Abstract
Homologous recombination is essential for DNA repair and generation of genetic diversity in all organisms. It occurs through a series of presynaptic steps where the substrate is presented to the recombinase (RecA in bacteria). Then, the recombinase nucleoprotein filament mediates synapsis by first promoting the formation of a D-loop and later of a Holliday junction (HJ) that is subsequently cleaved by the HJ resolvase. The coordination of the synaptic step with the late resolution step is poorly understood. Bacillus subtilis RecU catalyzes resolution of HJs, and biochemical evidence suggests that it might modulate RecA. We report here the isolation and characterization of two mutants of RecU (recU56 and recU71), which promote resolution of HJs, but do not promote RecA modulation. In vitro, the RecU mutant proteins (RecUK56A or RecUR71A) bind and cleave HJs and interact with RuvB. RecU interacts with RecA and inhibits its single-stranded DNA-dependent dATP hydrolysis, but RecUK56A and RecUR71A do not exert a negative effect on the RecA dATPase and fail to interact with it. Both activities are important in vivo since RecU mutants impaired only in RecA interaction are as sensitive to DNA damaging agents as a deletion mutant.
INTRODUCTION
All cells have evolved efficient mechanisms to promote genetic diversity, ensure proper chromosome segregation and restore genome integrity via homologous recombination (HR) (1,2). In bacteria, alternative avenues process the free duplex DNA end(s) at double-strand (ds) breaks and several modulators can recruit RecA onto the 3′ single-stranded (ss) DNA tailed duplex DNA (3,4–7). Then, the RecA nucleoprotein filaments search for homology and mediate invasion onto the homologous dsDNA region to form a three-strand intermediate (D-loop structure). The unidirectional movement of the strand transfer produces a Holliday junction (HJ) that is processed by the branch migration translocases, RuvAB or RecG (4,8–11). Finally, HJs are resolved by the HJ resolvase (Escherichia coli RuvC or Bacillus subtilis RecU) with a bias towards noncrossover products, which in circular genomes result in two monomeric chromosomes (12–14). Mutations in B. subtilis ruvA, ruvB, recU or recG genes lead to an 80- to 150-fold increase in anucleate cells in the absence of any externally added DNA damaging agent (11,14), suggesting that proper resolution of frequently formed HJs is essential for faithful segregation of chromosomes (note that unless stated otherwise, the indicated genes and products are of B. subtilis origin).
In bacteria, it is generally believed that strand exchange and resolution of HJs are two separated events. However, recent studies revealed that the RecU HJ resolvase, which is present in all Firmicutes, might also act early in HR. In vitro, RecU enhanced RecA loading onto ssDNA and stimulated RecA-catalyzed D-loop formation, yet inhibited ssDNA-dependent dATP or rATP hydrolysis and RecA-mediated DNA strand exchange (15). Is the involvement of a HJ resolvase at early stages of HR a unique phenomenon associated with the RecU resolvase? This has not been reported for other bacteria yet, but evidences exist in archaea and mammals. Pyrococcus furiosus RadB, which is a RadA paralog, interacts in vitro with RadA and the Hjc resolvase, and regulates the cleavage of HJs by Hjc (16). A pre- and postsynaptic role for some mammalian Rad51 orthologs has also been described (17–23). As an example, the Rad51C-Xrcc3 (CX3) complex is involved in the early stages of recombination by binding DNA, catalyzing DNA pairing and recruiting Rad51 onto RPA-coated ssDNA (17–20,22,24). On the other hand, the CX3 complex may play an essential role in the resolution of HJs, hence, it can also be associated with the late stages of HR (19,21,23).
Natural transformation, which is a genetically programmed mechanism for horizontal gene transfer in bacteria, provides a means to characterize RecA-dependent and RecA-independent pathways, and is another avenue to unravel the role of RecU. If taken up ssDNA shares homology with recipient dsDNA, it recombines in a RecA-dependent process (e.g. chromosomal transformation) (25,26). However, the role of RecU in transformation does not seem to be the cleavage of HJs, because the entering ssDNA should form a D-loop rather than a HJ intermediate, and RecU is not able to cleave D-loop structures (15). During natural plasmid transformation, if it shares no significant homology with recipient DNA, the internalized plasmid ssDNA is channelled into a RecA-independent yet RecU- and RecO-dependent pathway(s) (27–30). To gain insight into the mechanisms that couple early and late stages of HR in Firmicutes, and to further investigate the role of RecU in early stages of recombination, we searched for recU mutants proficient in chromosomal segregation (HJ resolution with a bias towards noncrossover products), while deficient in plasmid transformation, and studied their action. We report the isolation and characterization of two recU single mutants capable of promoting resolution of HJs and segregating chromosomes properly (late stage), but which have lost the capacity to modulate RecA activities and are impaired in plasmid transformation (early stage). In this study, we provide the first evidence that both RecU activities, which are essential for recombinational repair, can be genetically separated.
MATERIALS AND METHODS
Bacterial strains and plasmids
Escherichia coli strains BL21(DE3)[pLysS] and XL1-Blue were used (31). Bacillus subtilis BG214 and its isogenic derivatives are listed in Supplementary Table S1. pCB568-borne recU, pCB697-borne recU56 and pCB695-borne recU71 gene, under the control of a T7 promoter, were used to over-express RecU, RecUK56A and RecUR71A proteins in BL21(DE3)[pLysS] cells, respectively. pBT61-borne recA gene was used to over-express RecA. A plasmid-borne ruvB gene with six His codons at the C-terminal end was constructed (pCB737). pCB737-borne ruvB-His fully complemented the ΔruvB strain (data not shown), and the purified protein is therefore termed here as RuvB. pGEM-3Zf(+) is the source of circular ssDNA and linear dsDNA (Promega, Madison, WI, USA).
