligonucleotides form a stable triple helix in the absence of magnesium ions. Biochem. Biophys. Res. Commun. 2000;270:363–369. doi: 10.1006/bbrc.2000.2438. [DOI] [PubMed] [Google Scholar]
- 58.Grunweller A, Hartmann RK. Locked nucleic acid oligonucleotides: the next generation of antisense agents? BioDrugs. 2007;21:235–243. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 59.Lee JS, Woodsworth ML, Latimer LJ, Morgan AR. Poly(pyrimidine). poly(purine) synthetic DNAs containing 5-methylcytosine form stable triplexes at neutral pH. Nucleic Acids Res. 1984;12:6603–6614. doi: 10.1093/nar/12.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mills M, Arimondo PB, Lacroix L, Garestier T, Klump H, Mergny JL. Chemical modification of the third strand: differential effects on purine and pyrimidine triple helix formation. Biochemistry. 2002;41:357–366. doi: 10.1021/bi011122m. [DOI] [PubMed] [Google Scholar]
- 61.Froehler BC, Wadwani S, Terhorst TJ, Gerrard SR. Oligodeoxynucleotides containing C-5 propyne analogs of 2′-deoxyuridine and 2′-deoxycytidine. Tetrahedron Lett. 1992;33:5307–5310. [Google Scholar]
- 62.Bijapur J, Keppler MD, Bergqvist S, Brown T, Fox KR. 5-(1-propargylamino)-2′-deoxyuridine (UP): a novel thymidine analogue for generating DNA triplexes with increased stability. Nucleic Acids Res. 1999;27:1802–1809. doi: 10.1093/nar/27.8.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sollogoub M, Darby RA, Cuenoud B, Brown T, Fox KR. Stable DNA triple helix formation using oligonucleotides containing 2′-aminoethoxy,5-propargylamino-U. Biochemistry. 2002;41:7224–7231. doi: 10.1021/bi020164n. [DOI] [PubMed] [Google Scholar]
- 64.Brazier JA, Shibata T, Townsley J, Taylor BF, Frary E, Williams NH, Williams DM. Amino-functionalized DNA: the properties of C5-amino-alkyl substituted 2′-deoxyuridines and their application in DNA triplex formation. Nucleic Acids Res. 2005;33:1362–1371. doi: 10.1093/nar/gki254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alam MR, Majumdar A, Thazhathveetil AK, Liu ST, Liu JL, Puri N, Cuenoud B, Sasaki S, Miller PS, Seidman MM. Extensive sugar modification improves triple helix forming oligonucleotide activity in vitro but reduces activity in vivo. Biochemistry. 2007;46:10222–10233. doi: 10.1021/bi7003153. [DOI] [PubMed] [Google Scholar]
- 66.Rusling DA, Le Strat L, Powers VE, Broughton-Head VJ, Booth J, Lack O, Brown T, Fox KR. Combining nucleoside analogues to achieve recognition of oligopurine tracts by triplex-forming oligonucleotides at physiological pH. FEBS Lett. 2005;579:6616–6620. doi: 10.1016/j.febslet.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 67.Chan PP, Glazer PM. Triplex DNA: fundamentals, advances, and potential applications for gene therapy. J. Mol. Med. 1997;75:267–282. doi: 10.1007/s001090050112. [DOI] [PubMed] [Google Scholar]
- 68.Avino A, Cubero E, Gonzalez C, Eritja R, Orozco M. Antiparallel triple helices. Structural characteristics and stabilization by 8-amino derivatives. J. Am. Chem. Soc. 2003;125:16127–16138. doi: 10.1021/ja035039t. [DOI] [PubMed] [Google Scholar]
- 69.Spackova N, Cubero E, Sponer J, Orozco M. Theoretical study of the guanine - -> 6-thioguanine substitution in duplexes, triplexes, and tetraplexes. J. Am. Chem. Soc. 2004;126:14642–14650. doi: 10.1021/ja0468628. [DOI] [PubMed] [Google Scholar]
- 70.Guianvarc’h D, Benhida R, Fourrey JL, Maurisse R, Sun JS. Incorporation of a novel nucleobase allows stable oligonucleotide-directed triple helix formation at the target sequence containing a purine.pyrimidine interruption. Chem. Commun. 2001:1814–1815. doi: 10.1039/b103743a. [DOI] [PubMed] [Google Scholar]
- 71.Guianvarc’h D, Fourrey JL, Maurisse R, Sun JS, Benhida R. Synthesis, incorporation into triplex-forming oligonucleotide, and binding properties of a novel 2′-deoxy-C-nucleoside featuring a 6-(thiazolyl-5)benzimidazole nucleobase. Org. Lett. 2002;4:4209–4212. doi: 10.1021/ol026609h. [DOI] [PubMed] [Google Scholar]
- 72.Guianvarc’h D, Fourrey JL, Maurisse R, Sun JS, Benhida R. Design of artificial nucleobases for the recognition of the AT inversion by triple-helix forming oligonucleotides: a structure-stability relationship study and neighbour bases effect. Bioorg. Med. Chem. 2003;11:2751–2759. doi: 10.1016/s0968-0896(03)00229-3. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Rusling DA, Powers VE, Lack O, Osborne SD, Fox KR, Brown T. Stable recognition of TA interruptions by triplex forming oligonucleotides containing a novel nucleoside. Biochemistry. 2005;44:5884–5892. doi: 10.1021/bi050013v. [DOI] [PubMed] [Google Scholar]
- 74.de Bizemont T, Duval-Valentin G, Sun JS, Bisagni E, Garestier T, Helene C. Alternate strand recognition of double-helical DNA by (T,G)-containing oligonucleotides in the presence of a triple helix-specific ligand. Nucleic Acids Res. 1996;24:1136–1143. doi: 10.1093/nar/24.6.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Bizemont T, Sun JS, Garestier T, Helene C. New junction models for alternate-strand triple-helix formation. Chem. Biol. 1998;5:755–762. doi: 10.1016/s1074-5521(98)90667-6. [DOI] [PubMed] [Google Scholar]
