Abstract
Primary transcripts of certain microRNA (miRNA) genes (pri-miRNAs) are subject to RNA editing that converts adenosine to inosine (A→I RNA editing). However, the frequency of the pri-miRNA editing and the fate of edited pri-miRNAs remain largely to be determined. Examination of already known pri-miRNA editing sites indicated that adenosine residues of the UAG triplet sequence might be edited more frequently. In the present study, therefore, we conducted a large-scale survey of human pri-miRNAs containing the UAG triplet sequence. By direct sequencing of RT–PCR products corresponding to pri-miRNAs, we examined 209 pri-miRNAs and identified 43 UAG and also 43 non-UAG editing sites in 47 pri-miRNAs, which were highly edited in human brain. In vitro miRNA processing assay using recombinant Drosha-DGCR8 and Dicer-TRBP (the human immuno deficiency virus transactivating response RNA-binding protein) complexes revealed that a majority of pri-miRNA editing is likely to interfere with the miRNA processing steps. In addition, four new edited miRNAs with altered seed sequences were identified by targeted cloning and sequencing of the miRNAs that would be processed from edited pri-miRNAs. Our studies predict that ∼16% of human pri-miRNAs are subject to A→I editing and, thus, miRNA editing could have a large impact on the miRNA-mediated gene silencing.
INTRODUCTION
One type of RNA editing involves the conversion of adenosine residues into inosine (A→I RNA editing) specifically in double-stranded RNAs (dsRNAs) through the action of ADAR (adensosine deaminase acting on RNA) (1–3). The inosine residue converted from adenosine in RNA is detected as an A→G change of the cDNA sequence. The translation machinery reads inosine as guanosine, leading to alterations of codons. Three separate ADAR gene family members (ADAR1–3) are known in mammals. Both ADAR1 and ADAR2 are detected in many tissues, whereas ADAR3 is expressed only in brain (4–10). Involvement of both ADAR1 and ADAR2 in site-selective RNA editing has been demonstrated by in vitro editing experiments as well as by analysis of editing site selection changes in ADAR1 null and ADAR2 null mutant mice (7,11–15). When it occurs within the protein-coding sequence, A→I RNA editing results in the synthesis of proteins not directly encoded by the genome, as demonstrated with the glutamate receptors (GluR), the serotonin receptor 2C (5-HT2CR), the Kv1.1 potassium channel, and the α3 GABAA receptor, leading to diversification of these gene functions (11,14,16–18). Misregulated RNA editing may underlie certain human diseases and pathological processes. Underediting of GluR-B mRNAs at the Q/R site has been implicated in human amyotrophic lateral sclerosis (19), whereas inappropriate editing of 5-HT2CR mRNAs may have causative relevance to suicide, schizophrenia and depression (20–23). In addition, ADAR1 heterozygous mutations have been identified in dyschromatosis symmetrica hereditaria (DSH), a human disease of aberrant skin pigmentation with an autosomal dominant inheritance (24). However, the most common targets for A→I editing are non-coding RNAs that contain inversely oriented repeats of repetitive elements such as Alu and LINE (25–28). The biological significance of non-coding, repetitive RNA editing is largely unknown (29).
More recently, it has been reported that primary transcripts of certain microRNA (miRNA) genes (pri-miRNAs) are subject to A→I RNA editing (30–35). Pri-miRNAs are processed sequentially by two members of the RNase III superfamily, Drosha and Dicer (36). Nuclear Drosha, together with its essential partner, DGCR8, cleaves pri-miRNAs, releasing 60–70 nt precursor miRNAs (pre-miRNAs) (37–40). Recognition of correctly processed pre-miRNAs and their nuclear export is carried out by Exportin-5 and RanGTP (41). Cytoplasmic Dicer together with the dsRNA binding protein TRBP then processes the pre-miRNAs into 20–22 nt imperfect dsRNAs (42,43). One or both strands of the duplex may serve as the mature miRNA. Following integration into RISC (RNA-induced silencing complex), miRNAs block the translation of partially complementary targets located in the 3′ UTR of specific mRNAs or guide the degradation of target mRNAs as do siRNAs (3,36,44–46).
The presence of the post-transcriptional mechanisms that regulate processing of miRNAs at the Drosha or Dicer cleavage step has been reported (47–50). Editing of pri-miRNAs could alter their processing (31,35). For instance, editing of pri-miR151 suppresses the Dicer cleavage step (31), whereas editing of pri-miR142 inhibits the Drosha cleavage step and consequently suppresses the expression of the mature miRNA levels (35). Furthermore, editing of pri-miRNAs could also lead to the expression of edited mature miRNAs (32). For instance, editing of pri-miR-376 cluster RNAs resulted in expression of sequence altered miRNAs (edited miRNAs) and selection of a set of target genes different from those targeted by the unedited miRNAs (32).
Although several examples of pri-miRNA editing have been investigated (30–33,35), the frequency of the miRNA editing and its biological significance remains to be established. In this study, we conducted a large-scale survey and identified 86 A→I editing sites in 47 pri-miRNAs. We tested the significance of editing of select pri-miRNAs by performing an in vitro miRNA processing assay. Our studies revealed that a majority of pri-miRNA editing events appear to affect miRNA processing steps. In addition, we detected abundant expression of four new edited mature miRNAs with altered ‘seed’ sequences. These edited mature miRNAs are likely to repress a set of genes different from those targeted by the unedited miRNAs. Having established that ∼16% of human pri-miRNAs are edited, it is anticipated that the expression of a large number of genes is affected globally by A→I editing of miRNAs.
MATERIALS AND METHODS
Extraction of the potential UAG triplet from human pri-miRNAs
The 474 human miRNA hairpin structures available in miRBase 9.1 were retrieved and annotated with respect to mature sequence location using a Perl program. A second program parsed these annotated structures to extract each class of potentially edited sites. We divided 494 UAG triplet sites in dsRNA regions of 291 pri-miRNAs into four groups: W-C group; U and G in UAG triplet form Watson–Crick base pairing (201 sites in 157 pri-miRNAs). G-U group; Either U or G, or both in UAG triplet forms G-U wobble base pairing (122 sites in 105 pri-miRNAs). Mismatch group; Either U or G in UAG triplet forms mismatch base pairing (90 sites in 79 pri-miRNAs). Others group; Both U and G in UAG triplet form mismatch base pairing (81 sites in 76 pri-miRNAs). We excluded the Others group from further analysis, because it is very unlikely that RNA editing happens under this condition. Similarly, we also counted and divided all non-UAG triplet sites present in 474 pri-miRNAs into three categories: W-C group, G-U group and Mismatch group in order to estimate their editing frequency (Table 9).