Protein purification
RecA was purified from BG214 cells (15) and RecU, RecUK56A and RecUR71A proteins were purified from BL21(DE3)[pLysS] cells, as described for WT RecU (31,32). RuvB was purified from BG214 cells. Expression of RuvB was obtained by addition of 1 mM IPTG. Cells were lyzed by treatment with lysozyme and sonication, and the supernatant of the lysis, which contained RuvB, was loaded onto an Ni-column. RuvB, which eluted from the Ni-column at 60 mM imidazol, was further purified by Q Sepharose (GE Healthcare, Buckinghamshire, UK) chromatography. All proteins were purified to >98% homogeneity. The molar extinction coefficients for RecA, RecU and RuvB were calculated to be 15 200, 27 850 and 13 400 M−1 cm−1 at 280 nm, as described (15). The protein concentrations were determined using the above molar extinction coefficient, and RecA is expressed as moles of protein monomers, RecU and its variants as dimers and RuvB as hexamers.
Gene and protein manipulations
In vitro site-directed alanine scanning mutagenesis was performed using pCB568 DNA and the QuickChange mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the instructions of the supplier. The presence of the expected mutation and the absence of any other in the promoter region or in the recU gene were confirmed by sequencing. Protein cross-linking was used to study protein–protein interactions, as described (33). A constant amount of protein (2 μM) was mixed in buffer A [50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 50 μg/ml BSA, 1 mM DTT, 5% glycerol] containing 1 mM MgCl2. DSS was added to a final concentration of 50 μM and, after incubation for 10 min at 37°C, the reactions were stopped according to the manufacturer's instructions. Rabbit polyclonal anti-RecU or anti-RecA antibodies were obtained using standard techniques. For western blotting, proteins were separated on 10% SDS–PAGE. Blots were probed with antibodies against RecU or RecA.
Transformation and survival studies
Competent B. subtilis cells (34), were transformed with SB19 prototrophic chromosomal DNA or pUB110 plasmid DNA. The yield of met+ (chromosomal transformation) or kanamycin-resistant (KmR) transformants (plasmid transformation) was corrected for DNA uptake, and the values obtained were normalized relative to that of the rec+ strain, which are taken as 1 (35). Exponentially growing cells were exposed to 10 mM methylmethane sulfonate (MMS) for variable times, and serial dilutions were plated to measure survival rates, as described (35).
DNA manipulations
DNA concentrations are expressed as moles of nucleotides (nt). The binding of the HJ J3 (0.3 μM) was assayed for 15 min at 37°C in buffer A containing 1 mM MgCl2 by using 6% nondenaturing (nd) PAGE and autoradiography (36). Cleavage of HJ J3, labeled at one strand, was assayed for 30 min at 37°C in buffer A containing 10 mM MgCl2 and cleavage products were analyzed by 15% denaturing (d) PAGE and autoradiography (31). The strand exchange reactions were essentially performed as described (15). Linear duplex with 3′-ssDNA termini of >150 nt (3′-tailed dsDNA) was prepared as described (37). For DNA annealing, a 420-bp DraIII-EcoRI segment derived from pGEM-3Zf(+) was gel purified and labeled at the 3′-end with [α-32P]-dATP and Klenow enzyme. Labeled DNA (1 μM) was heat denatured by incubation at 95°C for 10 min and immediately placed on water ice. RecU variants (200 nM) were incubated with 0.3 μM denatured DNA (see above) in buffer A containing 1 mM MgCl2 for 15 min at 37°C, the reaction was stopped and the deproteinized products were analyzed by 6% ndPAGE and autoradiography (31).
dATPase activity
Circular ssDNA (10 μM) was preincubated with RecA (1.3 μM) or RecU variants (200 nM) and 2 mM dATP ([α-32P]-dATP, ratio 1 : 100 000) for 5 min at 37°C in buffer A containing 10 mM MgOAc2. Then the second protein was added, and incubation continued for 25 min. Aliquots were taken at time intervals and dATPase activity was determined, as described (38).
RESULTS
Isolation of recU mutants impaired in early stages of recombination
To test whether the putative protein–protein interaction between RecA (pI 4.9) and RecU (pI 9.5) is due to charged residues site-directed mutagenesis was used to generate recU mutants on highly conserved positively charged residues among Firmicutes RecU. The proficiency in chromosomal segregation and the impairment in plasmid transformation (see Introduction section) were used to search for mutants with a putative separation-of-function phenotype. From ∼25 genuine recU mutants, two [recU56 (lysine at position K56 replaced by alanine, RecUK56A) and recU71 (R71 by alanine, RecUR71A)] were selected for further analysis.