- 76.Escudé C, Sun JS, Rougée M, Garestier T, Hélène C. Stable triple helices are formed upon binding of RNA oligonucleotides and their 2′-O-methyl derivatives to double-helical DNA. C. R. Acad. Sci. Paris, Serie III. 1992;315:521–525. [PubMed] [Google Scholar]
- 77.Shimizu M, Konishi A, Shimada Y, Inoue H, Ohtsuka E. Oligo(2′-O-methyl)ribonucleotides. Effective probes for duplex DNA. FEBS Lett. 1992;302:155–158. doi: 10.1016/0014-5793(92)80428-j. [DOI] [PubMed] [Google Scholar]
- 78.Asensio JL, Brown T, Lane AN. Solution conformation of a parallel DNA triple helix with 5′ and 3′ triplex-duplex junctions. Structure. 1999;7:1–11. doi: 10.1016/s0969-2126(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 79.Seidman MM, Puri N, Majumdar A, Cuenoud B, Miller PS, Alam R. The development of bioactive triple helix-forming oligonucleotides. Ann. N Y Acad. Sci. 2005;1058:119–127. doi: 10.1196/annals.1359.020. [DOI] [PubMed] [Google Scholar]
- 80.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 81.Brunet E, Alberti P, Perrouault L, Babu R, Wengel J, Giovannangeli C. Exploring cellular activity of locked nucleic acid-modified triplex-forming oligonucleotides and defining its molecular basis. J. Biol. Chem. 2005;280:20076–20085. doi: 10.1074/jbc.M500021200. [DOI] [PubMed] [Google Scholar]
- 82.Sun BW, Babu BR, Sorensen MD, Zakrzewska K, Wengel J, Sun JS. Sequence and pH effects of LNA-containing triple helix-forming oligonucleotides: physical chemistry, biochemistry, and modeling studies. Biochemistry. 2004;43:4160–4169. doi: 10.1021/bi036064e. [DOI] [PubMed] [Google Scholar]
- 83.Rahman SM, Seki S, Obika S, Yoshikawa H, Miyashita K, Imanishi T. Design, synthesis, and properties of 2′,4′-BNA(NC): a bridged nucleic acid analogue. J. Am. Chem. Soc. 2008;130:4886–4896. doi: 10.1021/ja710342q. [DOI] [PubMed] [Google Scholar]
- 84.Michel T, Debart F, Heitz F, Vasseur JJ. Highly stable DNA triplexes formed with cationic phosphoramidate pyrimidine alpha-oligonucleotides. Chembiochem. 2005;6:1254–1262. doi: 10.1002/cbic.200400436. [DOI] [PubMed] [Google Scholar]
- 85.Michel T, Martinand-Mari C, Debart F, Lebleu B, Robbins I, Vasseur JJ. Cationic phosphoramidate alpha-oligonucleotides efficiently target single-stranded DNA and RNA and inhibit hepatitis C virus IRES-mediated translation. Nucleic Acids Res. 2003;31:5282–5290. doi: 10.1093/nar/gkg733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Besch R, Giovannangeli C, Degitz K. Triplex-forming oligonucleotides - sequence-specific DNA ligands as tools for gene inhibition and for modulation of DNA-associated functions. Curr. Drug Targets. 2004;5:691–703. doi: 10.2174/1389450043345100. [DOI] [PubMed] [Google Scholar]
- 87.Faria M, Giovannangeli C. Triplex-forming molecules: from concepts to applications. J. Gene Med. 2001;3:299–310. doi: 10.1002/jgm.192. [DOI] [PubMed] [Google Scholar]
- 88.Vasquez KM, Glazer PM. Triplex-forming oligonucleotides: principles and applications. Q Rev. Biophys. 2002;35:89–107. doi: 10.1017/s0033583502003773. [DOI] [PubMed] [Google Scholar]
- 89.Maurisse R, Feugeas JP, Biet E, Kuzniak I, Leboulch P, Dutreix M, Sun JS. A new method (GOREC) for directed mutagenesis and gene repair by homologous recombination. Gene Ther. 2002;9:703–707. doi: 10.1038/sj.gt.3301736. [DOI] [PubMed] [Google Scholar]
- 90.Giovannangeli C, Helene C. Progress in developments of triplex-based strategies. Antisense Nucleic Acid Drug Dev. 1997;7:413–421. doi: 10.1089/oli.1.1997.7.413. [DOI] [PubMed] [Google Scholar]
- 91.Maher LJ., III Prospects for the therapeutic use of antigene oligonucleotides. Cancer Invest. 1996;14:66–82. doi: 10.3109/07357909609018437. [DOI] [PubMed] [Google Scholar]
- 92.Praseuth D, Guieysse AL, Helene C. Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta. 1999;1489:181–206. doi: 10.1016/s0167-4781(99)00149-9. [DOI] [PubMed] [Google Scholar]
- 93.Vasquez KM, Wilson JH. Triplex-directed modification of genes and gene activity. Trends Biochem. Sci. 1998;23:4–9. doi: 10.1016/s0968-0004(97)01158-4. [DOI] [PubMed] [Google Scholar]
- 94.Hewett PW, Daft EL, Laughton CA, Ahmad S, Ahmed A, Murray JC. Selective inhibition of the human tie-1 promoter with triplex-forming oligonucleotides targeted to Ets binding sites. Mol. Med. 2006;12:8–16. doi: 10.2119/2005-00046.Hewett. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Svinarchuk F, Nagibneva I, Cherny D, Ait-Si-Ali S, Pritchard LL, Robin P, Malvy C, Harel-Bellan A, Chern D. Recruitment of transcription factors to the target site by triplex-forming oligonucleotides. Nucleic Acids Res. 1997;25:3459–3464. doi: 10.1093/nar/25.17.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karympalis V, Kalopita K, Zarros A, Carageorgiou H. Regulation of gene expression via triple helical formations. Biochemistry. 2004;69:855–860. doi: 10.1023/b:biry.0000040216.18119.d5. [DOI] [PubMed] [Google Scholar]
- 97.Giovannangeli C, Helene C. Triplex-forming molecules for modulation of DNA information processing. Curr. Opin. Mol. Ther. 2000;2:288–296. [PubMed] [Google Scholar]