Table 9.
Effects of the nucleotide complementary to the non-UAG site
| The complementary nucleotide |
||||||
|---|---|---|---|---|---|---|
| C | U | G | A | None | Total | |
| AAG | 0/13 | 10/185 | 0/11 | 0/16 | 0/6 | 10/231 |
| AAA | 3/15 | 5/205 | 0/31 | 0/33 | 0/12 | 8/296 |
| UAU | 1/13 | 4/240 | 0/8 | 1/20 | 0/5 | 6/286 |
| UAC | 0/10 | 5/157 | 0/8 | 0/17 | 0/9 | 5/201 |
| UAA | 0/6 | 4/164 | 0/10 | 0/27 | 0/11 | 4/218 |
| CAG | 1/19 | 1/238 | 0/16 | 0/15 | 0/7 | 2/295 |
| CAC | 2/10 | 0/170 | 0/6 | 0/9 | 0/9 | 2/204 |
| CAU | 1/5 | 1/221 | 0/4 | 0/15 | 0/13 | 2/258 |
| CAA | 1/7 | 0/166 | 0/10 | 0/18 | 0/7 | 1/208 |
| AAC | 0/9 | 1/135 | 0/13 | 0/10 | 0/7 | 1/174 |
| AAU | 0/16 | 1/190 | 0/21 | 0/17 | 0/10 | 1/254 |
| GAG | 1/14 | 0/202 | 0/11 | 0/17 | 0/9 | 1/253 |
| Totala | 10/137 (7.3%) | 32/2273 (1.4%) | 0/149 (0.0%) | 1/214 (0.5%) | 0/105 (0.0%) | 43/2878 (1.5%) |
aEdited and potential non-UAG sites identified in the 209 pri-miRNAs were counted (edited/potential).
Mice
All procedures involving mice were approved by the Institutional Animal Care and Use Committee of The Wistar Institute and performed in accordance to the US National Institutes of Health Guidelines.
RNA preparation
Human total brain RNA was obtained from Clontech (Palo Alto, CA, USA) and Ambion (Austin, TX, USA). Mouse total RNA was extracted from dissected total brains using TRIZOL reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA).
Determination of pri-miRNA editing
The method was previously described (31,32). Briefly, first-strand cDNA was synthesized using 1 μg of total RNA and each miRNA-specific RT primer (Tables S1, S2 and S3). The resultant cDNA was then amplified by PCR using each miRNA-specific PCR primer pairs (Table S1, S2 and S3). RT–PCR products were directly sequenced using the inward primer. If the editing frequency was <20%, RT–PCR products were sequenced from both directions to exclude the possibility of background noise. The editing frequency was determined as the % ratio of the ‘G’ peak over the sum of ‘G’ and ‘A’ peaks of the sequencing chromatogram. The editing frequency was given as round numbers of ±5%. When editing was found in certain human pri-miRNA, the corresponding mouse pri-miRNA was also amplified from wildtype and ADAR2 null brains as well as wildtype and ADAR1 null embryos to determine the conservation and the responsible editing enzyme in vivo.
Characterization of mature miRNAs
Small RNA (<200 nt) was extracted from 50 μg of total brain RNA using mirVana™ miRNA Isolation Kit (Ambion). Preparation of the cDNA library enriched in small RNA was described previously (31,32). Briefly, 1.5 μg of small RNA was polyadenylated using Poly(A) Tailing Kit (Ambion). A 5′ adaptor (5′-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3′) synthesized at Dharmacon (Lafayette, CO, USA) was ligated to poly(A)-tailed RNA using T4 RNA ligase followed by RT using an RT primer: 5′-ATTCTAGAGGCCGAGGCGGCCGACATG-d(T)30 (A, G, or C) (A, G, C, or, T)-3′. The resultant cDNA was then amplified by PCR using cFW primer (5′-CTGACATGGACTGAAGGA-3′) and each miRNA-specific DW primer (Table S5), or each miRNA-specific FW primer (Table S5) and cDW primer (5′-ATTCTAGAGGCCGAGGCGGCCGACATGT-3′). After gel purification, RT–PCR products were subcloned using the TOPO TA cloning kit (Invitrogen) followed by sequencing more than 50 cDNA isolates.
Analysis of in vitro edited and processed pri-, pre- and mature miRNAs
In vitro pri-miRNA processing assay was described previously (31,35). The DNA fragment encompassing each pri-miRNA region was amplified by PCR, and then it was cloned into TOPO vector (Invitrogen). Unedited and edited (containing ‘G’) versions were screened by sequencing.
In vitro transcription was performed with Riboprobe in vitro Transcription Systems (Promega, Madison, WI, USA) using 1 μg of linearized TOPO vector containing the fragment corresponding to each pri-miRNA in the presence of 50 μCi [α-32P] CTP according to the manufacturer's instructions. The labeled pri-miRNAs were purified using 6% (w/v) polyacrylamide, 8 M urea gel.
The labeled pri-miRNAs (2× 105 c.p.m.) were digested using 20 ng of the Drosha–DGCR8 complex at 37°C for 60 min and, in some experiments, then to the Dicer cleavage reaction using 20 ng of the Dicer–TRBP complex at 37°C for 90 min. The mixture was then incubated with Proteinase K at 37°C for 60 min and was recovered by phenol/chloroform extraction and ethanol precipitation. The precipitated RNA was loaded on 15% (w/v) polyacrylamide, 8 M urea gel. The radioactivity in the dried gels was quantified with a PhosphorImaging System (Molecular Dynamics, Sunnyvale, CA, USA).
