Abstract
The replication factors Cdt1 and Cdc6 are essential for origin licensing, a prerequisite for DNA replication initiation. Mechanisms to ensure that metazoan origins initiate once per cell cycle include degradation of Cdt1 during S phase and inhibition of Cdt1 by the geminin protein. Geminin depletion or overexpression of Cdt1 or Cdc6 in human cells causes rereplication, a form of endogenous DNA damage. Rereplication induced by these manipulations is however uneven and incomplete, suggesting that one or more mechanisms restrain rereplication once it begins. We find that both Cdt1 and Cdc6 are degraded in geminin-depleted cells. We further show that Cdt1 degradation in cells that have rereplicated requires the PCNA binding site of Cdt1 and the Cul4DDB1 ubiquitin ligase, and Cdt1 can induce its own degradation when overproduced. Cdc6 degradation in geminin-depleted cells requires Huwe1, the ubiquitin ligase that regulates Cdc6 after DNA damage. Moreover, perturbations that specifically disrupt Cdt1 and Cdc6 degradation in response to DNA damage exacerbate rereplication when combined with geminin depletion, and this enhanced rereplication occurs in both human cells and in Drosophila melanogaster cells. We conclude that rereplication-associated DNA damage triggers Cdt1 and Cdc6 ubiquitination and destruction, and propose that this pathway represents an evolutionarily conserved mechanism that minimizes the extent of rereplication.
One of the critical events in the cell division cycle is complete and precise duplication of the genome. In eukaryotic cells, origins of DNA replication acquire replication competence through the assembly of a prereplication complex (preRC)3 in the G1 phase of the cell cycle. PreRCs are assembled by the sequential origin binding of the origin recognition complex (ORC), Cdc6, Cdt1, and the minichromosome maintenance complex (MCM). Origins harboring preRCs are licensed for replication but do not initiate DNA synthesis until S phase begins and the Cdc7 and Cdk2 kinases are activated (1–3). The large genomes of metazoan cells necessitate the utilization of thousands of origins, but each origin that initiates DNA synthesis must do so only once. Failure to maintain this control has been linked to genome instability and oncogenesis (4, 5), but the cellular consequences of rereplication are not fully understood.
Multiple regulatory mechanisms operate to ensure that any origins that have “fired” do not fire again by blocking preRC assembly after the G1 to S phase transition. Among the most important of these mechanisms are degradation of Cdt1 during S phase and inhibition of Cdt1 by the geminin protein. Geminin depletion, overexpression of Cdt1, or overexpression of Cdc6 causes rereplication, which ultimately triggers a DNA damage response (6–11) and can promote tumorigenesis (4, 5). Rereplication induced by these manipulations is however incomplete, with the extent of rereplication varying widely among different cell lines. These observations suggest that in addition to the mechanisms that block rereplication in the first place, multiple events restrain rereplication once it begins (10, 12–17).
Previous investigators have noted that human or Drosophila cells depleted of geminin also become depleted of Cdt1 (14, 18), but the mechanism of that regulation has not been determined. Moreover, the effects of rereplication on the Cdc6 protein have not yet been explored. We hypothesized that DNA damage caused by rereplication is responsible for the observed Cdt1 degradation, and that a similar effect should result in degradation of Cdc6. In this study we provide evidence that rereplication-induced degradation of both Cdt1 and Cdc6 requires the same ubiquitin ligases that regulate Cdt1 and Cdc6 in response to exogenous DNA-damaging agents. Moreover, Cdt1 overexpression stimulates its own degradation by inducing rereplication-associated DNA damage. We further demonstrate that disrupting the degradation of either Cdt1 or Cdc6 combined with geminin depletion exacerbates rereplication. This study provides evidence for an evolutionarily conserved mechanism, which destroys essential replication licensing factors once rereplication begins as an important means to minimize the extent of rereplication.
EXPERIMENTAL PROCEDURES
Growth and Manipulation of Cells—HCT116 cells were cultured in McCoy's 5A medium (Cellgro, Mediatech) supplemented with 10% fetal bovine serum (Sigma), and 2 mm glutamine (Invitrogen). NHF1-hTert cells (NHF1) were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, and 2 mm glutamine. Cells were transfected at a final concentration of 100 nm siRNA using Dharmafect Reagent 1 (Dharmacon). siRNA sequences targeting geminin (18), DDB1 (19), Huwe1, or green fluorescent protein (as a control) (20) have been described. Drosophila S2 cells were grown in F-900 II SFM serum-free medium (Invitrogen) and treated with 20 μg of dsRNA/ml. Primers for generating dsRNA: cul-1 5′-CTGCTCAACGCAGACCG and 5′-TGTCCTGCAGTTGCTGG, cul-4 5′-TTGGCCAAACGATTACTTGTGGG and 5′-GAGAAGATTATGGCTCAGCG, geminin 5′-ATGTCTTCGAGCGCTGCC and 5′-GGCGTTGACCTTGTCCTCG, Cdt1Dup 5′-ACAAACCGCAAACGCGCCG and 5′-CCAGCACTGCCTTGAGTTCC, control (pBluescript SK sequence) 5′-ATGGATAAGTTGTCGATCG and 5′-ACCAGGTTCACATGCTTGCG.
The HA2-Cdt1 adenovirus has been described (21). C-terminally tagged versions of Cdt1 were constructed by eliminating the Cdt1 stop codon in pENTR-Cdt1 (21) followed by Clonase II recombination with pAD/CMV/V5-DEST or pcDNA-DEST40 according to the manufacturer's instructions (Invitrogen). Cdt1 was truncated after amino acid 321 using a naturally occurring NcoI site; the PIPm mutation was generated in a PCR-based strategy. Plasmids were introduced into HCT116 cells by polyethylenimine transfection.
Cell Cycle Analysis—Human cells were collected and processed for flow cytometric analysis by ethanol fixation and propidium iodide staining. Nuclei were analyzed using a CyAn FACScan (DakoCytomation), and cell cycle distributions were plotted with Summit v4.3 software (DakoCytomation). Drosophila cells were transfected then plated in concanavalin A-coated 24-well glass bottom dishes (MatTek) for 1 h prior to fixation as described (22), stained with DAPI at 5 μg/ml, and scanned with either an IC100 Image Cytometer (Beckman Coulter) or an Array Scan VTI (Cellomics) equipped with a 20× 0.5 NA objective and an ORCA-ER cooled CCD camera. Images of ∼5,000 cells per well were acquired and analyzed using CytoShop v2.1 (Beckman-Coulter) or vHCS View (Cellomics). Integrated fluorescence intensity measurements were determined from unsaturated images. p values were determined using an unpaired Student's t test.