- 98.Ebbinghaus SW, Fortinberry H, Gamper H.B., Jr Inhibition of transcription elongation in the HER-2/neu coding sequence by triplex-directed covalent modification of the template strand. Biochemistry. 1999;38:619–628. doi: 10.1021/bi980981g. [DOI] [PubMed] [Google Scholar]
- 99.Giovannangeli C, Perrouault L, Escude C, Gryaznov S, Helene C. Efficient inhibition of transcription elongation in vitro by oligonucleotide phosphoramidates targeted to proviral HIV DNA. J. Mol. Biol. 1996;261:386–398. doi: 10.1006/jmbi.1996.0471. [DOI] [PubMed] [Google Scholar]
- 100.Arimondo PB, Thomas CJ, Oussedik K, Baldeyrou B, Mahieu C, Halby L, Guianvarc'h D, Lansiaux A, Hecht SM, Bailly C, et al. Exploring the cellular activity of camptothecin-triple-helix-forming oligonucleotide conjugates. Mol. Cell. Biol. 2006;26:324–333. doi: 10.1128/MCB.26.1.324-333.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bailey C, Weeks DL. Understanding oligonucleotide-mediated inhibition of gene expression in Xenopus laevis oocytes. Nucleic Acids Res. 2000;28:1154–1161. doi: 10.1093/nar/28.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Faria M, Wood CD, Perrouault L, Nelson JS, Winter A, White MR, Helene C, Giovannangeli C. Targeted inhibition of transcription elongation in cells mediated by triplex-forming oligonucleotides. Proc. Natl Acad. Sci. USA. 2000;97:3862–3867. doi: 10.1073/pnas.97.8.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hacia JG, Dervan PB, Wold BJ. Inhibition of klenow fragment DNA polymerase on double-helical templates by oligonucleotide-directed triple-helix formation. Biochemistry. 1994;33:6192–6200. doi: 10.1021/bi00186a019. [DOI] [PubMed] [Google Scholar]
- 104.Napoli S, Negri U, Arcamone F, Capobianco ML, Carbone GM, Catapano CV. Growth inhibition and apoptosis induced by daunomycin-conjugated triplex-forming oligonucleotides targeting the c-myc gene in prostate cancer cells. Nucleic Acids Res. 2006;34:734–744. doi: 10.1093/nar/gkj473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carbone GM, McGuffie EM, Collier A, Catapano CV. Selective inhibition of transcription of the Ets2 gene in prostate cancer cells by a triplex-forming oligonucleotide. Nucleic Acids Res. 2003;31:833–843. doi: 10.1093/nar/gkg198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shahid KA, Majumdar A, Alam R, Liu ST, Kuan JY, Sui X, Cuenoud B, Glazer PM, Miller PS, Seidman MM. Targeted cross-linking of the human beta-globin gene in living cells mediated by a triple helix forming oligonucleotide. Biochemistry. 2006;45:1970–1978. doi: 10.1021/bi0520986. [DOI] [PubMed] [Google Scholar]
- 107.Rapozzi V, Cogoi S, Spessotto P, Risso A, Bonora GM, Quadrifoglio F, Xodo LE. Antigene effect in K562 cells of a PEG-conjugated triplex-forming oligonucleotide targeted to the bcr/abl oncogene. Biochemistry. 2002;41:502–510. doi: 10.1021/bi011314h. [DOI] [PubMed] [Google Scholar]
- 108.Aggarwal BB, Schwarz L, Hogan ME, Rando RF. Triple helix-forming oligodeoxyribonucleotides targeted to the human tumor necrosis factor (TNF) gene inhibit TNF production and block the TNF-dependent growth of human glioblastoma tumor cells. Cancer Res. 1996;56:5156–5164. [PubMed] [Google Scholar]
- 109.Marchand P, Resch K, Radeke HH. Selective inhibition of monocyte chemoattractant protein-1 gene expression in human embryonal kidney cells by specific triple helix-forming oligonucleotides. J. Immunol. 2000;164:2070–2076. doi: 10.4049/jimmunol.164.4.2070. [DOI] [PubMed] [Google Scholar]
- 110.Kochetkova M, Iversen PO, Lopez AF, Shannon MF. Deoxyribonucleic acid triplex formation inhibits granulocyte macrophage colony-stimulating factor gene expression and suppresses growth in juvenile myelomonocytic leukemic cells. J. Clin. Invest. 1997;99:3000–3008. doi: 10.1172/JCI119495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Besch R, Giovannangeli C, Kammerbauer C, Degitz K. Specific inhibition of ICAM-1 expression mediated by gene targeting with Triplex-forming oligonucleotides. J. Biol. Chem. 2002;277:32473–32479. doi: 10.1074/jbc.M203311200. [DOI] [PubMed] [Google Scholar]
- 112.Song J, Intody Z, Li M, Wilson JH. Activation of gene expression by triplex-directed psoralen crosslinks. Gene. 2004;324:183–190. doi: 10.1016/j.gene.2003.09.037. [DOI] [PubMed] [Google Scholar]
- 113.Xu XS, Glazer PM, Wang G. Activation of human gamma-globin gene expression via triplex-forming oligonucleotide (TFO)-directed mutations in the gamma-globin gene 5′ flanking region. Gene. 2000;242:219–228. doi: 10.1016/s0378-1119(99)00522-3. [DOI] [PubMed] [Google Scholar]
- 114.Ghosh MK, Katyal A, Chandra R, Brahmachari V. Targeted activation of transcription in vivo through hairpin-triplex forming oligonucleotide in Saccharomyces cerevisiae. Mol. Cell. Biochem. 2005;278:147–155. doi: 10.1007/s11010-005-7283-7. [DOI] [PubMed] [Google Scholar]