RESULTS AND DISCUSSION
Survey for A→I RNA editing of UAG triplets of pri-miRNA sequences
A→I editing of select pri-miRNAs was reported by several groups (30–32,34,35). By examining the editing frequency of the miRNA editing sites previously investigated by us (31,32), we noted that high-frequency editing often occurred at the adenosine residue within the UAG triplets (Table 1). The preference of the UAG triplet sequences for editing was reported previously in a small-scale survey of pri-miRNA editing sites (30) as well as statistical analysis of editing sites present in non-coding repetitive element sequences such as Alu and SINE (25–27,51). We reasoned that the adenosine in the UAG triplet of pri-miRNA sequences may be preferentially edited in vivo. The pri-miRNA sequences containing the UAG triplet were identified in 474 human pri-miRNAs registered at the Sanger Center miRBase site (miRBase 9.1). Since the UAG triplets in single-stranded regions are unlikely to be edited by ADARs, we selected 494 triplets in dsRNA regions of 291 pri-miRNAs. Among these candidates, UAG triplets that are mismatched at both U and G are also unlikely to be edited; therefore, 257 pri-miRNAs that contain 341 UAG triplet sequences in the dsRNA region were investigated. We attempted to amplify all of the 257 pri-miRNAs by RT–PCR, and 209 pri-miRNAs (∼80%) were successfully amplified from the total RNAs derived from human brain where pri-miRNAs are edited frequently (30–32,34). Direct cDNA sequencing of RT–PCR products identified 43 UAG triplets in 38 pri-miRNAs which were edited with frequency higher than 10% (Tables 2, 3 and Table S1, S2, S3). The 38 pri-miRNAs identified to contain one or more UAG editing sites accounted for 18.2% of 209 pri-miRNAs examined (Table 3). In addition, 43 non-UAG triplet editing sites were also identified while examining editing of UAG triplets (Table 8 and Table S4). Among those sites, 28 sites were detected in 11 pri-miRNAs which also contained at least a separate UAG triplet editing site, whereas 15 non-UAG triplet sites were found in 9 pri-miRNAs that did not contain the UAG triplet editing site (Table 3 and Table S4).
Table 1.
Known pri-miRNAs subject to A→I RNA editing
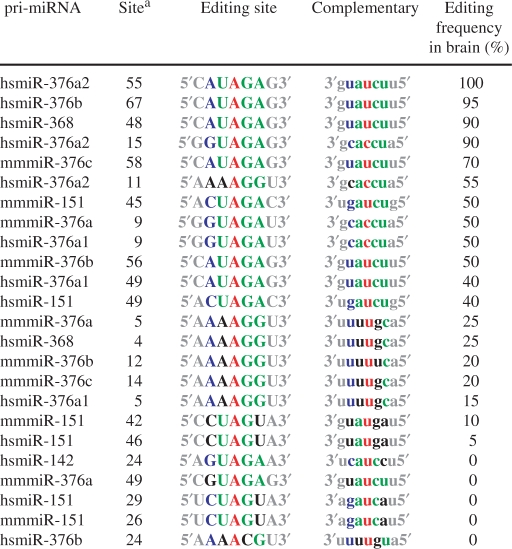 |
aThe 5′ end of the pri-miRNA sequence, registered at the Sanger Center miRBase site, is counted as 1.
Table 2.
Editing of UAG sites present in human pri-miRNAs
| W–C group (a) | G-U group (b) | Mismatch group (c) | Total of three groups (a+b+c) | |
|---|---|---|---|---|
| UAG sites examined | 172 | 96 | 73 | 341 |
| Edited at UAG sites | 34 | 4 | 5 | 43 |
| Editing frequency (%) | 19.8 | 4.1 | 6.8 | 12.6 |
Table 3.
Editing of pri-miRNAs containing UAG triplets
| W–C group (a) | G-U group (b) | Mismatch group (c) | Total of three groups (a+b+c)a | |
|---|---|---|---|---|
| miRNAs examined | 131 | 82 | 65 | 209 |
| Edited at UAG sites | 31 | 4 | 5 | 38 |
| Edited at non-UAG sites | 4 | 3 | 4 | 9 |
| All edited pri-miRNAs | 35 | 7 | 9 | 47 |
| Frequency (%) | 23.7 (26.7)b | 4.9 (8.5)b | 7.7 (13.8)b | 18.2 (22.5)b |
aMany pri-miRNAs contained multiple UAG triplets of different types, leading to the total pri-miRNA number that is less than the summation of three types.
bFrequency for all pri-miRNAs edited at both UAG and non-UAG sites.
Table 8.
Non-UAG editing sites
| The number | The mean editing efficiency (%) | |
|---|---|---|
| AAG | 10 | 49.5 |
| AAA | 8 | 50.0 |
| UAU | 6 | 23.3 |
| UAC | 5 | 20.0 |
| UAA | 4 | 43.8 |
| CAG | 2 | 75.0 |
| CAC | 2 | 20.0 |
| CAU | 2 | 22.5 |
| CAA | 1 | 40.0 |
| AAC | 1 | 20.0 |
| AAU | 1 | 10.0 |
| GAG | 1 | 30.0 |
| Total | 43 | 38.3 |
Expression of edited mature miRNAs
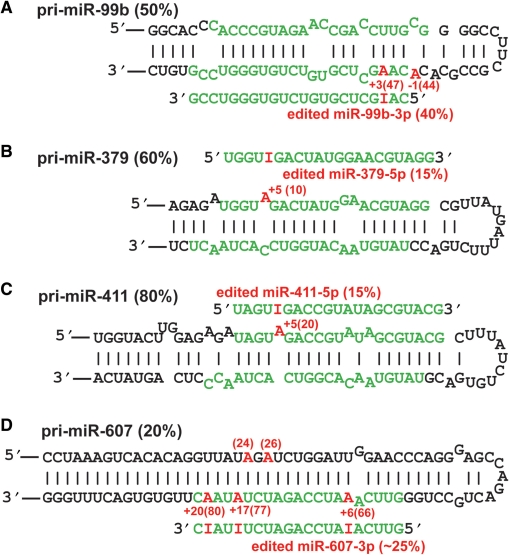
Many pri-miRNA editing sites were located within the mature miRNA sequence or corresponding partner miRNA sequence in the opposite strand (Table S5), which could be processed to edited mature miRNAs as we previously reported for miR-376 cluster RNAs (32). We applied the targeted cloning strategy as described previously (31,32), attempting to isolate cDNA clones corresponding to all of these potential edited mature miRNAs. Only four mature miRNAs containing UAG and/or non-UAG editing sites not previously reported were identified; miR-379-5p, miR-411-5p, miR-607-3p and miR-99b-3p (Figure 1). Interestingly, both miR-379 and miR-411 are members of the miR-379 cluster. Simultaneous expression of several edited mature miRNAs of the miR-376 cluster has been also reported (32), indicating the possibility of simultaneous editing of clustered miRNAs. The A→I editing sites identified in these mature miRNAs were located in the seed sequence known to play a major role in the hybridization of a miRNA with the target site, thus affecting selection of the target genes. As we have demonstrated for edited miR-376 RNAs (32), application of target prediction programs is likely to reveal that these newly identified edited mature miRNAs target genes that are different from those targeted by the unedited miRNAs.