Immunoblotting—Anti-Cdt1 is described in Cook et al. (23); anti-geminin (FL-209), and anti-HA (Y-11) were purchased from Santa Cruz Biotechnology. Cul4 antibody was raised in rabbits against the peptide sequence MSAAKKYKPMDTTELHEN (Pocono Farms). Antibodies to p53 phosphorylated at Ser-15 and Chk2 phosphorylated at Thr-68 were purchased from Cell Signaling Technologies. Anti-V5 antibody was purchased from Invitrogen. Antibody to Drosophila Cul1 (ZL18) was purchased from Zymed Laboratories Inc., and antibodies to both Drosophila and human tubulin were purchased from Sigma. Cdt1Dup antibody was a gift from T. Orr-Weaver, anti-Huwe1 was a gift from S. Wing, and anti-DDB1 was a gift from Y. Xiong. Co-immunoprecipitations were performed essentially as described in Ref. 20, except that whole cell lysates rather than nuclear lysates were used as the starting material.
RESULTS
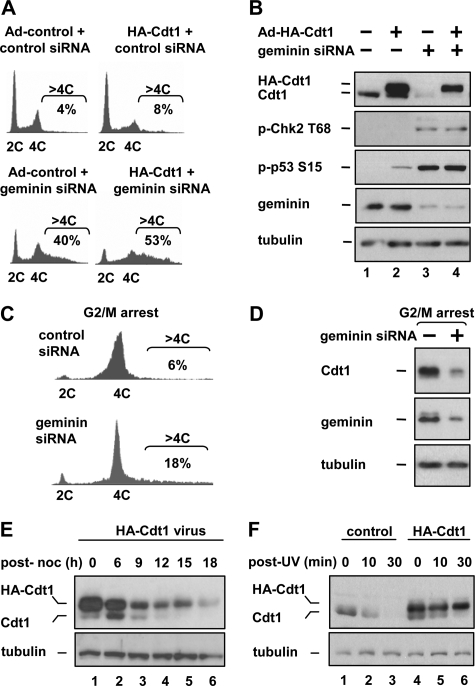
Rereplication Induces the Degradation of Cdt1 in Human Cells—We and others have previously observed that rereplication induced by RNAi-mediated geminin depletion is incomplete. To explore the mechanisms that restrain rereplication in these cells, we first manipulated the expression of both geminin and Cdt1 in HCT116 colon carcinoma cells and analyzed the cells by flow cytometry to determine DNA content. Depletion of geminin by siRNA transfection for 48 h resulted in a population of cells with a heterogeneous DNA content greater than 4C, which we defined as cells that had undergone rereplication (Fig. 1A, left histograms). Rereplication can also be induced by overproduction of Cdt1 to levels that overwhelm the ability of geminin to inhibit Cdt1 activity (10). Infecting cells with a recombinant adenovirus that directs moderate overexpression of an N-terminally HA-tagged Cdt1 induced rereplication after 24 h, although not as extensively as geminin siRNA transfection (Fig. 1A, top right histogram). Combining Cdt1 expression with geminin depletion induced more extensive rereplication than either single treatment, as determined by the number of cells with DNA content greater than 4C (Fig. 1A, bottom right histogram). Thus, geminin depletion and Cdt1 expression cooperate to induce rereplication.
FIGURE 1.
Cdt1, but not N-terminally tagged Cdt1, is degraded in geminin-depleted cells. A, HCT116 cells were transfected with geminin siRNA or a control sequence for 24 h. Cells were then infected with recombinant adenovirus expressing HA-Cdt1 or control virus at a multiplicity of infection (m.o.i.) of 200 for an additional 24 h. Cells were fixed, stained with propidium iodide, and analyzed for DNA content by flow cytometry. The percentage of the cell population harboring DNA content greater than 4C is reported under the brackets for each plot. B, portions of the cells from A were assayed by immunoblotting with the indicated antibodies. Both ectopic and endogenous Cdt1 were detected with anti-Cdt1 antibody; phosphorylation of Chk2 at Thr-68 and p53 at Ser-15 were detected with phosphospecific antibodies. C, HCT116 cells transfected with geminin siRNA for 24 h were treated with 100 ng/μl nocodazole for an additional 24 h to arrest them in prometaphase (G2/M) and evaluated for DNA content by flow cytometry. D, portions of the cells from C were assayed by immunoblotting with the indicated antibodies. E, HCT116 cells were infected with recombinant adenovirus as in A. Cells were then treated with 100 ng/μl nocodazole for 18 h to arrest cells at G2/M. Arrested cells were collected by “mitotic shake-off,” re-plated in nocodazole-free medium and collected at the indicated times after release. Endogenous (lower band) and HA-Cdt1 (upper band) were assayed by immunoblotting with anti-Cdt1 antibody. F, HCT116 cells were infected with recombinant adenovirus as in A for 24 h. Cells were then treated with 20 J/m2 UV and collected at the indicated times post-irradiation and assayed by immunoblotting with anti-Cdt1 and tubulin antibodies.
Recent evidence suggests that the forks derived from refiring origins collapse to generate double-strand breaks that can trigger activation of the DNA damage checkpoint (24, 25). As had been observed by others (12, 13, 26), geminin-depleted cells acquired molecular markers associated with DNA damage, including phosphorylation of both p53 and Chk2 (Fig. 1B compare lanes 1 and 3). Cdt1 overproduction also resulted in p53 phosphorylation (Fig. 1B, lane 2). Seemingly paradoxically and similar to previous observations (14, 18), endogenous Cdt1 levels were quite low in geminin-depleted cells despite the fact that Cdt1 is required for replication (Fig. 1B top, compare lanes 1 and 3).