- 115.Kuznetsova S, Ait-Si-Ali S, Nagibneva I, Troalen F, Le Villain JP, Harel-Bellan A, Svinarchuk F. Gene activation by triplex-forming oligonucleotide coupled to the activating domain of protein VP16. Nucleic Acids Res. 1999;27:3995–4000. doi: 10.1093/nar/27.20.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pesce CD, Bolacchi F, Bongiovanni B, Cisotta F, Capozzi M, Diviacco S, Quadrifoglio F, Mango R, Novelli G, Mossa G, et al. Anti-gene peptide nucleic acid targeted to proviral HIV-1 DNA inhibits in vitro HIV-1 replication. Antiviral Res. 2005;66:13–22. doi: 10.1016/j.antiviral.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 117.Rocher C, Dalibart R, Letellier T, Precigoux G, Lestienne P. Initiation of DNA replication by DNA polymerases from primers forming a triple helix. Nucleic Acids Res. 2001;29:3320–3326. doi: 10.1093/nar/29.16.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Guieysse AL, Praseuth D, Francois JC, Helene C. Inhibition of replication initiation by triple helix-forming oligonucleotides. Biochem. Biophys. Res. Commun. 1995;217:186–194. doi: 10.1006/bbrc.1995.2762. [DOI] [PubMed] [Google Scholar]
- 119.Diviacco S, Rapozzi V, Xodo L, Helene C, Quadrifoglio F, Giovannangeli C. Site-directed inhibition of DNA replication by triple helix formation. FASEB J. 2001;15:2660–2668. doi: 10.1096/fj.01-0440com. [DOI] [PubMed] [Google Scholar]
- 120.Nagatsugi F, Sasaki S, Miller PS, Seidman MM. Site-specific mutagenesis by triple helix-forming oligonucleotides containing a reactive nucleoside analog. Nucleic Acids Res. 2003;31:e31. doi: 10.1093/nar/gng031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Birg F, Praseuth D, Zerial A, Thuong NT, Asseline U, Le Doan T, Helene C. Inhibition of simian virus 40 DNA replication in CV-1 cells by an oligodeoxynucleotide covalently linked to an intercalating agent. Nucleic Acids Res. 1990;18:2901–2908. doi: 10.1093/nar/18.10.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ziemba AJ, Reed MW, Raney KD, Byrd AB, Ebbinghaus SW. A bis-alkylating triplex forming oligonucleotide inhibits intracellular reporter gene expression and prevents triplex unwinding due to helicase activity. Biochemistry. 2003;42:5013–5024. doi: 10.1021/bi0273112. [DOI] [PubMed] [Google Scholar]
- 123.Guieysse AL, Praseuth D, Helene C. Identification of a triplex DNA-binding protein from human cells. J. Mol. Biol. 1997;267:289–298. doi: 10.1006/jmbi.1997.0884. [DOI] [PubMed] [Google Scholar]
- 124.Jimenez-Garcia E, Vaquero A, Espinas ML, Soliva R, Orozco M, Bernues J, Azorin F. The GAGA factor of Drosophila binds triple-stranded DNA. J. Biol. Chem. 1998;273:24640–24648. doi: 10.1074/jbc.273.38.24640. [DOI] [PubMed] [Google Scholar]
- 125.Musso M, Nelson LD, Van Dyke MW. Characterization of purine-motif triplex DNA-binding proteins in HeLa extracts. Biochemistry. 1998;37:3086–3095. doi: 10.1021/bi9717486. [DOI] [PubMed] [Google Scholar]
- 126.Nelson LD, Musso M, Van Dyke MW. The yeast STM1 gene encodes a purine motif triple helical DNA-binding protein. J. Biol. Chem. 2000;275:5573–5581. doi: 10.1074/jbc.275.8.5573. [DOI] [PubMed] [Google Scholar]
- 127.Guillonneau F, Guieysse AL, Le Caer JP, Rossier J, Praseuth D. Selection and identification of proteins bound to DNA triple-helical structures by combination of 2D-electrophoresis and MALDI-TOF mass spectrometry. Nucleic Acids Res. 2001;29:2427–2436. doi: 10.1093/nar/29.11.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Strobel SA, Doucette-Stamm LA, Riba L, Housman DE, Dervan PB. Site-specific cleavage of human chromosome 4 mediated by triple-helix formation. Science. 1991;254:1639–1642. doi: 10.1126/science.1836279. [DOI] [PubMed] [Google Scholar]
- 129.Takasugi M, Guendouz A, Chassignol M, Decout JL, Lhomme J, Thuong NT, Helene C. Sequence-specific photo-induced cross-linking of the two strands of double-helical DNA by a psoralen covalently linked to a triple helix-forming oligonucleotide. Proc. Natl Acad. Sci. USA. 1991;88:5602–5606. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vasquez KM, Wensel TG, Hogan ME, Wilson JH. High-efficiency triple-helix-mediated photo-cross-linking at a targeted site within a selectable mammalian gene. Biochemistry. 1996;35:10712–10719. doi: 10.1021/bi960881f. [DOI] [PubMed] [Google Scholar]
- 131.Barre FX, Ait-Si-Ali S, Giovannangeli C, Luis R, Robin P, Pritchard LL, Helene C, Harel-Bellan A. Unambiguous demonstration of triple-helix-directed gene modification. Proc. Natl Acad. Sci. USA. 2000;97:3084–3088. doi: 10.1073/pnas.050368997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Christensen LA, Conti CJ, Fischer SM, Vasquez KM. Mutation frequencies in murine keratinocytes as a function of carcinogenic status. Mol. Carcinog. 2004;40:122–133. doi: 10.1002/mc.20026. [DOI] [PubMed] [Google Scholar]
- 133.Majumdar A, Khorlin A, Dyatkina N, Lin FL, Powell J, Liu J, Fei Z, Khripine Y, Watanabe KA, George J, et al. Targeted gene knockout mediated by triple helix forming oligonucleotides. Nat. Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 134.Datta HJ, Chan PP, Vasquez KM, Gupta RC, Glazer PM. Triplex-induced recombination in human cell-free extracts. Dependence on XPA and HsRad51. J. Biol. Chem. 2001;276:18018–18023. doi: 10.1074/jbc.M011646200. [DOI] [PubMed] [Google Scholar]