Figure 1.
Identification of edited mature miRNAs. Four pri-miRNA structures and edited mature miRNAs are presented (A–D). Editing efficiency of pri-miRNA and mature miRNAs is indicated in parentheses in black and red, respectively. The 5′ end of the mature miRNA is counted as +1, and the editing site is indicated by the number highlighted in red. The position of the pri-miRNA editing site is also indicated in parenthesis. The 5′ end of the pri-miRNA structure registered at the Sanger Center miRBase site is counted as 1.
A recent global survey for miRNAs from various human and mouse tissues identified a large number of miRNAs that had nucleotide changes within the mature miRNA sequence, including those already reported previously as edited miRNAs such as miR-368, miR-376a1, miR-376a2 (32). However, four edited miRNAs identified in the present study as well as those identified previously (hs-miR-376b, mu-miR-376a, mu-miR-376b and mu-miR-376c) by the targeted cloning strategy were not included in the list of the miRNAs with A→G changes determined by the unbiased cloning strategy (33), indicating a limitation of the unbiased cloning strategy for detection of certain miRNA editing sites. Nevertheless, this deep sequencing study for unbiased expression profiling identified a few new miRNAs that had A→G nucleotide changes (potential A→I editing) at a frequency higher than 5%, i.e. hs-19b-2, hs-miR-512-1 and hs-miR-512-2, mu-miR-17, mu-miR-450b and mu-miR-483 (33). We tested some of these miRNAs containing the A→G sequence changes independently to see whether they are A→I edited miRNAs. Through analysis of the pri-miRNA sequence corresponding to the mature miRNAs with A→G changes, we found that at least the reported sequence heterogeneity of hs-miR-19b-2 and mu-miR-17 is not due to A→I editing (data not shown), indicating that they are most likely sequencing errors. Together, expression of edited mature miRNAs appears to be relatively rare, although a large fraction of human pri-miRNAs undergoes editing at the sequence corresponding to mature miRNAs. Thus, processing of a majority of edited pri-miRNAs must be suppressed as we previously reported for miR-142 (35) and miR-151 (31).
Processing of edited pri-miRNAs
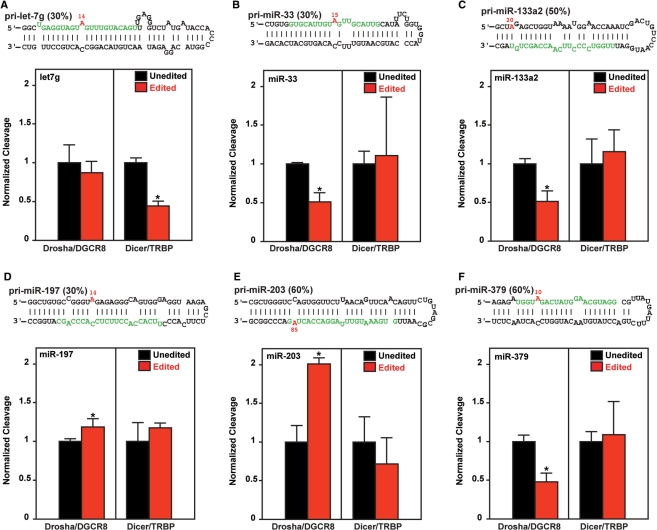
We then investigated the effects of miRNA editing on processing of pri- to pre- or pre- to mature miRNAs by subjecting unedited and edited pri-miRNAs to an in vitro processing assay using purified Drosha–DGCR8 or Dicer–TRBP complexes as described previously (31,35). We randomly selected six pri-miRNAs, i.e. pri-let-7g, pri-miR-33, pri-miR-133a2, pri-miR-197, pri-miR-203 and pri-miR-379, among 43 pri-miRNAs that have at least one UAG triplet editing site (Figure 2). None of the edited mature miRNAs expected to be processed from pri-let-7g and pri-miR-33 were detected, although editing sites were located within the mature miRNA sequences of these pri-miRNAs. Although the edited miR-379 RNAs were detected (Figure 1B), the frequency of detection of edited mature miR-379 (15%) versus unedited miR-379 was much lower than that expected from the editing frequency of pri-miR-379 (60%), indicating that processing of edited pri-miR-379 RNAs may not be as efficient as that of unedited pri-miR-379 RNAs. We also noted this proportionally less frequent detection of edited mature miR-411 RNAs (Figure 1C). Furthermore, neither unedited nor edited miR-203 was detected in human brain (Table S5), perhaps due to their low expression levels. We tested a set of unedited and ‘pre-edited’ pri-miRNAs for in vitro processing assays. In the ‘pre-edited’ pri-miRNA used for the assay, the adenosine residue of the editing site was replaced with guanosine. We previously demonstrated that the A→G alteration of pri-miRNA is treated the same as A→I editing by Drosha–DGCR8 complexes and also by the Dicer–TRBP complexes (31,35).
Figure 2.