Cdt1 is most abundant in G1 cells, and geminin depletion results in fewer G1 cells (Fig. 1A). To determine if the down-regulation of Cdt1 in geminin-depleted cells could be explained simply by this cell cycle effect, we treated cells with nocodazole after siRNA transfection to arrest them in G2/prometaphase. Geminin-depleted G2/M cells rereplicated as before (Fig. 1C), but also had dramatically less Cdt1 than control G2/M cells (Fig. 1D). A similar observation was made in G2-arrested cells by Ballabeni et al. (18), and the loss of Cdt1 was attributed to ubiquitin-mediated degradation. Geminin binding to Cdt1 may physically block interaction with a Cdt1 ubiquitin ligase, or geminin may regulate Cdt1 stability by some replication-independent mechanism. An alternative possibility we considered is that the DNA damage induced by rereplication results in Cdt1 degradation.
Surprisingly, the ectopically expressed N-terminally tagged HA-Cdt1 protein was largely resistant to degradation in geminin-depleted cells, whereas the endogenous Cdt1 was readily degraded (Fig. 1B compare lanes 2 and 4). Persistence of HA-Cdt1 may have contributed to the increased rereplication we observed when HA-Cdt1 was expressed in geminin-depleted cells (Fig. 1A). The resistance of N-terminally tagged Cdt1 to degradation in geminin-depleted cells suggested that the Cdt1 N terminus is important for regulating Cdt1 stability in rereplicating cells. In S phase, Cdt1 ubiquitination and destruction is mediated by two ubiquitin ligase complexes, Cul1Skp2 and Cul4DDB1 (27). Cul4DDB1 is also responsible for Cdt1 ubiquitination in response to DNA damage (28, 29). Cul1Skp2-mediated ubiquitination requires phosphorylation of human Cdt1 on T29, whereas DNA damage-induced ubiquitination of Cdt1 requires Cdt1 binding to PCNA (proliferating cell nuclear antigen), which also binds close to the Cdt1 N terminus (27, 30, 31). We hypothesized that N-terminally tagged Cdt1 was resistant to degradation in geminin-depleted cells because one or both of these two ubiquitin ligases could not access Cdt1. To determine which mode of Cdt1 degradation was blocked by the HA tag, we monitored the stability of HA-Cdt1 as cells progressed through the cycle or in response to DNA damage. Upon entry into S phase the HA-Cdt1 protein was degraded at the same cell cycle point as endogenous Cdt1 (Fig. 1E). The fact that HA-Cdt1 is sensitive to cell cycle-dependent degradation but not degradation in geminin-depleted cells (Fig. 1B) further demonstrates that the Cdt1 degradation in geminin-depleted cells is not due to a passive cell cycle effect. Unlike endogenous Cdt1, HA-Cdt1 was not degraded in response to ultraviolet (UV) irradiation (Fig. 1F); a similar observation had been made with N-terminally Myc-tagged Cdt1 in UV-treated cells (30). These results suggest that the degradation of Cdt1 in geminin-depleted cells is accomplished by a mechanism similar to Cdt1 degradation in response to DNA damage.
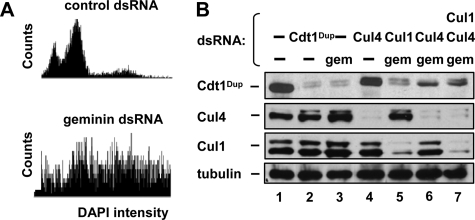
Drosophila Cdt1Dup Is Degraded in Geminin-depleted Cells through a Cul4-dependent Mechanism—To determine if loss of Cdt1 in response to rereplication is conserved in other species we examined the effects of geminin depletion in Drosophila-melanogaster cells. Transfection of cultured Drosophila S2 cells with dsRNA targeting geminin for 96 h resulted in a dramatic increase in the number of cells with greater than 4C DNA content. In this experiment, DNA content per cell was assessed by a novel microscopy-based method to measure DAPI-staining intensity using an IC100 Image Cytometer (Fig. 2A). Geminin depletion resulted in the degradation of the Cdt1 ortholog double-parked (Cdt1Dup) as first reported by Mihaylov et al. (Fig. 2B, lane 3) (14). To determine which ubiquitin ligase mediates Cdt1Dup degradation, we co-transfected dsRNA targeting either Cul1 or Cul4 with the geminin dsRNA. When S2 cells were depleted of Cul4 and geminin, Cdt1Dup was partially stabilized compared with geminin depletion alone (Fig. 2B, compare lanes 3 and 6). The inability of Cul4 depletion to completely rescue Cdt1Dup levels may have been due to the reported down-regulation of Cdt1Dup mRNA in geminin-depleted S2 cells (14). Depletion of Cul1 had little effect on Cdt1Dup levels in geminin-depleted cells (Fig. 2B, compare lanes 3 and 5). Triple depletion of geminin, Cul1, and Cul4 resulted in Cdt1Dup levels close to the levels in geminin and Cul4-depleted cells (Fig. 2B, lane 7), indicating that Cul4 is more important than Cul1 for regulating Cdt1Dup under these conditions. Interestingly, the small amount of Cdt1Dup that accumulated in the Cul1-depleted cells had a slower mobility than the Cdt1Dup in Cul4-depleted cells, suggesting that Cdt1Dup may have been differentially modified, perhaps by phosphorylation (43).
FIGURE 2.
Geminin depletion in Drosophila S2 cells induces Cul4-dependent Cdt1Dup degradation. A, S2 cells were treated with dsRNA targeting control sequence or geminin for 4 days. DNA content was determined by integrated fluorescence intensity of DAPI-stained nuclei. B, S2 cells were transfected with the indicated dsRNA for 96 h then evaluated for Cdt1Dup, Cul4, Cul1, and tubulin by immunoblotting.