- 135.Faruqi AF, Datta HJ, Carroll D, Seidman MM, Glazer PM. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol. Cell. Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kalish JM, Glazer PM. Targeted genome modification via triple helix formation. Ann. N Y Acad. Sci. 2005;1058:151–161. doi: 10.1196/annals.1359.023. [DOI] [PubMed] [Google Scholar]
- 138.Kuan JY, Glazer PM. Targeted gene modification using triplex-forming oligonucleotides. Methods Mol. Biol. 2004;262:173–194. doi: 10.1385/1-59259-761-0:173. [DOI] [PubMed] [Google Scholar]
- 139.Wang G, Levy DD, Seidman MM, Glazer PM. Targeted mutagenesis in mammalian cells mediated by intracellular triple helix formation. Mol. Cell. Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Perrouault L, Asseline U, Rivalle C, Thuong NT, Bisagni E, Giovannangeli C, Le Doan T, Helene C. Sequence-specific artificial photo-induced endonucleases based on triple helix-forming oligonucleotides. Nature. 1990;344:358–360. doi: 10.1038/344358a0. [DOI] [PubMed] [Google Scholar]
- 141.Strobel SA, Dervan PB. Site-specific cleavage of a yeast chromosome by oligonucleotide-directed triple-helix formation. Science. 1990;249:73–75. doi: 10.1126/science.2195655. [DOI] [PubMed] [Google Scholar]
- 142.Francois JC, Saison-Behmoaras T, Barbier C, Chassignol M, Thuong NT, Helene C. Sequence-specific recognition and cleavage of duplex DNA via triple-helix formation by oligonucleotides covalently linked to a phenanthroline-copper chelate. Proc. Natl Acad. Sci. USA. 1989;86:9702–9706. doi: 10.1073/pnas.86.24.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Francois JC, Saison-Behmoaras T, Chassignol M, Thuong NT, Helene C. Sequence-targeted cleavage of single- and double-stranded DNA by oligothymidylates covalently linked to 1,10-phenanthroline. J. Biol. Chem. 1989;264:5891–5898. [PubMed] [Google Scholar]
- 144.Bigey P, Pratviel G, Meunier B. Cleavage of double-stranded DNA by ‘metalloporphyrin-linker-oligonucleotide’ molecules: influence of the linker. Nucleic Acids Res. 1995;23:3894–3900. doi: 10.1093/nar/23.19.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Landgraf R, Chen CH, Sigman DS. Oligonucleotide-directed nucleic acid scission by micrococcal nuclease. Biochemistry. 1994;33:10607–10615. doi: 10.1021/bi00201a006. [DOI] [PubMed] [Google Scholar]
- 146.Pei D, Corey DR, Schultz PG. Site-specific cleavage of duplex DNA by a semisynthetic nuclease via triple-helix formation. Proc. Natl Acad. Sci. USA. 1990;87:9858–9862. doi: 10.1073/pnas.87.24.9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hearst JE. Photochemistry of the psoralens. Chem. Res. Toxicol. 1989;2:69–75. doi: 10.1021/tx00008a001. [DOI] [PubMed] [Google Scholar]
- 148.Kean JM, Miller PS. Detection of psoralen cross-link sites in DNA modified by psoralen-conjugated oligodeoxyribonucleoside methylphosphonates. Bioconjug. Chem. 1993;4:184–187. doi: 10.1021/bc00020a012. [DOI] [PubMed] [Google Scholar]
- 149.Ziemba A, Derosier LC, Methvin R, Song CY, Clary E, Kahn W, Milesi D, Gorn V, Reed M, Ebbinghaus S. Repair of triplex-directed DNA alkylation by nucleotide excision repair. Nucleic Acids Res. 2001;29:4257–4263. doi: 10.1093/nar/29.21.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Faruqi AF, Egholm M, Glazer PM. Peptide nucleic acid-targeted mutagenesis of a chromosomal gene in mouse cells. Proc. Natl Acad. Sci. USA. 1998;95:1398–1403. doi: 10.1073/pnas.95.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rogers FA, Manoharan M, Rabinovitch P, Ward DC, Glazer PM. Peptide conjugates for chromosomal gene targeting by triplex-forming oligonucleotides. Nucleic Acids Res. 2004;32:6595–6604. doi: 10.1093/nar/gkh998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rogers FA, Vasquez KM, Egholm M, Glazer PM. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc. Natl Acad. Sci. USA. 2002;99:16695–16700. doi: 10.1073/pnas.262556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Armitage B, Koch T, Frydenlund H, Orum H, Batz HG, Schuster GB. Peptide nucleic acid-anthraquinone conjugates: strand invasion and photoinduced cleavage of duplex DNA. Nucleic Acids Res. 1997;25:4674–4678. doi: 10.1093/nar/25.22.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Okamoto A, Tanabe K, Saito I. Synthesis and properties of peptide nucleic acids containing a psoralen unit. Org. Lett. 2001;3:925–927. doi: 10.1021/ol015549x. [DOI] [PubMed] [Google Scholar]
- 155.Ross GF, Smith PM, McGregor A, Turnbull DM, Lightowlers RN. Synthesis of trifunctional PNA-benzophenone derivatives for mitochondrial targeting, selective DNA binding, and photo-cross-linking. Bioconjug. Chem. 2003;14:962–966. doi: 10.1021/bc034050f. [DOI] [PubMed] [Google Scholar]
- 156.Carpenter M, Divvela P, Pingoud V, Bujnicki J, Bhagwat AS. Sequence-dependent enhancement of hydrolytic deamination of cytosines in DNA by the restriction enzyme PspGI. Nucleic Acids Res. 2006;34:3762–3770. doi: 10.1093/nar/gkl545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Strobel SA, Dervan PB. Single-site enzymatic cleavage of yeast genomic DNA mediated by triple helix formation. Nature. 1991;350:172–174. doi: 10.1038/350172a0. [DOI] [PubMed] [Google Scholar]