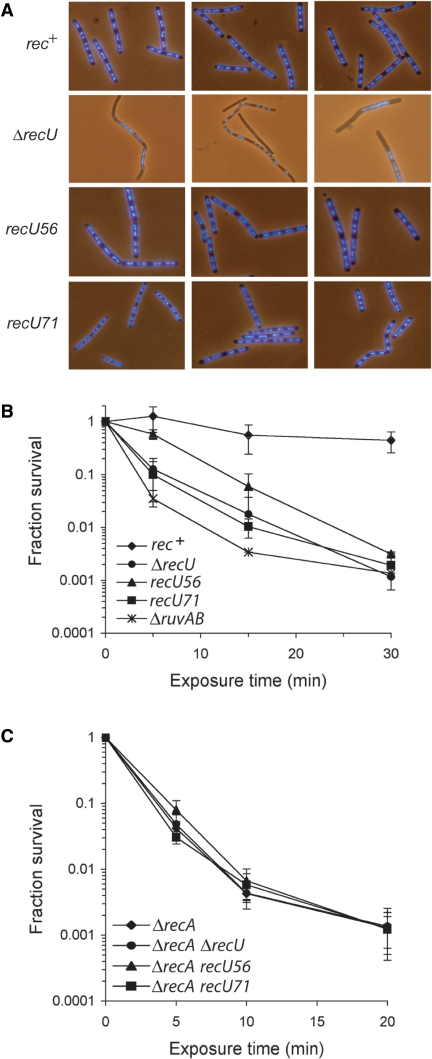
Anucleate cells (measured as absence of DAPI-stained material) were rare in exponentially growing wild-type (WT) cells (0.05% of total cells). The frequency of anucleates (or accumulation of nonsegregated chromosomes) increased 80- and 100-fold in ΔruvAB (9) and in ΔrecU cells, respectively [(14,39), Figure 1A]. In exponentially growing ΔrecU cells, 4.1% of total cells (719 cells analyzed) were anucleates, whereas in recU56 or recU71 cells the frequency of anucleates was significantly reduced (Figure 1A). Four anucleates were observed in 846 recU56 cells analyzed and two anucleates in 738 recU71 cells. These results suggest that unlike in ΔruvAB or ΔrecU cells, in recU56 or recU71 cells HJs were resolved efficiently with bias towards noncrossover products. We next measured the frequency of genetic exchange. The frequency of chromosomal transformation in ΔrecU, recU56 or recU71 cells only decreased <3-fold, whereas plasmid establishment was reduced ∼30-, 6- or 50-fold, respectively, relative to the rec+ values (Table 1). If during plasmid transformation RecU is only required at a late stage of HR, ΔrecU cells should have a similar defect as ΔruvAB cells. However, the frequency of plasmid transformants only decreased <3-fold in ΔruvAB cells relative to rec+ values (Table 1). One hypothesis is that the presence of RecA, which is not required for plasmid transformation, might lead to the accumulation of ‘toxic intermediates’, and RecU might modulate such an accumulation during plasmid transformation (15). If this hypothesis is correct, the absence of RecA should overcome the requirement of RecU for plasmid transformation. Indeed, the absence of RecA overcame the RecU requirement during plasmid transformation (Table 1), and as expected still blocked chromosomal transformation in the recU context [(25,40), Table 1]. It is likely that in the absence of RecU the RecA nucleoprotein filament opens an unproductive avenue that is deleterious to plasmid transformation. Alternatively, in the absence of both proteins an uncharacterized recombination pathway becomes operative (see below).
Figure 1.
Segregation and DNA repair defect of ΔrecU, recU56 or recU71 cells. (A) Exponentially growing WT, ΔrecU, recU56 or recU71 cells in LB were fixed, stained with DAPI, and analyzed by fluorescence microscopy to visualize the nucleoid. (B and C) Survival of strains after exposure to MMS. Exponentially growing cells in LB were exposed to 10 mM MMS for a variable time, and serial dilutions were plated to measure survival rates. (B) rec+, ΔrecU, recU56, recU71 and ΔruvAB cells; (C) ΔrecA, ΔrecA ΔrecU, ΔrecA recU56 and ΔrecA recU71 cells.
Table 1.
The absence of RecA suppresses the RecU defect in plasmid transformation
| Relevant genotype | Normalized plasmid transformation | Normalized chromosomal transformation |
|---|---|---|
| rec+ | 1 | 1 |
| ΔrecUa | 0.03 | 0.40 |
| recU56 | 0.15 | 0.43 |
| recU71 | 0.02 | 0.36 |
| ΔruvAB | 0.30 | 0.81 |
| ΔrecAa | 0.96 | <0.0001 |
| ΔrecA ΔrecU | 0.42 | <0.0001 |
| ΔrecA recU56 | 0.59 | <0.0001 |
| ΔrecA recU71 | 0.40 | <0.0001 |
The yield of met+ transformants (chromosomal transformation) and KmR transformants (plasmid pUB110 transformation) was corrected for DNA uptake and cell viability and the values obtained normalized relative to that of the rec+ strain, taken as 1. The results are the average of at least five independent experiments and are within a 10% standard error. aThe transformation frequencies of ΔrecA and ΔrecU cells have been reported elsewhere (27,30,33) and brought here for direct comparison.
The capacity of recU56 or recU71 cells to catalyze recombinational repair was also analyzed. MMS is a DNA alkylating agent that causes various DNA lesions, that if unrepaired lead to a severe survival phenotype in ΔrecU cells (29). Following acute exposure to 10 mM MMS, the survival of rec+ cells was slightly affected [lethal dose to allow 10% survival (LD10) of >70 min]. However, the recU56 cells were sensitive (LD10 was ∼14 min) and ΔruvAB, ΔrecU or recU71 cells were very sensitive (LD10 = 4, 6 and 5 min, respectively) to 10 mM MMS (Figure 1B). To address whether DNA repair was RecA-dependent in the recU56 or recU71 context the double mutant strains were examined. Inactivation of ΔrecA in the recU context rendered cells extremely sensitive (LD10 ∼ 3 min) to 10 mM MMS (Figure 1C). These results indicate that RecU action is RecA-dependent, because the double mutants were as sensitive as the ΔrecA strain, and discard the hypothesis of opening a new recombination avenue.
RecUK56A or RecUR71A binds and cleaves HJs
RecU and variants RecUK56A and RecUR71A were over-expressed and purified towards homogeneity using a similar purification protocol. Far-ultraviolet circular dichroism spectra revealed that the proportion of α-helices and β-sheets of RecU was similar to that of the mutant proteins (data not shown). The dimeric state of the RecU (31,36) and mutant RecUK56A and RecUR71A proteins was confirmed by protein cross-linking (see below) and gel filtration chromatography (data not shown). It is likely, therefore, that the structural organization was not affected by the mutations.