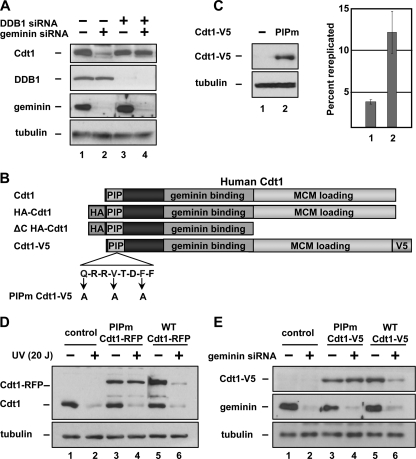
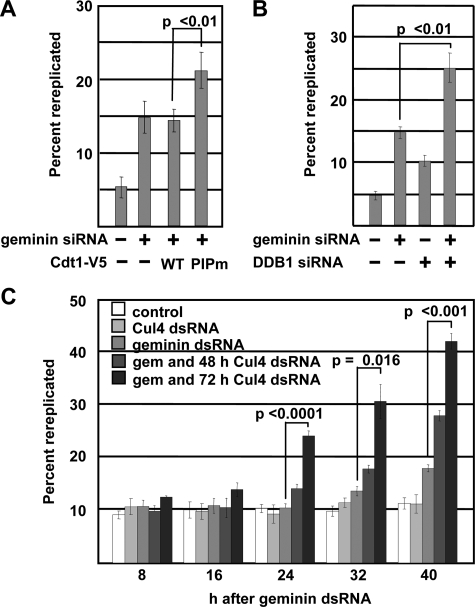
The Rereplication-associated Degradation of Human Cdt1 Requires the PCNA Binding Site and Cul4DDB1—To determine if Cul4 is similarly required for degradation of human Cdt1 in rereplicating cells, we co-transfected HCT116 cells with siRNA targeting both geminin and the Cul4 adaptor DDB1. As in Drosophila cells depleted of Cul4, depletion of DDB1 prevented the Cdt1 degradation induced by geminin depletion (Fig. 3A, compare lanes 1 and 2 with lanes 3 and 4). Because Cul4DDB1 is required for Cdt1 degradation both after DNA damage and after geminin depletion, and because rereplication induces DNA damage, we tested other aspects of DNA damage-induced Cdt1 degradation for similar involvement in Cdt1 degradation in geminin-depleted cells. DNA damage-induced degradation of Cdt1 requires an association between the Cdt1 PIP (PCNA-interacting protein) motif and PCNA. We therefore asked whether this association is also required for Cdt1 degradation in geminin-depleted cells. Conserved PIP motif residues at positions 3, 6, and 9 were altered to alanines (PIPm-Cdt1, Fig. 3B); similar mutations are sufficient to disrupt PCNA binding and inhibit Cdt1 degradation after UV damage (30, 31) (including the single mutation F9A in human Cdt1 (30)). A V5 epitope tag (or alternatively an RFP tag) was then inserted at the C terminus of both normal and PIPm-Cdt1. This mutation did not block the ability of Cdt1 to induce rereplication (Fig. 3C). A similar PIPm-Cdt1 (31) and even truncation of the first 242 Cdt1 residues (32) complemented Cdt1-depleted Xenopus laevis extracts for MCM loading and DNA replication, indicating that PIPm-Cdt1 retains both replication and rereplication-inducing functions.
FIGURE 3.
Rereplication-induced human Cdt1 degradation requires DDB1 and the PIP motif of Cdt1. A, HCT116 cells were transfected with DDB1 siRNA for 72 h and geminin siRNA for 48 h as indicated. Proteins in whole cell lysates were detected by immunoblotting with the indicated antibodies. B, schematic diagram of full-length Cdt1 and derivatives used in this study. C, HCT116 cells were infected with adenovirus expressing PIPm Cdt1-V5 or control virus at an m.o.i of 150. 48 h after infection, cells were harvested and probed for ectopic Cdt1 with anti-V5 antibody or for rereplication by flow cytometry. D, HCT116 cells were transfected with plasmids encoding C-terminally RFP-tagged Cdt1, Cdt1-RFP PIPm or control vectors. 24 h after transfection, cells were irradiated as in 1F and collected 30-min post-irradiation. Endogenous and Cdt1-RFP were detected by immunoblotting with anti-Cdt1 antibody. E, HCT116 cells were infected with adenovirus expressing WT Cdt1-V5 or PIPm Cdt1-V5 at an m.o.i. of 40 4 h prior to transfection with siRNA targeting geminin or control sequence. Cells were collected 32 h after transfection and assayed by immunoblotting with anti-V5, geminin, and tubulin antibodies.
To verify that the PIPm mutation blocks Cdt1 degradation in UV-treated cells, we expressed normal Cdt1 and PIPm-Cdt1 in HCT116 cells then treated with UV. As expected, wild-type Cdt1 is readily degraded after UV treatment, but PIPm-Cdt1 is not (Fig. 3D, compare lanes 3 and 4 with lanes 5 and 6). To test if the PIP motif is required for Cdt1 degradation in geminin-depleted cells, WT Cdt1-V5 or PIPm-Cdt1-V5 were expressed from recombinant adenoviruses in HCT116 cells that had also been transfected with control or geminin siRNA. Similar to Cdt1 degradation in UV-treated cells, the PCNA binding site was required for Cdt1 degradation in cells that had been induced to rereplicate by geminin depletion (Fig. 3E, compare lanes 3 and 4 to lanes 5 and 6).
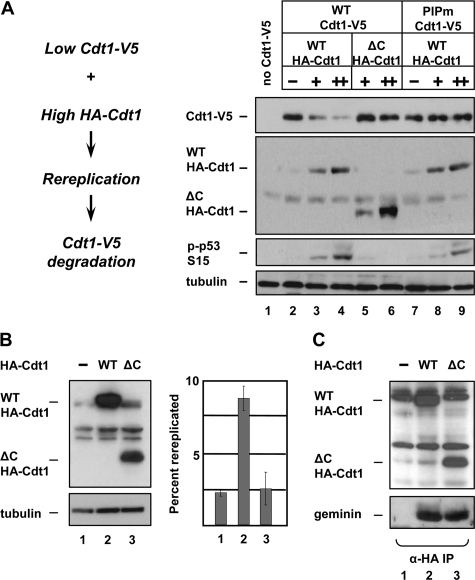
The results described above are consistent with the model in which rereplication generates DNA damage which then stimulates the PCNA- and Cul4DDB1-dependent ubiquitination and degradation of Cdt1. An alternative model is that geminin protects Cdt1 from Cul4DDB1 binding through direct competition, and in the absence of geminin Cdt1 is more accessible to Cul4DDB1. To distinguish between degradation of Cdt1 as a result of rereplication per se and degradation as a consequence of competition between geminin and Cul4 for Cdt1 binding, we designed an experiment in which rereplication was induced by overproduction of the N-terminally tagged HA-Cdt1 protein instead of geminin siRNA transfection (as in Fig. 1), and Cdt1 degradation was monitored by expression of low levels of the C-terminally tagged Cdt1-V5. With increasing amounts of HA-Cdt1, WT Cdt1-V5 was degraded (Fig. 4A, lanes 2–4), whereas PIPm-Cdt1-V5 was stable (Fig. 4A, lanes 7–9). To distinguish whether the excess HA-Cdt1 induced Cdt1-V5 degradation by inducing rereplication or by sequestering geminin from Cdt1-V5, we overproduced a form of HA-Cdt1 truncated after amino acid 321 (ΔC-HA-Cdt1, Fig. 3B). The corresponding truncation of the X. laevis Cdt1 fails to load the MCM complex or initiate replication but retains geminin binding (32). We confirmed that this mutant fails to induce rereplication (Fig. 4B) but that it binds geminin equally as well as full-length Cdt1 (Fig. 4C). HCT116 cells expressing high levels of HA-Cdt1-ΔC failed to induce either the degradation of Cdt1-V5 or the phosphorylation of p53, though it accumulated to similar levels (Fig. 4A, lanes 5 and 6). These results indicate that it is indeed the rereplication induced by geminin depletion that causes Cdt1 degradation.