- 158.Kalish JM, Seidman MM, Weeks DL, Glazer PM. Triplex-induced recombination and repair in the pyrimidine motif. Nucleic Acids Res. 2005;33:3492–3502. doi: 10.1093/nar/gki659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dobbelstein M. Viruses in therapy- -royal road or dead end? Virus Res. 2003;92:219–221. doi: 10.1016/s0168-1702(02)00355-6. [DOI] [PubMed] [Google Scholar]
- 160.Hermiston TW, Kuhn I. Armed therapeutic viruses: strategies and challenges to arming oncolytic viruses with therapeutic genes. Cancer Gene Ther. 2002;9:1022–1035. doi: 10.1038/sj.cgt.7700542. [DOI] [PubMed] [Google Scholar]
- 161.Boletta A, Benigni A, Lutz J, Remuzzi G, Soria MR, Monaco L. Nonviral gene delivery to the rat kidney with polyethylenimine. Hum. Gene. Ther. 1997;8:1243–1251. doi: 10.1089/hum.1997.8.10-1243. [DOI] [PubMed] [Google Scholar]
- 162.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kircheis R, Wightman L, Wagner E. Design and gene delivery activity of modified polyethylenimines. Adv. Drug Deliv. Rev. 2001;53:341–358. doi: 10.1016/s0169-409x(01)00202-2. [DOI] [PubMed] [Google Scholar]
- 164.Haensler J, Szoka F.C., Jr Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjug. Chem. 1993;4:372–379. doi: 10.1021/bc00023a012. [DOI] [PubMed] [Google Scholar]
- 165.Zinselmeyer BH, Mackay SP, Schatzlein AG, Uchegbu IF. The lower-generation polypropylenimine dendrimers are effective gene-transfer agents. Pharm. Res. 2002;19:960–967. doi: 10.1023/a:1016458104359. [DOI] [PubMed] [Google Scholar]
- 166.Santhakumaran LM, Thomas T, Thomas TJ. Enhanced cellular uptake of a triplex-forming oligonucleotide by nanoparticle formation in the presence of polypropylenimine dendrimers. Nucleic Acids Res. 2004;32:2102–2112. doi: 10.1093/nar/gkh526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vijayanathan V, Thomas T, Antony T, Shirahata A, Thomas TJ. Formation of DNA nanoparticles in the presence of novel polyamine analogues: a laser light scattering and atomic force microscopic study. Nucleic Acids Res. 2004;32:127–134. doi: 10.1093/nar/gkg936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cheng K, Ye Z, Guntaka RV, Mahato RI. Enhanced hepatic uptake and bioactivity of type alpha1(I) collagen gene promoter-specific triplex-forming oligonucleotides after conjugation with cholesterol. J. Pharmacol. Exp. Ther. 2006;317:797–805. doi: 10.1124/jpet.105.100347. [DOI] [PubMed] [Google Scholar]
- 169.Besch R, Giovannangeli C, Schuh T, Kammerbauer C, Degitz K. Characterization and quantification of triple helix formation in chromosomal DNA. J. Mol. Biol. 2004;341:979–989. doi: 10.1016/j.jmb.2004.05.079. [DOI] [PubMed] [Google Scholar]
- 170.Brunet E, Corgnali M, Cannata F, Perrouault L, Giovannangeli C. Targeting chromosomal sites with locked nucleic acid-modified triplex-forming oligonucleotides: study of efficiency dependence on DNA nuclear environment. Nucleic Acids Res. 2006;34:4546–4553. doi: 10.1093/nar/gkl630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Giovannangeli C, Diviacco S, Labrousse V, Gryaznov S, Charneau P, Helene C. Accessibility of nuclear DNA to triplex-forming oligonucleotides: the integrated HIV-1 provirus as a target. Proc. Natl Acad. Sci. USA. 1997;94:79–84. doi: 10.1073/pnas.94.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Macris MA, Glazer PM. Transcription dependence of chromosomal gene targeting by triplex-forming oligonucleotides. J. Biol. Chem. 2003;278:3357–3362. doi: 10.1074/jbc.M206542200. [DOI] [PubMed] [Google Scholar]
- 173.Majumdar A, Puri N, McCollum N, Richards S, Cuenoud B, Miller P, Seidman MM. Gene targeting by triple helix-forming oligonucleotides. Ann. N Y Acad. Sci. 2003;1002:141–153. doi: 10.1196/annals.1281.013. [DOI] [PubMed] [Google Scholar]
- 174.Oh DH, Hanawalt PC. Triple helix-forming oligonucleotides target psoralen adducts to specific chromosomal sequences in human cells. Nucleic Acids Res. 1999;27:4734–4742. doi: 10.1093/nar/27.24.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ye Z, Guntaka RV, Mahato RI. Sequence-specific triple helix formation with genomic DNA. Biochemistry. 2007;46:11240–11252. doi: 10.1021/bi700580y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Majumdar A, Puri N, Cuenoud B, Natt F, Martin P, Khorlin A, Dyatkina N, George AJ, Miller PS, Seidman MM. Cell cycle modulation of gene targeting by a triple helix-forming oligonucleotide. J. Biol. Chem. 2003;278:11072–11077. doi: 10.1074/jbc.M211837200. [DOI] [PubMed] [Google Scholar]
- 177.Carbone GM, McGuffie E, Napoli S, Flanagan CE, Dembech C, Negri U, Arcamone F, Capobianco ML, Catapano CV. DNA binding and antigene activity of a daunomycin-conjugated triplex-forming oligonucleotide targeting the P2 promoter of the human c-myc gene. Nucleic Acids Res. 2004;32:2396–2410. doi: 10.1093/nar/gkh527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Stierle V, Duca M, Halby L, Senamaud-Beaufort C, Capobianco ML, Laigle A, Jolles B, Arimondo PB. Targeting MDR1 gene: synthesis and cellular study of modified daunomycin-TFO conjugates able to inhibit gene expression in resistant cell lines. Mol. Pharmacol. 2008;73:1568–1577. doi: 10.1124/mol.107.042010. [DOI] [PubMed] [Google Scholar]