In vitro processing of edited pri-miRNAs. In vitro cleavage of pri-miRNAs by Drosha/DGCR8 complexes and pre-miRNAs by Dicer/TRBP were determined separately (A–F). Editing efficiency of pri-miRNA is indicated in parentheses. The editing site is indicated by the number highlighted in red. The 5′ end of the pri-miRNA structure registered at the Sanger Center miRBase site is counted as 1. Three independent assays were done (n = 3). Mann–Whitney U-test, *P < 0.05.
Both +11 and +14 sites of pri-let-7g were edited at 10 and 30% in vivo in human brain (Table S1). The effects of editing at the major +14 site were tested for in vitro processing (Figure 2A). Although the editing at the +14 site did not affect the Drosha cleavage step, it suppressed the Dicer cleavage step, leading to 2-fold reduction in the mature miRNA levels. In contrast, editing of pri-miR-33 at the +15 site, pri-miR-133a2 at the +20 site and pri-miR-379 at the +10 site inhibited the Drosha cleavage step and reduced the generation of their precursor forms (pre-miRNAs) down to a 50% level as compared to their unedited controls (Figure 2B, C and F). This is likely the reason for less frequent detection of edited mature miR-379 RNAs (Figure 1B). Editing of these three pri-miRNAs had no effect on the Dicer cleavage step (Figure 2B, C and F). Interestingly, editing of pri-miR-203 at the +85 site enhanced its processing by the Drosha–DGCR8 complex, leading to 2-fold increase in generation of pre-miR-203 RNAs (Figure 2E). This is the first example of pri-miRNA editing that results in promotion of processing. Similarly, a slight increase in processing of pri-miR-197 edited at the +14 site by the Drosha–DGCR8 complex was detected (Figure 2D). Editing of both pri-miR-203 and pri-miR-197 did not affect the Dicer cleavage step (Figure 2D and E). The editing site of pri-miR-203 is located within the mature miRNA sequence (Figure 2E). Thus, although not detected in human brain investigated in this study, it is possible that edited miR-203 RNAs may be detected in the tissues where they are highly expressed. Interestingly, it has been recently reported that blockage of pri-let-7g processing by Lin28 RNA binding protein plays an important role in the miRNA-mediated differentiation in stem cells (50). Furthermore, a critical role played by miR-203 and miR-133a2 in differentiation and proliferation of skin progenitor cells and cardiomyocytes, respectively, has been reported (52–55). It remains to be investigated whether A→I editing of let-7g, miR-133a2 and miR-203 affects the cellular process proposed to be regulated by these miRNAs. Although only a limited number of pri-miRNA editing sites were tested, all six examples investigated were found to affect the processing of miRNAs, indicating that the major function of pri-miRNA editing is modulation of miRNA biogenesis and expression levels rather than generation of edited mature miRNAs and redirection of target gene selection. Finally, editing of pri-mRNAs may affect the processing steps not tested in this study such as nuclear export (36,41) as well as degradation of edited pri-miRNAs by the ribonuclease specific to inosine-containing RNAs (35,56).
Most preferred UAG triplet editing sites
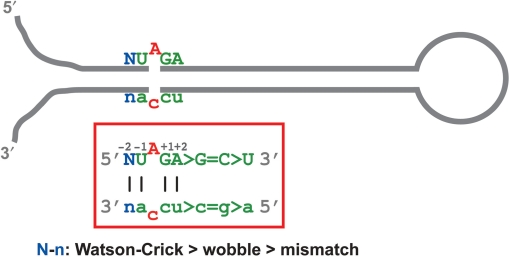
While analyzing the sequence and dsRNA structure of the UAG triplets, we noted that the editing frequency varies dependent on the complementary strand sequence that form the duplex structure with the UAG triplet (Figure 3 and Table 1). Therefore, we subgrouped 341 UAG triplets of 209 pri-miRNAs into three groups based on the nucleotide sequence complementary to U and G residues of the UAG triplet. (a) Both U and G are Watson–Crick base paired. (b) Either one of U and G, or both, are wobble base paired. (c) Either one of U and G is mismatched (Figure 1, Tables 2, 3 and Tables S1, S2, S3). We detected editing of 34 sites (19.8%) of 31 pri-miRNAs (23.7%) with both U and G residues base paired with A and C, respectively, in the Watson–Crick base pairing, i.e. group (a) (Tables 2, 3 and Table S1), four sites (4.1%) of four pri-miRNAs (4.9%) with either U and G residues or both base paired in the wobble pairing, i.e. group (b) (Tables 2, 3 and Table S2) and five sites (6.8%) in five pri-miRNAs (7.7%) with either U or G residues in a mismatched base pairing, i.e. group (c) (Tables 2, 3 and Table S3). The results revealed that the two neighboring nucleotides of the UAG triplet need to be stably base paired preferentially in the Watson–Crick base pairing in order to be edited efficiently (Figure 3). Finally, the editing frequency was plotted against the effect of the nucleotide complementary to the adenosine residue of the UAG triplet. The presence of cytosine in the opposite strand resulted in the most frequent (15/27 or 55.6%) and highest mean of the editing efficiency (57.3%) followed by uridine (27/275 or 9.8%) and absence of the complementary nucleotide (1/15 or 6.7%) (Table 4). The preference of A·C mismatched pairs for editing was noted through mutagenesis of known protein coding pre-mRNA (57). Furthermore, analysis of non-coding Alu and SINE sequences once again revealed much higher frequency for A·C mismatched editing sites, revealing its common preference for editing of pre-mRNAs, non-coding RNAs containing inversely oriented repeat elements and also miRNAs (25–27,30,51,58).
Figure 3.
The most preferred sequence and dsRNA structure for A→I editing of UAG triplets. The most frequently edited UAG triplet of pri-miRNA is indicated. The consensus pri-miRNA sequence with a ‘preferred’ editing site is derived by sequence comparison of 43 UAG triplet editing sites in 47 pri-miRNAs which we experimentally examined for in vivo editing.
Table 4.