FIGURE 4.
Rereplication-induced Cdt1 degradation is independent of geminin sequestration. A, HCT116 cells were infected with WT Cdt1-V5, PIPm Cdt1-V5, or control adenovirus at an m.o.i of 40 and with WT HA-Cdt1 or ΔC HA-Cdt1 at a m.o.i of 100 (+) or 300 (++) for 24 h followed by immunoblot analysis using the indicated antibodies. B, HCT116 cells were infected with control, WT HA-Cdt1, or ΔC HA-Cdt1 recombinant adenovirus at an m.o.i of 150. 24 h after infection, cells were harvested and probed for ectopic Cdt1 with anti-HA antibody or for rereplication by flow cytometry. C, HCT116 cells were infected as in B for 24 h. Cell lysates were probed for ectopic Cdt1 with anti-HA antibody (top panel) or were subjected to immunoprecipitation with anti-HA antibody and probed for endogenous geminin (bottom panel).
Degradation of Cdt1 Limits the Extent of Rereplication—We hypothesized that the early stages of rereplication might generate sufficient DNA damage to trigger Cdt1 degradation, and that the loss of Cdt1 would then inhibit further origin relicensing and rereplication. If Cdt1 degradation is an important mechanism to restrain rereplication, then manipulations that interfere with Cdt1 degradation are predicted to exacerbate rereplication. To test that prediction, we infected geminin-depleted HCT116 cells with a moderate dose of the adenoviruses producing the stabilized PIPm-Cdt1-V5 (as in Fig. 3E). Quantification of the number of cells with greater than 4C DNA content revealed that PIPm-Cdt1-V5 expression promoted significantly more rereplication than normal Cdt1 (WT) did. Normal Cdt1 was degraded in geminin-depleted cells (Fig. 3E) and thus had little additive effect on rereplication (Fig. 5A; representative histograms are provided as supplemental Fig. S1A). Furthermore, a similar significant increase in rereplication was observed when endogenous Cdt1 was stabilized by co-depletion of DDB1 with geminin (Fig. 5B and supplemental Fig. S1B). While depletion of DDB1 induced a small amount of rereplication, and this rereplication has been shown to depend on Cdt1 (33), the number of cells that had rereplicated was highest in cells transfected with both geminin and DDB1 siRNA compared with cells transfected with either siRNA alone (Fig. 5B).
FIGURE 5.
Cdt1 degradation limits the extent of rereplication. A, HCT116 cells depleted of geminin and infected with the indicated adenoviruses exactly as in Fig. 3E were evaluated for rereplication by flow cytometry as in Fig. 1A. The percentage of the total population harboring DNA content greater than 4C is reported. Representative histograms are available as supplemental Fig. S1A. B, HCT116 cells depleted of DDB1 and geminin exactly as in Fig. 3A were evaluated for rereplication by flow cytometry as in Fig. 1A. Representative histograms are available as supplemental Fig. S1B. C, Drosophila S2 cells were treated with Cul4 dsRNA every 24 h for either 48 or 72 h prior to treatment with geminin dsRNA. Samples were collected every 8 h post-geminin dsRNA treatment. DNA content was measured as in Fig. 2A, and the percentage of the total population harboring DNA content greater than 4C is reported. Error bars indicate S.D. from three independent experiments. Representative histograms are available as supplemental Fig. S1C.
We performed similar rereplication assays to determine if Cul4-dependent Cdt1Dup degradation also restricts rereplication in Drosophila S2 cells. Because depletion of the Cul4 protein takes longer than geminin depletion, S2 cells were treated with Cul4 dsRNA 48 or 72 h prior to geminin dsRNA treatment. Cells were harvested at various times after the geminin dsRNA treatment and analyzed for DNA content as in Fig. 2A. Compared with treatment with geminin dsRNA alone, the combination of Cul4 and geminin dsRNAs significantly increased the number of cells that had rereplicated, particularly when the Cul4 depletion was carried out for 72 h prior to geminin depletion (Fig. 5C and supplemental Fig. S1C). Unlike DDB1 depletion in human cells, Cul4 depletion in Drosophila S2 cells did not induce rereplication detectable by this assay, perhaps because Cul4 depletion results in a G1 arrest in S2 cells.4 Taken together, these findings suggest that rereplication in geminin-depleted cells is limited by Cdt1 degradation.