- 179.Christensen LA, Finch RA, Booker AJ, Vasquez KM. Targeting oncogenes to improve breast cancer chemotherapy. Cancer Res. 2006;66:4089–4094. doi: 10.1158/0008-5472.CAN-05-4288. [DOI] [PubMed] [Google Scholar]
- 180.Wu Q, Gaddis SS, MacLeod MC, Walborg EF, Thames HD, DiGiovanni J, Vasquez KM. High-affinity triplex-forming oligonucleotide target sequences in mammalian genomes. Mol. Carcinog. 2007;46:15–23. doi: 10.1002/mc.20261. [DOI] [PubMed] [Google Scholar]
- 181.Matteucci M, Lin KY, Huang T, Wagner R, Sternbach DD, Mehrotra M, Besterman JM. Sequence-specific targeting of duplex DNA using a camptothecin-triple helix forming oligonucleotide conjugate and topoisomerase I. J. Am. Chem. Soc. 1997;119:6939. [Google Scholar]
- 182.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 183.Arimondo PB, Boutorine A, Baldeyrou B, Bailly C, Kuwahara M, Hecht SM, Sun JS, Garestier T, Helene C. Design and optimization of camptothecin conjugates of triple helix-forming oligonucleotides for sequence-specific DNA cleavage by topoisomerase I. J. Biol. Chem. 2002;277:3132–3140. doi: 10.1074/jbc.M110181200. [DOI] [PubMed] [Google Scholar]
- 184.Arimondo PB, Helene C. Design of new anti-cancer agents based on topoisomerase poisons targeted to specific DNA sequences. Curr. Med. Chem. Anticancer Agents. 2001;1:219–235. doi: 10.2174/1568011013354642. [DOI] [PubMed] [Google Scholar]
- 185.Arimondo PB, Laco GS, Thomas CJ, Halby L, Pez D, Schmitt P, Boutorine A, Garestier T, Pommier Y, Hecht SM, et al. Activation of camptothecin derivatives by conjugation to triple helix-forming oligonucleotides. Biochemistry. 2005;44:4171–4180. doi: 10.1021/bi048031k. [DOI] [PubMed] [Google Scholar]
- 186.Arimondo PB, Moreau P, Boutorine A, Bailly C, Prudhomme M, Sun JS, Garestier T, Helene C. Recognition and cleavage of DNA by rebeccamycin- or benzopyridoquinoxaline conjugated of triple helix-forming oligonucleotides. Bioorg. Med. Chem. 2000;8:777–784. doi: 10.1016/s0968-0896(00)00012-2. [DOI] [PubMed] [Google Scholar]
- 187.Arimondo PB, Bailly C, Boutorine AS, Moreau P, Prudhomme M, Sun JS, Garestier T, Helene C. Triple helix-forming oligonucleotides conjugated to indolocarbazole poisons direct topoisomerase I-mediated DNA cleavage to a specific site. Bioconjug. Chem. 2001;12:501–509. doi: 10.1021/bc000162k. [DOI] [PubMed] [Google Scholar]
- 188.Koster DA, Czerwinski F, Halby L, Crut A, Vekhoff P, Palle K, Arimondo PB, Dekker NH. Single-molecule observations of topotecan-mediated TopIB activity at a unique DNA sequence. Nucleic Acids Res. 2008;36:2301–2310. doi: 10.1093/nar/gkn035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Capobianco ML, De Champdore M, Arcamone F, Garbesi A, Guianvarc’h D, Arimondo PB. Improved synthesis of daunomycin conjugates with triplex-forming oligonucleotides. The polypurine tract of HIV-1 as a target. Bioorg. Med. Chem. 2005;13:3209–3218. doi: 10.1016/j.bmc.2005.02.040. [DOI] [PubMed] [Google Scholar]
- 190.Garbesi A, Bonazzi S, Zanella S, Capobianco ML, Giannini G, Arcamone F. Synthesis and binding properties of conjugates between oligodeoxynucleotides and daunorubicin derivatives. Nucleic Acids Res. 1997;25:2121–2128. doi: 10.1093/nar/25.11.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Duca M, Guianvarc'h D, Oussedik K, Halby L, Garbesi A, Dauzonne D, Monneret C, Osheroff N, Giovannangeli C, Arimondo PB. Molecular basis of the targeting of topoisomerase II-mediated DNA cleavage by VP16 derivatives conjugated to triplex-forming oligonucleotides. Nucleic Acids Res. 2006;34:1900–1911. doi: 10.1093/nar/gkl126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Duca M, Oussedik K, Ceccaldi A, Halby L, Guianvarc’h D, Dauzonne D, Monneret C, Sun JS, Arimondo PB. Triple helix-forming oligonucleotides conjugated to new inhibitors of topoisomerase II: synthesis and binding properties. Bioconjug. Chem. 2005;16:873–884. doi: 10.1021/bc050031p. [DOI] [PubMed] [Google Scholar]
- 193.Vasquez KM, Christensen J, Li L, Finch RA, Glazer PM. Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proc. Natl Acad. Sci. USA. 2002;99:5848–5853. doi: 10.1073/pnas.082193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 195.Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry. 2005;44:4188–4195. doi: 10.1021/bi047902n. [DOI] [PubMed] [Google Scholar]
- 196.Wu Q, Christensen LA, Legerski RJ, Vasquez KM. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO Rep. 2005;6:551–557. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ji H, Smith LM. Rapid purification of double-stranded DNA by triple-helix-mediated affinity capture. Anal. Chem. 1993;65:1323–1328. doi: 10.1021/ac00058a005. [DOI] [PubMed] [Google Scholar]
- 198.Ito T, Smith CL, Cantor CR. Sequence-specific DNA purification by triplex affinity capture. Proc. Natl Acad. Sci. USA. 1992;89:495–498. doi: 10.1073/pnas.89.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Noonberg SB, Scott GK, Hunt CA, Benz CC. Detection of triplex-forming RNA oligonucleotides by triplex blotting. Biotechniques. 1994;16:1070–1072. 1074. [PubMed] [Google Scholar]