Effects of the nucleotide complementary to the UAG editing site
| The complementary nucleotide |
||||||
|---|---|---|---|---|---|---|
| C | U | G | A | None | Total | |
| Edited/total (%) | 15/27 (55.6) | 27/275 (9.8) | 0/11 (0.0) | 0/13 (0.0) | 1/15 (6.7) | 43/341 (12.6) |
| Mean editing efficiency (%) | 57.3 | 50.3 | – | – | 20.0 | 52.0 |
The effects of the −2 and +2 positions
We then examined the effects of base pairings at the −2 and +2 positions on editing efficiency at UAG triplets that already had Watson–Crick base pairings at the −1 and +1 sites (Table 5). When both the −2 and the +2 positions were stabilized by Watson–Crick base pairings, editing efficiency was 29.0%. In contrast, the replacement of even a single Watson–Crick base pairing with a U-G wobble base pairing or a mismatched base pairing reduced the overall editing efficiency to 10.3 and 8.0%, respectively, indicating the importance of the stable dsRNA structure locked by Watson–Crick base pairings not only at the −1 and +1 positions but also at the −2 and +2 sites (Tables 2, 3 and 5). We also examined the influence of the −3 and +3 positions, which had no significant effects on the editing efficiency (data not shown).
Table 5.
The effects of the basepairing at the −2 and +2 positions
| W–C group | G-U group | Mismatch group | Total | |
|---|---|---|---|---|
| UAG sites examined | 93 | 29 | 50 | 172 |
| Edited at UAG sites | 27 | 3 | 4 | 34 |
| Mean editing efficiency (%) | 29.0 | 10.3 | 8.0 | 19.8 |
Effects of a specific nucleotide at the −2 and +2 positions on the editing efficiency were also investigated. No significant effects were detected for the editing frequency of UAG triplets with regard to the nucleotide at the −2 position, although the average editing efficiency was slightly higher for the triplets with A or G compared to those U or C at the −2 position (Table 6). In contrast, the presence of the adenosine residue at the +2 position resulted in the highest rate of the UAG triplet editing (59.3%), followed by guanosine (20.0%) or cytidine (21.4%), and uridine (11.1%) (Table 7). The +2 nucleotide appeared to influence also the mean editing frequency; adenosine the highest (69.1%) followed by uridine (50.0%), guanosine (24.0%) and cytidine (10.0%) (Table 7).
Table 6.
The effect of the nucleotide −2 position
| The nucleotide at −2 position |
|||||
|---|---|---|---|---|---|
| A | G | U | C | Total | |
| Editing frequency, edited/total (%) | 10/27 (37.0) | 7/21 (33.3) | 4/16 (25.0) | 6/29 (20.7) | 27/93 (29.0) |
| Mean editing efficiency (%) | 69.5 | 58.6 | 30.0 | 30.0 | 52.0 |
Table 7.
The effect of the nucleotide +2 position
| The nucleotide at +2 position |
|||||
|---|---|---|---|---|---|
| A | G | U | C | Total | |
| Editing frequency, edited/total (%) | 16/27 (59.3) | 5/25 (20.0) | 3/27 (11.1) | 3/14 (21.4) | 27/93 (29.0) |
| Mean editing efficiency (%) | 69.1 | 24.0 | 50.0 | 10.0 | 52.0 |
Together, our statistical analysis revealed the most preferred sequence and structure of the UAG triplet editing site (Figure 3). The adenosine residue of the UAG triplet with a partner nucleotide of cytidine stably locked with four Watson–Crick base pairings and the presence of adenosine at the +2 position appears to be the most frequently and highly edited site of pri-miRNAs in human brain.
Non-UAG editing sites
In this study, 43 non-UAG triplet editing sites were also identified in 209 pri-miRNAs examined (Table 8 and Table S4). Among 43 non-UAG triplets, 32 sites had Watson–Crick base pairings at both the −1 and the +1 sites, indicating once again the importance of the stable structure surrounding the adenosine residue to be edited (Table S4). The presence of cytidine in the opposite strand resulted in most frequent editing (10/137 sites, 7.3%) in comparison to the presence of other partner nucleotides, e.g. uridine (32/2273 sites, 1.4%), indicating that mismatched A·C pairs are also the most favored editing sites even among non-UAG triplets (Table 9). AAG, AAA, UAU, UAC and UAA were frequently edited. In contrast, editing of triplets having guanosine as the 5′ neighbor, e.g. GAU, was extremely rare. The preference of 5′-A and U neighbors and disfavor of 5′-G for A→I editing by ADAR1 and ADAR2 was first reported through studies on in vitro editing of a synthetic long dsRNA substrate (59). Although these non-UAG triplet editing sites identified were in some cases highly edited, 10 AAG editing sites out of a total of 231 AAG triplets present in 209 pri-miRNAs, among the most frequently edited non-UAG triplets, form a marked contrast to 43 UAG triplet sites (Tables 4 and 9). The 209 pri-miRNAs were selected unbiased for non-UAG triplet sites. According to non-UAG triplets located within the dsRNA region of 209 pri-miRNAs (examined), we anticipate proportionally that an additional 11 AAG triplet editing sites might be identified among 253 AAG sites (data not shown) present in 217 pri-miRNAs not examined in this study, or a total of 21 AAG editing sites, far less than the 43 UAG triplets. Thus, the results of our survey indicate that the UAG triplet is likely the most favored triplet of pri-miRNAs for A→I editing. A statistical analysis of a large number of editing sites identified in Alu and mouse SINE repeat sequences also revealed the preference of U and C as the 5′-nearest neighbor and G preference for the 3′-nearest neighbor and favored UAG triplet sequences (25–27,51). In vitro and in vivo editing analysis of 16 sites located within intron 4 of ADAR2 pre-mRNAs had previously revealed the 5′-nearest neighbor preference (U = A>G) (58), again consistent with our analysis of pri-miRNA editing sites.