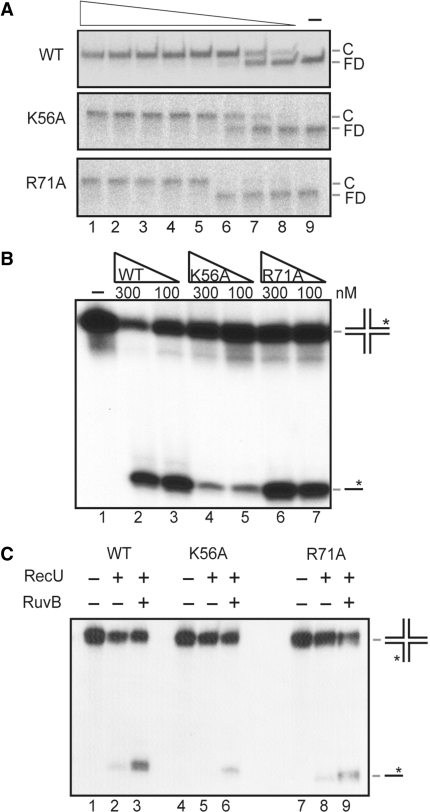
RecU binds HJs with high affinity, with an apparent binding constant (KDapp) of 0.6 nM [(31), Figure 2A]. The KDapp for RecUK56A was 0.8 nM and comparable to that of the WT protein, whereas RecUR71A binds HJs with slightly higher cooperativity, albeit with a slightly lower affinity (KDapp 1.2 nM) (Figure 2A).
Figure 2.
RecUK56A or RecUR71A binds and cleaves HJs and interacts with RuvB. (A) [γ-32P]-HJ DNA was incubated with RecU, RecUK56A or RecUR71A (0.25–36 nM) for 15 min and the complexes were separated by ndPAGE and autoradiographed. FD: free DNA, C: protein–DNA complex. (B) [γ-32P]-HJ was incubated with RecU variants (100 and 300 nM) for 30 min at 37°C and the cleavage products analyzed. (C) The cleavage of the HJ by RecU variants (40 nM) was analyzed with RuvB (40 nM) in the presence of 1 mM ATPγ S. In (B) and (C), the position of the asterisk indicates the strand of the HJ, which was labelled in each experiment.
The RecUK56A and RecUR71A mutants were tested for their ability to cleave HJs. Both mutants cleaved HJs and rendered products with mobility similar to the one generated by WT RecU (Figure 2B), suggesting that cleavage occurred preferentially at the consensus sequence, albeit RecUK56A was ∼5-times less active. One explanation for this difference might be that the K56 residue is located in the vicinity of the catalytic centre, which is located at the dimer interface, whereas the R71 residue is far apart of it (36, see below).
Previously, it was shown that a specific interaction between E. coli RuvC (RuvCEco) and RuvBEco (33) stimulated RuvCEco cleavage in the presence of ATPγS (41). RecU variants were tested for their ability to interact with RuvB by testing RuvB stimulation in a HJ cleavage assay in the presence of ATPγS. RuvB stimulated the resolution of HJs by RecU or its variants (Figure 2C). The stimulation of RecUK56A activity by RuvB under the conditions assayed was significant, albeit less manifest than the stimulation observed with WT or RecUR71A variant. Furthermore, the three RecU variants interacted with RuvB as detected by protein cross-linking (data not shown). These data suggest that the action of RecU variants at HJs is ∼5-times affected (RecUK56A) or not at all (RecUR71A), and in none of the case the coordination with the RuvB helicase is impaired. This is consistent with the observation in vivo that the recU56 and recU71 mutations did not affect the bias of HJ resolution and chromosomal segregation (Figure 1A).
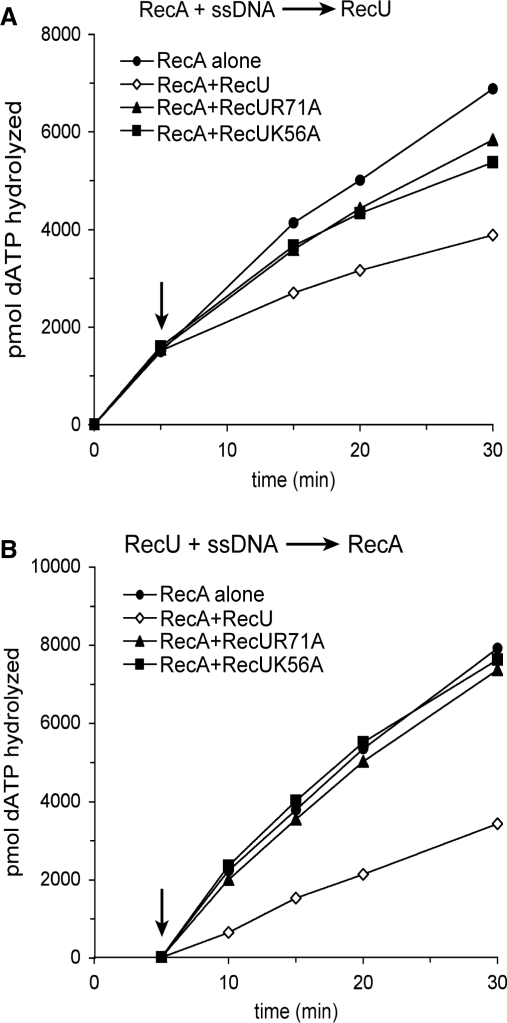
RecUK56A or RecUR71A fails to inhibit the ssDNA-dependent RecA dATPase activity
Previously, it was shown that independently of the order of addition, RecU partially inhibited the hydrolysis of dATP or rATP by RecA (15). Since RecA hydrolyzes dATP in preference to rATP both in the absence or presence of SsbA (32), the ability of mutants to inhibit RecA-catalyzed ssDNA-dependent dATP hydrolysis was measured in the presence of an excess of ssDNA over proteins. Addition of RecU to the preformed ssDNA·RecA·dATP·Mg2+ complex partially inhibited the dATPase activity of RecA, but RecUK56A or RecUR71A failed to exert the same extent of inhibition on the RecA dATPase (Figure 3A). When RecA·dATP·Mg2+ was added to the preformed ssDNA·RecU complex the inhibition caused by RecU was higher, while the mutants did not affect the dATPase activity of RecA (Figure 3B). One possibility could be that RecUK56A and RecUR71A fail to inhibit the dATPase activity of RecA because they do not bind ssDNA. Previously, it was shown that RecU binds ssDNA and catalyzes renaturation of two homologous ssDNAs (31). As revealed in Supplementary Figure S1, RecUK56A or RecUR71A binds and anneals complementary ssDNAs with similar efficiency as WT RecU.