- 200.Schluep T, Cooney CL. Purification of plasmids by triplex affinity interaction. Nucleic Acids Res. 1998;26:4524–4528. doi: 10.1093/nar/26.19.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Takabatake T, Asada K, Uchimura Y, Ohdate M, Kusukawa N. The use of purine-rich oligonucleotides in triplex-mediated DNA isolation and generation of unidirectional deletions. Nucleic Acids Res. 1992;20:5853–5854. doi: 10.1093/nar/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Maine IP, Kodadek T. Efficient unwinding of triplex DNA by a DNA helicase. Biochem. Biophys. Res. Commun. 1994;204:1119–1124. doi: 10.1006/bbrc.1994.2578. [DOI] [PubMed] [Google Scholar]
- 203.Wang Z, Rana TM. DNA damage-dependent transcriptional arrest and termination of RNA polymerase II elongation complexes in DNA template containing HIV-1 promoter. Proc. Natl Acad. Sci. USA. 1997;94:6688–6693. doi: 10.1073/pnas.94.13.6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Spitzner JR, Chung IK, Muller MT. Determination of 5′ and 3′ DNA triplex interference boundaries reveals the core DNA binding sequence for topoisomerase II. J. Biol. Chem. 1995;270:5932–5943. doi: 10.1074/jbc.270.11.5932. [DOI] [PubMed] [Google Scholar]
- 205.Firman K, Szczelkun MD. Measuring motion on DNA by the type I restriction endonuclease EcoR124I using triplex displacement. EMBO J. 2000;19:2094–2102. doi: 10.1093/emboj/19.9.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.McClelland SE, Dryden DT, Szczelkun MD. Continuous assays for DNA translocation using fluorescent triplex dissociation: application to type I restriction endonucleases. J. Mol. Biol. 2005;348:895–915. doi: 10.1016/j.jmb.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 207.Cherny DI, Fourcade A, Svinarchuk F, Nielsen PE, Malvy C, Delain E. Analysis of various sequence-specific triplexes by electron and atomic force microscopies. Biophys. J. 1998;74:1015–1023. doi: 10.1016/S0006-3495(98)74026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cherny DI, Malkov VA, Volodin AA, Frank-Kamenetskii MD. Electron microscopy visualization of oligonucleotide binding to duplex DNA via triplex formation. J. Mol. Biol. 1993;230:379–383. doi: 10.1006/jmbi.1993.1154. [DOI] [PubMed] [Google Scholar]
- 209.Pfannschmidt C, Schaper A, Heim G, Jovin TM, Langowski J. Sequence-specific labeling of superhelical DNA by triple helix formation and psoralen crosslinking. Nucleic Acids Res. 1996;24:1702–1709. doi: 10.1093/nar/24.9.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Grimm GN, Boutorine AS, Lincoln P, Norden B, Helene C. Formation of DNA triple helices by an oligonucleotide conjugated to a fluorescent ruthenium complex. Chembiochem. 2002;3:324–331. doi: 10.1002/1439-7633(20020402)3:4<324::AID-CBIC324>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 211.Escude C, Garestier T, Helene C. Padlock oligonucleotides for duplex DNA based on sequence-specific triple helix formation. Proc. Natl Acad. Sci. USA. 1999;96:10603–10607. doi: 10.1073/pnas.96.19.10603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Roulon T, Coulaud D, Delain E, Le Cam E, Helene C, Escude C. Padlock oligonucleotides as a tool for labeling superhelical DNA. Nucleic Acids Res. 2002;30:E12. doi: 10.1093/nar/30.3.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Roulon T, Helene C, Escude C. Coupling of a targeting peptide to plasmid DNA using a new type of padlock oligonucleotide. Bioconjug. Chem. 2002;13:1134–1139. doi: 10.1021/bc025551o. [DOI] [PubMed] [Google Scholar]
- 214.Geron-Landre B, Roulon T, Desbiolles P, Escude C. Sequence-specific fluorescent labeling of double-stranded DNA observed at the single molecule level. Nucleic Acids Res. 2003;31:e125. doi: 10.1093/nar/gng125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Geron-Landre B, Roulon T, Escude C. Stem-loop oligonucleotides as tools for labelling double-stranded DNA. FEBS J. 2005;272:5343–5352. doi: 10.1111/j.1742-4658.2005.04932.x. [DOI] [PubMed] [Google Scholar]
- 216.Hausmann M, Winkler R, Hildenbrand G, Finsterle J, Weisel A, Rapp A, Schmitt E, Janz S, Cremer C. COMBO-FISH: specific labeling of nondenatured chromatin targets by computer-selected DNA oligonucleotide probe combinations. Biotechniques. 2003;35:564–570. doi: 10.2144/03353rr03. 572–567. [DOI] [PubMed] [Google Scholar]
- 217.Van Daele I, Bomholt N, Filichev VV, Van Calenbergh S, Pedersen EB. Triplex formation by pyrene-labelled probes for nucleic acid detection in fluorescence assays. Chembiochem. 2008;9:791–801. doi: 10.1002/cbic.200700533. [DOI] [PubMed] [Google Scholar]
- 218.Biroccio A, Leonetti C, Zupi G. The future of antisense therapy: combination with anticancer treatments. Oncogene. 2003;22:6579–6588. doi: 10.1038/sj.onc.1206812. [DOI] [PubMed] [Google Scholar]
- 219.Li W, Cha L. Predicting siRNA efficiency. Cell. Mol. Life Sci. 2007;64:1785–1792. doi: 10.1007/s00018-007-7057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]