Editing site location and ADAR enzymes responsible for editing of UAG triplets
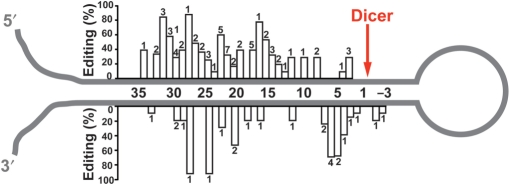
We investigated whether the editing frequency of each site correlates with the distance from the end loop structure. The size and the structure of the loop varied significantly, relative to the position of the editing site. Therefore, the distance from the Dicer cleavage site and editing frequency of 86 editing sites (UAG and non-UAG triplets) were plotted, revealing that the pri-miRNA editing sites were more often found on the sense strand (5p strand) than the antisense strand (3p strand) and more distal to the Dicer cleavage site. The region near the end loop appeared to be devoid of editing (Figure 4). This trend of preference of 5p strand and more distal to the Dicer cleavage site was noted also for analysis of UAG triplet editing sites (Figure 3) (data not shown).
Figure 4.
Summary of UAG and non-UAG triplet editing sites identified in pri-miRNA hairpin structures. The mean editing efficiency of 86 UAG and non-UAG triplets is plotted against the distance relative to the Dicer cleavage site (position 0). The number of pri-miRNAs edited at the specific site is also indicated for each bar.
In order to determine the ADAR enzyme responsible for editing of a specific site, we examined mouse homologues of human pri-miRNAs in wild-type and mutant mice with inactivated ADAR1 or ADAR2 genes (Table 10). The editing frequency of these conserved pri-miRNAs was determined with the total RNAs extracted from ADAR1−/− embryos or ADAR2−/− mouse brains (Table 10). ADAR2−/− mice are viable (14), whereas ADAR1−/− mouse embryos die at the embryonic day 12 (E12) (13,15). We used ADAR1−/− and wild-type embryos collected at E11, whereas adult brains of wild-type and ADAR2−/− mice were utilized. We confirmed that pri-miR-99b, -411 and -423 are edited by ADAR1, as we reported previously for miR-376b and miR-151 (31,32), since editing efficiency of these two pri-miRNAs remained the same in the wild-type and ADAR2−/− mouse brains as well as wild-type embryos but lost in the ADAR1−/− embryos. ADAR2 appeared to be responsible for editing of pri-let-7g, pri-miR-27a, -99a, -203 and -379 RNAs, as we previously reported for miR-376a (32), because editing of these sites is present in wild-type and ADAR1−/− embryos as well as wild-type mouse brains, but lost in ADAR2−/− mouse brains (Table 10). No specific relationship for the responsible ADAR enzyme and the position of the editing site relative to the end loop or Drosha and Dicer cleavage sites was detected (data not shown).
Table 10.
Conservation of miRNA editing between human and mouse
| Location in humana | Human (%) | Mouse (%) | Responsible ADARs | ||
|---|---|---|---|---|---|
| let-7g | 11, UAG | 10 | 0 | – | |
| let-7g | 14, UAG | 30 | 20 | ADAR2 | |
| miR-7-2 | 41, UAG | 10 | 0 | – | |
| miR-27a | 4, GAG | 30 | 0 | – | |
| miR-27a | 10, CAG | 50 | 20 | ADAR2 | |
| miR-27a | 17, UAG | 10 | 0 | – | |
| miR-33 | 15, UAG | 30 | 0 | – | |
| miR-99a | 13, AAA | 20 | 20 | ADAR2 | |
| miR-99b | 44, CAC | 10 | 10 | ADAR1 | |
| miR-99b | 47, AAG | 50 | 10 | ADAR1 | |
| miR-151 | 49, UAG | 40 | 30 | ADAR1 | |
| miR-153 | 60, UAG; 59, UAG | 30, 10 | 0 | – hsa-miR-153-1, -153-2 | |
| miR-203 | 85, UAG | 60 | 30 | ADAR2 | |
| miR-376a | 9, UAG; 15, UAG | 50, 90 | 50 | ADAR2 hsa-miR-376a1, -376a2 | |
| miR-376a | 49, UAG; 55, UAG | 40, 100 | 0 | – hsa-miR-376a1, -376a2 | |
| miR-376b | 67, UAG | 95 | 50 | ADAR1 | |
| miR-379 | 10, UAG | 60 | 20 | ADAR2 | |
| miR-411 | 20, UAG | 80 | 60 | ADAR1 | |
| miR-423 | 13, UAG | 40 | 20 | ADAR1 | |
| miR-532 | 34, UAG | 10 | 0 | – | |
| miR-652 | 11, UAU | 40 | 0 | – | |
aThe 5′ end of the pri-miRNA sequence, registered at the Sanger Center miRbase, is counted as 1.
During comparison of conserved sets of human and mouse pri-miRNAs, we noted that the editing frequency of mouse pri-miRNAs is often lower than that of the corresponding human pri-miRNA (Table 10). It has been reported that A→I editing of non-coding RNAs consisting of inverted repeats of human Alu repetitive sequences is at least an order of magnitude higher compared with SINE repeats (mouse equivalent of Alu) (27,60). Furthermore, a 4-fold lower frequency of A→I editing of mature miRNAs in mouse compared to human was recently reported (33). In contrast, certain editing sites of protein coding genes, e.g. GluR-B Q/R site, are highly edited in both human and mouse (61). Together, the overall A→I RNA editing frequency of non-coding RNAs, i.e. repetitive RNAs and pri-miRNAs, is set higher in human compared to mouse by a currently unknown mechanism.
Frequency of pri-miRNA editing
A small-scale survey for A→I editing of pri-miRNAs in human brain was conducted before, revealing 6 out of 99 pri-miRNAs examined and thus the estimate of 6% of pri-miRNAs to be subject to editing (30). In this study, we noted that many pri-miRNAs reported previously not subject to A→I RNA editing (30) were determined to be in fact highly edited, e.g. pri-miR-368, pri-miR-376a-2 and pri-miR-411. This is likely due to the relatively low frequency of successful amplification of pri-miRNA sequences (99 pri-miRNAs amplified from 231 investigated) in the previous study (30). For analysis of UAG triplet editing sites, we preselected 257 out of 474 human pri-miRNAs (miRBase 9.1) that contained 341 UAG triplets in the dsRNA region. In this study, 209 pri-miRNAs out of the 257 pri-miRNAs were successfully RT–PCR amplified from human brain RNAs, revealing 47 pri-miRNAs that indeed contained UAG and/or non-UAG editing sites. Among the 47 pri-miRNAs, 20 pri-miRNAs contained non-UAG editing sites regardless of the presence or absence of UAG editing sites (Table S4). Although 209 pri-miRNAs were selected for those containing UAG triplets, they were unbiased for non-UAG triplets. Therefore, we proportionally anticipate an additional 21 pri-miRNAs to contain the non-UAG triplet site in 217 pri-miRNAs excluded in this study. Together, we expect 68 (47 + 21) out of 426 (209 + 217) pri-miRNAs, or ∼16% of all human pri-miRNAs to contain UAG and/or non-UAG editing sites. Our updated estimate of ∼16% certainly changes the global impact of A→I editing on the miRNA-based silencing of mammalian genes.