Figure 3.
Effect of RecUK56A or RecUR71A on RecA catalyzed dATP hydrolysis. Circular ssDNA (10 μM) was preincubated with RecA (1.3 μM, in (A)) or with RecU variants (200 nM, in (B)) in buffer A containing 2 mM dATP for 5 min at 37°C. Then an RecU variant was added to RecA (A) or RecA was added to the RecU variants (B), and the reaction mixtures were incubated for further 25 min at 37°C. Aliquots were taken and the extent of RecA catalyzed hydrolysis of dATP is shown in pmols. The arrow indicates the addition of the second protein. Values are averages of four independent experiments.
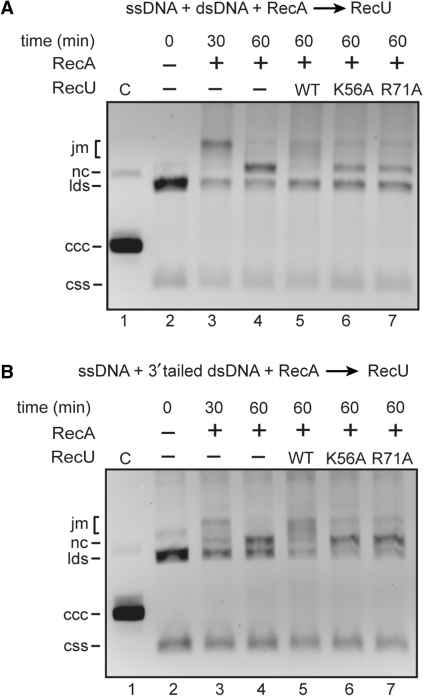
RecUK56A or RecUR71A does not inhibit dATP-dependent RecA-mediated DNA strand exchange
The effect of RecU mutant proteins in RecA-mediated dATP-dependent DNA strand exchange was then analyzed. Addition of RecU inhibited the accumulation of RecA-mediated final recombination products (nicked circular DNA, nc) between circular ssDNA and homologous linear dsDNA, but RecUK56A or RecUR71A did not inhibit RecA-mediated accumulation of nc products (Figure 4A). Similar results were observed when circular and homologous linear 3′-tailed duplex DNA were used (Figure 4B).
Figure 4.
Effect of RecUK56A or RecUR71A on RecA-mediated DNA strand exchange. (A) Circular ssDNA (10 μM) and homologous linear dsDNA (20 μM) were incubated with RecA (1.3 μM) in buffer A containing 10 mM MgOAc2 and 2 mM dATP for 30 min at 37°C. Then a RecU variant (200 nM) was added, and the reaction mixture was incubated for further 30 min at 37°C. After deproteinization, the reaction mixtures were analyzed by 0.8% agarose gel electrophoresis. In lane 3, the reaction was stopped after the first 30 min of incubation as indicated. (B) The same experiment was performed with circular ssDNA and homologous 3′-tailed dsDNA. C, partially nicked dsDNA; css, circular ssDNA; lds, linear dsDNA; nc, nicked circular DNA; jm, joint molecules; ccc, covalently closed circular DNA.
RecUK56A or RecUR71A fails to interact with RecA
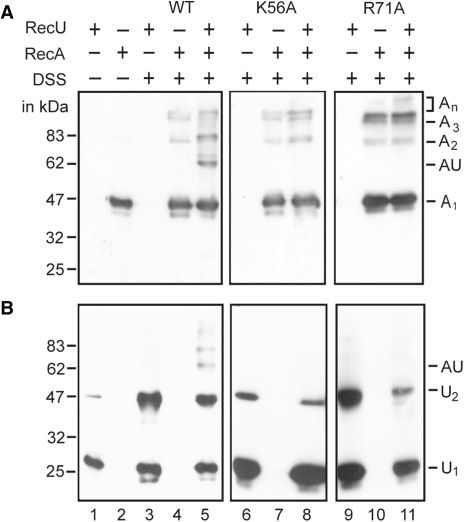
The RecU, RecUK56A or RecUR71A proteins were tested for physical interaction with RecA in the presence of the cross-linking agent bisdisuccinimidyl suberate (DSS). RecA (38.0 kDa) ran as a homogeneous band of ∼42 kDa (A1), whereas RecU (23.9 kDa) ran as ∼24 kDa (U1) under SDS–PAGE (Figure 5).
Figure 5.
RecUK56A or RecUR71A does not interact with RecA. (A and B), RecUWT, RecUK56A, RecUR71A (2 μM) or RecA (2 μM) or combinations of them were incubated with 50 μM DSS for 10 min. Immunoblot of cross-linked proteins using anti-RecA (A), or anti-RecU antibodies (B). The novel 62 kDa band is indicated by AU. An and Un denote the predicted state of the given protein (e.g. A1, A2, A3 indicates the oligomeric state of RecA). Plus and minus denote the presence and absence of the indicated protein, and/or cross-linking agent. The position of the molecular mass marker (in kDa) is shown.