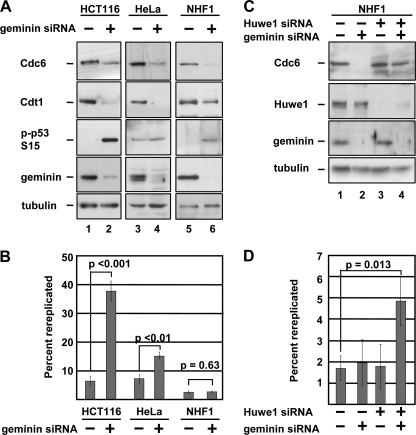
Rereplication-induced Degradation of Cdc6 Requires the Huwe1 Ubiquitin Ligase—The preceding results demonstrated that rereplication-induced Cdt1 degradation occurs by the same mechanism as DNA damage-induced Cdt1 degradation. The only other component of the preRC that has been shown to be degraded in response to DNA damage is Cdc6 (20, 34, 35). It thus seemed possible that the rereplication-induced DNA damage could also trigger Cdc6 degradation in geminin-depleted cells. To test this idea, we monitored the levels of Cdc6 in several cell lines transfected with geminin siRNA. Consistent with the presence of DNA damage as indicated by phosphorylation of p53 (Fig. 6A), Cdc6 was markedly down-regulated in geminin-depleted HCT116 cells. We made similar observations in two other cell lines, HeLa and NHF1 (36). NHF1 cells, like many other non-transformed cell lines, did not display an overt rereplication profile by flow cytometry (Fig. 6B and supplemental Fig. S2A) although geminin depletion was robust and p53 was phosphorylated (Fig. 6A, compare lanes 5 and 6).
FIGURE 6.
Geminin depletion induces Huwe1-dependent Cdc6 degradation. A, HCT116, HeLa, and NHF1 cells were transfected with geminin siRNA for 48 h and evaluated by immunoblotting with the indicated antibodies. To prevent premature mitosis, HeLa cells were treated with nocodazole for 24 h prior to harvest. B, percentage of cells from A with greater than 4C DNA content determined by flow cytometry as in Fig. 1A. Representative histograms are available as supplemental Fig. S2A. C, NHF1 cells were transfected with Huwe1 siRNA for 24 h then transfected with geminin siRNA for an additional 48 h. Cell lysates were probed with the indicated antibodies. D, percentage of cells from C with greater than 4C DNA content determined by flow cytometry as in Fig. 1A. Representative histograms are available as supplemental Fig. S2B.
DNA damage-dependent ubiquitination of Cdc6 has been attributed to two ubiquitin ligases, depending on the cell type and source of DNA damage. In p53-proficient cells treated with ionizing radiation, Cdc6 is down-regulated through ubiquitination by the Cdh1-associated form of the anaphase promoting complex, APC/C (35). In response to other forms of DNA damage, Cdc6 is ubiquitinated by the Huwe1 enzyme irrespective of the p53 status of the cells (20). Because we observed Cdc6 degradation after geminin siRNA transfection in a variety of cell lines, including HeLa cells which have severely compromised p53 expression, we hypothesized that Huwe1 is required for Cdc6 degradation in geminin-depleted cells. To test this hypothesis, we transfected NHF1 cells with siRNA targeting Huwe1 and geminin either singly or in combination, and then evaluated those cells for Cdc6 protein levels. As before, geminin depletion induced a marked down-regulation of Cdc6, but in cells co-transfected with Huwe1 siRNA, Cdc6 degradation was prevented (Fig. 6C, compare lanes 2 and 4). Strikingly, the combination of Huwe1 and geminin depletion induced overt rereplication in NHF1 cells whereas neither treatment alone was sufficient to cause DNA to accumulate to levels greater than 4C (Fig. 6D).
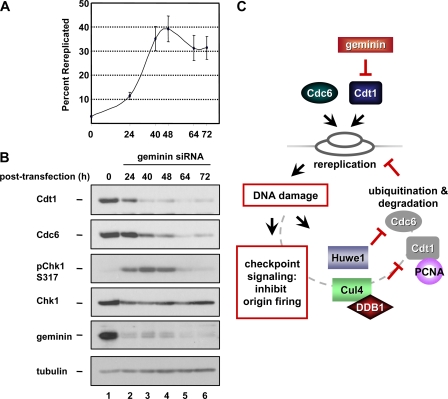
Rereplication Is Limited after Cdt1 and Cdc6 Degradation—How can a cell rereplicate when Cdt1 and Cdc6, two proteins required for replication initiation, have been degraded? Presumably, the inappropriate relicensing occurred before Cdc6 and Cdt1 were degraded. To test that notion, we transfected HCT116 cells with siRNA targeting geminin and collected cells at various times after transfection. Those cells were then evaluated for rereplication and the abundance of Cdt1 and Cdc6. Geminin was maximally depleted by 24 h after siRNA transfection, but significant degradation of Cdt1 and Cdc6 did not occur until 40 h after transfection (Fig. 7B). Rereplication was detectable by flow cytometric analysis beginning at 24-h post-transfection with the greatest number of cells harboring a DNA content of greater than 4C peaking at 48 h (Fig. 7A). The initial checkpoint response to rereplication-associated DNA damage has been linked to activation of Chk1 (15). In these time courses, we observed that the activating Ser-317 phosphorylation of Chk1 coincided with the time when Cdt1 and Cdc6 had been degraded and the number of rereplicated cells had peaked and would increase no further (Fig. 7, 40 and 48 h). At the latest time points, phospho-Chk1 had disappeared either due to signal quenching or to the induction of apoptosis, which we observed as a population with sub-G1 DNA content (data not shown). These results indicate that once rereplication has begun, Cdc6 and Cdt1 are degraded, and their loss limits the extent of rereplication even in cells that no longer contain activated Chk1.
FIGURE 7.
Rereplication is blocked after degradation of Cdt1 and Cdc6. A, HCT116 cells were transfected with geminin siRNA, harvested at the indicated times following transfection and evaluated by flow cytometry. The percentage of the population containing DNA content greater than 4C at the indicated time points from three independent experiments is plotted. B, immunoblot analysis of lysates from a representative experiment in A. C, model. Rereplication induced by either geminin depletion (or the overproduction of Cdt1 or Cdc6) results in DNA damage. The DNA damage triggers the ubiquitination of PCNA-bound Cdt1 by Cul4-DDB1 and ubiquitination of Cdc6 by Huwe1 as well as Chk1 activation. Degradation of Cdt1 and Cdc6 prevents further rounds of relicensing, and Chk1 activation inhibits origin firing, thus limiting the extent of rereplication.