CONCLUSION
In this study, we examined 209 pri-miRNAs out of 474 pri-miRNAs registered at the Sanger Center miRBase site (9.1) and identified 43 UAG triplet and 43 non-UAG triplet editing sites in 47 pri-miRNAs that are highly edited in human brain. We expect that ∼16% of human pri-miRNAs contain UAG and/or non-UAG triplets that undergo A→I RNA editing. We show that editing and consequent alterations of the dsRNA structure most frequently affects the processing of pri-miRNAs to functional mature miRNAs. However, four edited pri-miRNAs were processed to generate edited mature miRNAs with altered sequence at high levels. Their predicted target genes are likely to be different from those targeted by unedited mature miRNAs. Our study suggests that A→I editing of mammalian pri-miRNAs has a large impact on processing and function of miRNAs and thus affects expression (silencing) of many genes on a global scale.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank M. Higuchi and P.H. Seeburg for ADAR2−/− mice and J.M. Murray and Boris Zinshteyn for reading and comments. We also thank the Wistar Animal, Protein Expression and Mouse Genetics Core facilities for their services. This work was supported by grants from the National Institutes of Health (CA72764, GM-40536, GM079091 and P30 CA10815), the Juvenile Diabetes Research Foundation (1-2006-210) and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health. Funds received to L.V. are the National Institutes of Health Postdoctoral Supplemental Award (L070045) and the NCI National Institutes of Health Postdoctoral Training Grant (CA09171). Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jepson JE, Reenan RA. RNA editing in regulating gene expression in the brain. Biochim. 2007 doi: 10.1016/j.bbagrm.2007.11.009. Biophys. Acta, December 3 [Epub ahead of print 10.1016/j.bbagrm.2007.1011.1009] [DOI] [PubMed] [Google Scholar]
- 3.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerber A, O’Connell MA, Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 6.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl Acad. Sci. USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai F, Chen CX, Carter KC, Nishikura K. Editing of glutamate receptor B subunit ion channel RNAs by four alternatively spliced DRADA2 double-stranded RNA adenosine deaminases. Mol. Cell. Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 9.Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 10.O’Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol. Cell. Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 12.Dabiri GA, Lai F, Drakas RA, Nishikura K. Editing of the GLuR-B ion channel RNA in vitro by recombinant double-stranded RNA adenosine deaminase. EMBO J. 1996;15:34–45. [PMC free article] [PubMed] [Google Scholar]
- 13.Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 14.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol. Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 16.Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- 17.Ohlson J, Pedersen JS, Haussler D, Ohman M. Editing modifies the GABA(A) receptor subunit alpha3. RNA. 2007;13:698–703. doi: 10.1261/rna.349107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rula EY, Lagrange AH, Jacobs MM, Hu N, Macdonald RL, Emeson RB. Developmental modulation of GABA(A) receptor function by RNA editing. J. Neurosci. 2008;28:6196–6201. doi: 10.1523/JNEUROSCI.0443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 20.Dracheva S, Patel N, Woo DA, Marcus SM, Siever LJ, Haroutunian V. Increased serotonin 2C receptor mRNA editing: a possible risk factor for suicide. Mol. Psychiatr. 2007 doi: 10.1038/sj.mp.4002081. September 11 [Epub ahead of print 10.1038/sj.mp.4002081] [DOI] [PubMed] [Google Scholar]
- 21.Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- 22.Niswender CM, Herrick-Davis K, Dilley GE, Meltzer HY, Overholser JC, Stockmeier CA, Emeson RB, Sanders-Bush E. RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology. 2001;24:478–491. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 23.Sodhi MS, Burnet PW, Makoff AJ, Kerwin RW, Harrison PJ. RNA editing of the 5-HT(2C) receptor is reduced in schizophrenia. Mol. Psychiatry. 2001;6:373–379. doi: 10.1038/sj.mp.4000920. [DOI] [PubMed] [Google Scholar]
- 24.Miyamura Y, Suzuki T, Kono M, Inagaki K, Ito S, Suzuki N, Tomita Y. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am. J. Hum. Genet. 2003;73:693–699. doi: 10.1086/378209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 29.Nishikura K. Editing the message from A to I. Nat. Biotechnol. 2004;22:962–963. doi: 10.1038/nbt0804-962. [DOI] [PubMed] [Google Scholar]
- 30.Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of microRNA-151 blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 37.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 38.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 39.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 42.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 45.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 46.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 47.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piskounova E, Viswanathan SR, Janas M, Lapierre RJ, Daley GQ, Sliz P, Gregory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008 doi: 10.1074/jbc.C800108200. June 12 [Epub ahead of print 10.1074/jbc.C800108200] [DOI] [PubMed] [Google Scholar]
- 49.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riedmann EM, Schopoff S, Hartner JC, Jantsch MF. Specificity of ADAR-mediated RNA editing in newly identified targets. RNA. 2008;14:1110–1118. doi: 10.1261/rna.923308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl Acad. Sci. USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 56.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 57.Wong SK, Sato S, Lazinski DW. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–858. doi: 10.1017/s135583820101007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dawson TR, Sansam CL, Emeson RB. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J. Biol. Chem. 2004;279:4941–4951. doi: 10.1074/jbc.M310068200. [DOI] [PubMed] [Google Scholar]
- 59.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 60.Eisenberg E, Nemzer S, Kinar Y, Sorek R, Rechavi G, Levanon EY. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 61.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.