In the presence of DSS, RecU bands of ∼24 (U1) and ∼47 kDa (U2) were observed. This is consistent with the observations that RecU is mainly a dimer in solution and in the crystal (36). New minor bands also appeared in RecA after DSS treatment corresponding to ∼84 (A2), ∼126 (A3) and >160 kDa (An), which indicated that RecA may form high order oligomers (42). When RecA was incubated with RecU and DSS, a novel band of ∼62 kDa was observed using anti-RecA antibodies (Figure 5A, lane 5). The ∼62 kDa protein was also observed when the membrane was revealed with anti-RecU antibodies (Figure 5B, lane 5). We have ruled out protein aggregation because 2 μM BSA was present in all reactions, and bands corresponding to nonspecific cross-linking (e.g. RecA or RecU cross-linked with BSA) were not detected. The gel-purified 62 kDa protein was subjected to partial proteolysis followed by MALDI-TOF analysis, and polypeptides from both proteins were present (data not shown). It is likely that this novel ∼62 kDa protein might be composed of one monomer of RecU (∼24 kDa) and one of RecA (∼38 kDa) cross-linked, and that RecU and RecA interact even in the absence of dATP and/or ssDNA. When RecU was replaced by RecUK56A, or by RecUR71A the 62 kDa band was not detected (Figure 5, lanes 8 and 11).
DISCUSSION
In this work, we have isolated and characterized two ‘separation-of-function’ recU mutants (recU56 and recU71) competent to resolve HJs with a bias towards noncrossover products (monomeric chromosomes in circular genomes), while they have lost the capacity to promote recombinational repair and plasmid transformation. Since plasmid transformation is a RuvAB-independent event, whereas it requires RecU (when RecA is present), we propose that RecU might also act outside of the resolvasome context. RecU is a RecA mediator regulating the formation of RecA presynaptic filaments, that otherwise might be deleterious during plasmid establishment in a ΔrecU context. This is in agreement with in vitro data showing that (i) RecU prevents RecA nucleoprotein filament formation and RecA-mediated DNA strand exchange activities and interacts with RecA in the absence of ssDNA and/or dATP, whereas RecUK56A or RecUR71A fails to show these inhibitory activities, and (ii) RecU, RecUK56A or RecUR71A binds and cleaves HJs, and this activity is stimulated by the addition of RuvB. Altogether, these findings indicate that the mutant alleles are proficient at late stages of HR, but they are deficient at early stages because they fail to exert a negative effect on RecA activities.
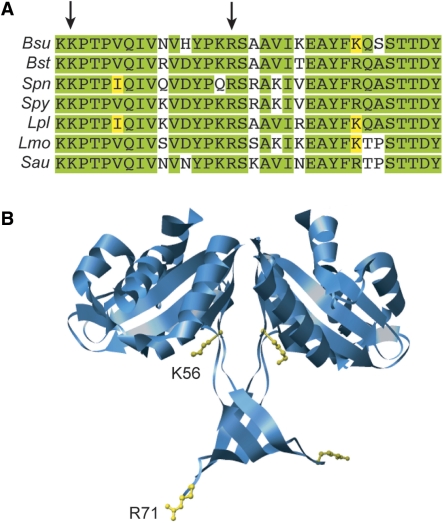
The RecU homodimer contains a rigid mushroom-like cap (residues 1–55 and 90–206) and a flexible stalk region (Figure 6). The stalk contains two extended loops (residues 56–89), whose ends are disordered in the RecU structure (36), and the RecUK56A and RecUR71A mutations map in this region. RecU shows an overall 59% sequence identity with B. stearothermophis RecU (RecUBst) and this identity increases to 88% within the region under analysis (Figure 6A). The full structure of the stalk region was visible in the electron density map of RecUBst (PDB 1Y10) that crystallized in a different space group than RecU (36). Positions equivalent to RecUK56 and RecUR71 in RecUBst are clustered in the outer region of the stalk, whereas the catalytic triad (Asp88, Asp99 and Glu101) is located at the dimer interface [(36), Figure 6B)].
Figure 6.
Conservation of mutated residues and their localization in the RecUBst X-ray structure. (A) sequence alignment of the tail region from several RecU proteins from Firmicutes. Bsu, B. subtilis; Bst, B. stearotermophilus; Spn, Streptococcus pneumoniae; Spy, S. pyogenes; Lpl, Lactobacillus plantarum; Lmo, Lysteria monocitogenes; Sau, Staphylococcus aureus. The tail region comprises residues 56–89 in RecU (PDB: 1ZP7). Arrows indicate the mutated residues. Identical residues are highlighted in green and conserved residues in yellow. (B) localization of mutated residues in the X-ray structure of RecUBst (PDB: 1Y1O, Osipiuk,J., Li,H., Moy,S., Collart,F. and Joachmiak,A., deposited in 2004). Residues are localized in the structure of RecUBst whose tail region could be resolved, although residue numbers correspond to the RecU protein.