DISCUSSION
In this report we provide evidence that DNA damage that results from rereplication induces the degradation of both Cdt1 and Cdc6. This phenomenon is conserved in both human and Drosophila cells with respect to Cdt1 regulation, and perturbations that interfere with DNA damage-dependent degradation exacerbate rereplication. It is worth noting that the enhanced rereplication we observed is not likely the result simply of an initial higher rate of rereplication for two reasons. First, the stabilizing mutations introduced into Cdt1, the N-terminal epitope tag and the PIP motif mutant, did not perturb the normal S phase-dependent degradation of Cdt1, and the overall abundance of both the normal and DNA damage-resistant forms of Cdt1 were equivalent in the absence of rereplication in all experiments. Second, stabilization of Cdc6 by Huwe1 depletion does not induce Cdc6 accumulation or rereplication in the absence of geminin depletion (Fig. 6C and Ref. 20). Thus, the effects of preventing DNA damage-dependent degradation of Cdt1 and Cdc6 on the overall extent of rereplication in these assays are manifest only in the context of rereplication.
Origin relicensing and consequent rereplication can be induced acutely by transfection with geminin siRNA or with high levels of ectopically expressed Cdt1. These events may also occur under more physiological circumstances should cells lose regulation of Cdt1 or geminin, as is the case for many tumor cells (37, 38). The ability of Cdt1 to cause rereplication when it is overproduced combined with the rereplication-induced degradation of Cdt1 indicates that Cdt1-overproducing cells are actively working to degrade Cdt1 once rereplication begins. In other words, excess Cdt1 bears the seeds of its own destruction. As a consequence, rereplication from inappropriately relicensed origins prevents additional origin relicensing because both Cdc6 and Cdt1 are absent (Fig. 7, A and B).
The DNA damage that results from rereplication induces not only Cdt1 and Cdc6 degradation, but also activation of the ATR-Chk1 checkpoint pathway (10, 12, 13, 16, 26, 39). Recent studies have shown that components of this DNA damage checkpoint signaling pathway also play an important role in restricting the extent of rereplication when geminin is depleted or Cdt1 is overproduced (15, 16, 40). One of the principal outcomes of checkpoint activation is the inhibition of cyclin/Cdk activity. The relationship of Cdk activity to events at replication origins is complex as Cdks are not only required for origin firing but they also act to prevent new origin licensing after the G1/S transition by phosphorylating preRC components (2, 3, 8, 9, 14, 41–43). The accumulation of supraphysiological DNA contents for a short time after Chk1 activation and the onset of Cdc6 and Cdt1 degradation (Fig. 7) could be attributed to fork elongation from relicensed origins that had already fired, but we presume that both origin licensing and origin firing are already inhibited during that period.
Activation of Chk1 is predicted to block origin firing by inducing Cdk inhibition, but this Chk1-dependent Cdk inhibition would not prevent origin relicensing. In fact, inactivation of Cdks in rereplicating cells as a consequence of DNA damage checkpoint signaling has the potential to promote even further origin licensing. To prevent the relicensing of origins during the period of low Cdk activity, Cdk-independent mechanisms to block preRC assembly are required. Degradation of Cdt1 in response to double-strand breaks induced by ionizing radiation does not require phosphorylation by Cdk2 (27, 30) or signaling through the ATM/ATR checkpoint pathway (29, 44). In that regard, it is important to note that although Chk1 is phosphorylated as Cdt1 is degraded (Fig. 7B), checkpoint activation does not itself stimulate Cdt1 degradation; these events are independent of one another. Huwe1-mediated Cdc6 degradation is similarly unaffected by Cdk2 phosphorylation (20), though the effects of the checkpoint signaling pathway on Huwe1 activity have yet to be determined. Furthermore, we have previously demonstrated that the DNA damage-dependent degradation of Cdt1 and Cdc6 are independent of one another (20). Thus, the degradation of Cdt1 and Cdc6 in rereplicating cells represent two separate mechanisms that block further origin licensing when Cdk activity is low, and these responses reinforce the checkpoint restraints on rereplication.
Many laboratories have noted cell line-associated diversity in the effects of geminin depletion on the final cellular DNA content. Components of the DNA damage checkpoint pathway are frequently targets for mutational inactivation or dysregulation during carcinogenesis, particularly the p53 branch of the pathway. It has been suggested that differences in the status of this checkpoint contribute to the propensity of cells to extensively rereplicate their DNA (10, 15). We propose that efficient degradation of Cdt1 and Cdc6 to block licensing, combined with robust DNA damage checkpoint activation to prevent origin firing, provides an effective restriction of rereplication (Fig. 7C). The diversity of rereplication phenotypes associated with different cell lines is then a reflection of differences in the genetic alterations that impact both the checkpoint response and the efficiency of Cdt1 and Cdc6 degradation. In that regard, one of the many differences between individual cancers may be the ability to induce Cdt1 and Cdc6 degradation to limit rereplication and the genome instability that it promotes.
Supplementary Material
Acknowledgments
We thank T. Orr-Weaver (Massachusetts Institute of Technology), Y. Xiong (University of North Carolina at Chapel Hill, and S. Wing (McGill University) for gifts of antibodies and W. Kaufmann (University of North Carolina at Chapel Hill) for NHF1-hTert cells. We thank the laboratory of Steve Rogers for assistance with S2 cell imaging.
This work was supported, in whole or in part, by National Institutes of Health Grant K01-CA094907 (to J. G. C.). This work was also supported by the American Cancer Society GMC-111880 (to J. G. C.) and RSG-04–179-01-DDC (to R. J. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: preRC, prereplication complex; ORC, origin recognition complex; MCM, minichromosome maintenance complex; m.o.i., multiplicity of infection; PIP, PCNA-interacting protein; HA, hemagglutinin; DAPI, 4′,6-diamidino-2-phenylindole; WT, wild type.