These results and previous ones (15,31) indicate that RecU has three activities in vitro: (i) to mediate DNA annealing, (ii) to catalyze HJ resolution, and (iii) to modulate RecA activities. The first RecU activity, which is unlikely to be the main activity required for plasmid transformation because the absence of RecA suppresses the RecU requirement, remains elusive and will not be further discussed. Our results show that the second and third activities, which are important in vivo, can be genetically separated. Since the MMS sensitivity of RecUK56A or RecUR71A is similar to the one of a deletion mutant, but both mutants are able to cleave the HJ and interact with RuvB, it is likely that the third activity plays an essential role in repair-by-recombination. Is RecU action at the early stages of HR a unique event? The RuvABEco complex, at relative high protein concentration, promotes rapid dissociation of RecAEco nucleoprotein filaments formed on supercoiled dsDNA (43–45), and removes RecAEco from recombination intermediates in vitro (33), but the genetics provides only limited support for this role in vivo. Pyrococcus furiosus RadB, which regulates RadA activities, was shown to interact with RadA and the Hjc resolvase. The activity of the Hjc resolvase is modulated by RadB in an ATP-dependent manner (16). The mammalian CX3 complex (see Introduction section) is involved in the early stages of HR by binding DNA, catalyzing DNA pairing and recruiting Rad51 (19) and also plays an essential but unknown role in the resolution of recombination intermediates (19,21,23). It is tempting to speculate that a player coupling recombinase action with HJ branch migration and resolution is ubiquitous. In archaea and higher eukaryotes, this would be a Rad51 paralog, and in bacteria this might be accomplished by a direct interaction between RecA and a component of the resolvasome. How does RecU modulate RecA activities, even in the presence of an excess of free ssDNA? Previously, it was shown that: i) RecA·dATP forms filaments and the RecU protein forms blobs in ssDNA, and ii) discrete RecU blobs are embedded in the RecA nucleoprotein filaments, under a condition in which the RecA dATPase is inhibited (15). It is likely that RecU, upon interacting with RecA at the end of a filament might cap it and control RecA nucleoprotein filament dynamics. Then RuvAB would promote translocation, and RecU resolution of HJs. By this action, the early and late HR stages will be two rapid and coordinated events.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for Open Access publication charge: Ministerio de Ciencia e Innovación (MCI). This work was financed by MCI grants. BFU2006-01062 to J.C.A., and BFU 2006-02907 to S.A. C.C. was a recipient of a Fellowship of the Ministerio de Ciencia e Innovación and B.C. was supported by the Juan de la Cierva program.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank J. Gómez-Gutiérrez for helping them with circular dichroism experiments.
REFERENCES
- 1.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 2.Pâques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amundsen SK, Smith GR. Interchangeable parts of the Escherichia coli recombination machinery. Cell. 2003;112:741–744. doi: 10.1016/s0092-8674(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 4.Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc. Natl Acad. Sci. USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox MM. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez H, Kidane D, Cozar MC, Graumann PL, Alonso JC. Recruitment of Bacillus subtilis RecN to DNA double-strand breaks in the absence of DNA end processing. J. Bacteriol. 2006;188:353–360. doi: 10.1128/JB.188.2.353-360.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez H, Carrasco B, Ayora S, Alonso JC. Dynamics of DNA double-strand break repair in Bacillus subtilis. In: Graumann PL, editor. Bacillus: Cellular and Molecular Biology. Caister Academic Press; 2007. pp. 43–66. [Google Scholar]
- 8.McGlynn P, Lloyd RG. Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 2002;18:413–419. doi: 10.1016/s0168-9525(02)02720-8. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez H, Kidane D, Reed P, Curtis FA, Cozar MC, Graumann PL, Sharples GJ, Alonso JC. The RuvAB branch migration translocase and RecU Holliday junction resolvase are required for double-stranded DNA break repair in Bacillus subtilis. Genetics. 2005;171:873–883. doi: 10.1534/genetics.105.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen Q, Mahdi AA, Briggs GS, Sharples GJ, Lloyd RG. Conservation of RecG activity from pathogens to hyperthermophiles. DNA Repair. 2005;4:23–31. doi: 10.1016/j.dnarep.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez H, Carrasco B, Cozar MC, Alonso JC. Bacillus subtilis RecG branch migration translocase is required for DNA repair and chromosomal segregation. Mol. Microbiol. 2007;65:920–935. doi: 10.1111/j.1365-2958.2007.05835.x. [DOI] [PubMed] [Google Scholar]
- 12.Cromie GA, Leach DR. Control of crossing over. Mol. Cell. 2000;6:815–826. doi: 10.1016/s1097-2765(05)00095-x. [DOI] [PubMed] [Google Scholar]
- 13.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 14.Carrasco B, Cozar MC, Lurz R, Alonso JC, Ayora S. Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction processing functions in chromosome segregation. J. Bacteriol. 2004;186:5557–5566. doi: 10.1128/JB.186.17.5557-5566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrasco B, Ayora S, Lurz R, Alonso JC. Bacillus subtilis RecU Holliday-junction resolvase modulates RecA activities. Nucleic Acids Res. 2005;33:3942–3952. doi: 10.1093/nar/gki713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komori K, Miyata T, DiRuggiero J, Holley-Shanks R, Hayashi I, Cann IK, Mayanagi K, Shinagawa H, Ishino Y. Both RadA and RadB are involved in homologous recombination in Pyrococcus furiosus. J. Biol. Chem. 2000;275:33782–33790. doi: 10.1074/jbc.M004557200. [DOI] [PubMed] [Google Scholar]
- 17.Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurumizaka H, Ikawa S, Nakada M, Eda K, Kagawa W, Takata M, Takeda S, Yokoyama S, Shibata T. Homologous-pairing activity of the human DNA-repair proteins Xrcc3 Rad51C. Proc. Natl Acad. Sci. USA. 2001;98:5538–5543. doi: 10.1073/pnas.091603098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuznetsov S, Pellegrini M, Shuda K, Fernandez-Capetillo O, Liu Y, Martin BK, Burkett S, Southon E, Pati D, Tessarollo L, et al. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J. Cell Biol. 2007;176:581–592. doi: 10.1083/jcb.200608130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Masson JY, Shah R, O’Regan P, West SC. RAD51C is required for Holliday junction processing in mammalian cells. Science. 2004;303:243–246. doi: 10.1126/science.1093037. [DOI] [PubMed] [Google Scholar]
- 22.Forget AL, Bennett BT, Knight KL. Xrcc3 is recruited to DNA double strand breaks early and independent of Rad51. J. Cell Biochem. 2004;93:429–436. doi: 10.1002/jcb.20232. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Tarsounas M, O’Regan P, West SC. Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J. Biol. Chem. 2007;282:1973–1979. doi: 10.1074/jbc.M609066200. [DOI] [PubMed] [Google Scholar]
- 24.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubnau D, Davidoff-Abelson R, Scher B, Cirigliano C. Fate of transforming deoxyribonucleic acid after uptake by competent Bacillus subtilis: phenotypic characterization of radiation-sensitive recombination-deficient mutants. J. Bacteriol. 1973;114:273–286. doi: 10.1128/jb.114.1.273-286.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonso JC, Luder G, Tailor RH. Characterization of Bacillus subtilis recombinational pathways. J. Bacteriol. 1991;173:3977–3980. doi: 10.1128/jb.173.13.3977-3980.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canosi U, Morelli G, Trautner TA. The relationship between molecular structure and transformation efficiency of some S. aureus plasmids isolated from B. subtilis. Mol. Gen. Genet. 1978;166:259–267. doi: 10.1007/BF00267617. [DOI] [PubMed] [Google Scholar]
- 28.de Vos WM, Venema G, Canosi U, Trautner TA. Plasmid transformation in Bacillus subtilis: fate of plasmid DNA. Mol. Gen. Genet. 1981;181:424–433. doi: 10.1007/BF00428731. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez S, Sorokin A, Alonso JC. Genetic recombination in Bacillus subtilis 168: effects of recU and recS mutations on DNA repair and homologous recombination. J. Bacteriol. 1998;180:3405–3409. doi: 10.1128/jb.180.13.3405-3409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez S, Kobayashi Y, Ogasawara N, Alonso JC. Analysis of the Bacillus subtilis recO gene: RecO forms part of the RecFLOR function. Mol. Gen. Genet. 1999;261:567–573. doi: 10.1007/s004380051002. [DOI] [PubMed] [Google Scholar]
- 31.Ayora S, Carrasco B, Doncel E, Lurz R, Alonso JC. Bacillus subtilis RecU protein cleaves Holliday junctions and anneals single-stranded DNA. Proc. Natl Acad. Sci. USA. 2004;101:452–457. doi: 10.1073/pnas.2533829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrasco B, Manfredi C, Ayora S, Alonso JC. Bacillus subtilis SsbA and dATP regulate RecA nucleation onto single-stranded DNA. DNA Repair. 2008;7:990–996. doi: 10.1016/j.dnarep.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 33.Eggleston AK, Mitchell AH, West SC. In vitro reconstitution of the late steps of genetic recombination in. E. coli. Cell. 1997;89:607–617. doi: 10.1016/s0092-8674(00)80242-1. [DOI] [PubMed] [Google Scholar]
- 34.Rottlander E, Trautner TA. Genetic and transfection studies with B. subtilis phage SP50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol. Gen. Genet. 1970;108:47–60. doi: 10.1007/BF00343184. [DOI] [PubMed] [Google Scholar]
- 35.Alonso JC, Tailor RH, Luder G. Characterization of recombination-deficient mutants of Bacillus subtilis. J. Bacteriol. 1988;170:3001–3007. doi: 10.1128/jb.170.7.3001-3007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGregor N, Ayora S, Sedelnikova S, Carrasco B, Alonso JC, Thaw P, Rafferty J. The structure of Bacillus subtilis RecU holliday junction resolvase and its role in substrate selection and sequence-specific cleavage. Structure. 2005;13:1341–1351. doi: 10.1016/j.str.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 37.Ayora S, Missich R, Mesa P, Lurz R, Yang S, Egelman EH, Alonso JC. Homologous-pairing activity of the Bacillus subtilis bacteriophage SPP1 replication protein G35P. J. Biol. Chem. 2002;277:35969–35979. doi: 10.1074/jbc.M204467200. [DOI] [PubMed] [Google Scholar]
- 38.Ayora S, Weise F, Mesa P, Stasiak A, Alonso JC. Bacillus subtilis bacteriophage SPP1 hexameric DNA helicase, G40P, interacts with forked DNA. Nucleic Acids Res. 2002;30:2280–2289. doi: 10.1093/nar/30.11.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedersen LB, Setlow P. Penicillin-binding protein-related factor A is required for proper chromosome segregation in Bacillus subtilis. J. Bacteriol. 2000;182:1650–1658. doi: 10.1128/jb.182.6.1650-1658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceglowski P, Luder G, Alonso JC. Genetic analysis of recE activities in Bacillus subtilis. Mol. Gen. Genet. 1990;222:441–445. doi: 10.1007/BF00633853. [DOI] [PubMed] [Google Scholar]
- 41.van Gool AJ, Shah R, Mezard C, West SC. Functional interactions between the Holliday junction resolvase and the branch migration motor of Escherichia coli. EMBO J. 1988;17:1838–1845. doi: 10.1093/emboj/17.6.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brenner SL, Zlotnick A, Griffith JD. RecA protein self-assembly. Multiple discrete aggregation states. J. Mol. Biol. 1988;204:959–972. doi: 10.1016/0022-2836(88)90055-1. [DOI] [PubMed] [Google Scholar]
- 43.Iwasaki H, Takahagi M, Nakata A, Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992;6:2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- 44.Tsaneva IR, Muller B, West SC. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992;69:1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]
- 45.Adams DE, Tsaneva IR, West SC. Dissociation of RecA filaments from duplex DNA by the RuvA and RuvB DNA repair proteins. Proc. Natl Acad. Sci. USA. 1994;91:9901–9905. doi: 10.1073/pnas.91.21.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.