H. Lee and G. Rogers, unpublished observation.
References
- 1.Sclafani, R. A., and Holzen, T. M. (2007) Annu. Rev. Genet. 41 237-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, S. P., and Dutta, A. (2002) Annu. Rev. Biochem. 71 333-374 [DOI] [PubMed] [Google Scholar]
- 3.Diffley, J. F. (2004) Curr. Biol. 14 R778-R786 [DOI] [PubMed] [Google Scholar]
- 4.Arentson, E., Faloon, P., Seo, J., Moon, E., Studts, J. M., Fremont, D. H., and Choi, K. (2002) Oncogene 21 1150-1158 [DOI] [PubMed] [Google Scholar]
- 5.Liontos, M., Koutsami, M., Sideridou, M., Evangelou, K., Kletsas, D., Levy, B., Kotsinas, A., Nahum, O., Zoumpourlis, V., Kouloukoussa, M., Lygerou, Z., Taraviras, S., Kittas, C., Bartkova, J., Papavassiliou, A. G., Bartek, J., Halazonetis, T. D., and Gorgoulis, V. G. (2007) Cancer Res. 67 10899-10909 [DOI] [PubMed] [Google Scholar]
- 6.Fujita, M. (2006) Cell Div. 1 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arias, E. E., and Walter, J. C. (2007) Genes Dev. 21 497-518 [DOI] [PubMed] [Google Scholar]
- 8.Machida, Y. J., Hamlin, J. L., and Dutta, A. (2005) Cell 123 13-24 [DOI] [PubMed] [Google Scholar]
- 9.Blow, J. J., and Dutta, A. (2005) Nat. Rev. Mol. Cell. Biol. 6 476-486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaziri, C., Saxena, S., Jeon, Y., Lee, C., Murata, K., Machida, Y., Wagle, N., Hwang, D. S., and Dutta, A. (2003) Mol. Cell 11 997-1008 [DOI] [PubMed] [Google Scholar]
- 11.Saxena, S., and Dutta, A. (2005) Mutat. Res. 569 111-121 [DOI] [PubMed] [Google Scholar]
- 12.Melixetian, M., Ballabeni, A., Masiero, L., Gasparini, P., Zamponi, R., Bartek, J., Lukas, J., and Helin, K. (2004) J. Cell Biol. 165 473-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu, W., Chen, Y., and Dutta, A. (2004) Mol. Cell. Biol. 24 7140-7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mihaylov, I. S., Kondo, T., Jones, L., Ryzhikov, S., Tanaka, J., Zheng, J., Higa, L. A., Minamino, N., Cooley, L., and Zhang, H. (2002) Mol. Cell. Biol. 22 1868-1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, E., Lee, A. Y., Chiba, T., Olson, E., Sun, P., and Wu, X. (2007) J. Cell Biol. 179 643-657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, A. Y., Liu, E., and Wu, X. (2007) J. Biol. Chem. 282 32243-32255 [DOI] [PubMed] [Google Scholar]
- 17.Tatsumi, Y., Sugimoto, N., Yugawa, T., Narisawa-Saito, M., Kiyono, T., and Fujita, M. (2006) J. Cell Sci. 119 3128-3140 [DOI] [PubMed] [Google Scholar]
- 18.Ballabeni, A., Melixetian, M., Zamponi, R., Masiero, L., Marinoni, F., and Helin, K. (2004) EMBO J. 23 3122-3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schrofelbauer, B., Hakata, Y., and Landau, N. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 4130-4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall, J. R., Kow, E., Nevis, K. R., Lu, C. K., Luce, K. S., Zhong, Q., and Cook, J. G. (2007) Mol. Biol. Cell 18 3340-3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braden, W. A., Lenihan, J. M., Lan, Z., Luce, K. S., Zagorski, W., Bosco, E., Reed, M. F., Cook, J. G., and Knudsen, E. S. (2006) Mol. Cell. Biol. 26 7667-7681 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Rogers, S. L., Rogers, G. C., Sharp, D. J., and Vale, R. D. (2002) J. Cell Biol. 158 873-884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook, J. G., Chasse, D. A., and Nevins, J. R. (2004) J. Biol. Chem. 279 9625-9633 [DOI] [PubMed] [Google Scholar]
- 24.Archambault, V., Ikui, A. E., Drapkin, B. J., and Cross, F. R. (2005) Mol. Cell. Biol. 25 6707-6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson, I. F., Li, A., and Blow, J. J. (2006) Mol. Cell 24 433-443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGarry, T. J. (2002) Mol. Biol. Cell 13 3662-3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishitani, H., Sugimoto, N., Roukos, V., Nakanishi, Y., Saijo, M., Obuse, C., Tsurimoto, T., Nakayama, K. I., Nakayama, K., Fujita, M., Lygerou, Z., and Nishimoto, T. (2006) EMBO J. 25 1126-1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, J., McCall, C. M., Ohta, T., and Xiong, Y. (2004) Nat. Cell Biol. 6 1003-1009 [DOI] [PubMed] [Google Scholar]
- 29.Higa, L. A., Mihaylov, I. S., Banks, D. P., Zheng, J., and Zhang, H. (2003) Nat. Cell Biol. 5 1008-1015 [DOI] [PubMed] [Google Scholar]
- 30.Senga, T., Sivaprasad, U., Zhu, W., Park, J. H., Arias, E. E., Walter, J. C., and Dutta, A. (2006) J. Biol. Chem. 281 6246-6252 [DOI] [PubMed] [Google Scholar]
- 31.Arias, E. E., and Walter, J. C. (2006) Nat. Cell Biol. 8 84-90 [DOI] [PubMed] [Google Scholar]
- 32.Ferenbach, A., Li, A., Brito-Martins, M., and Blow, J. J. (2005) Nucleic Acids Res. 33 316-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovejoy, C. A., Lock, K., Yenamandra, A., and Cortez, D. (2006) Mol. Cell. Biol. 26 7977-7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blanchard, F., Rusiniak, M. E., Sharma, K., Sun, X., Todorov, I., Castellano, M. M., Gutierrez, C., Baumann, H., and Burhans, W. C. (2002) Mol. Biol. Cell 13 1536-1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duursma, A., and Agami, R. (2005) Mol. Cell. Biol. 25 6937-6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heffernan, T. P., Unsal-Kacmaz, K., Heinloth, A. N., Simpson, D. A., Paules, R. S., Sancar, A., Cordeiro-Stone, M., and Kaufmann, W. K. (2007) J. Biol. Chem. 282 9458-9468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xouri, G., Lygerou, Z., Nishitani, H., Pachnis, V., Nurse, P., and Taraviras, S. (2004) Eur. J. Biochem. 271 3368-3378 [DOI] [PubMed] [Google Scholar]
- 38.Saxena, S., and Dutta, A. (2003) Cell Cycle 2 283-286 [PubMed] [Google Scholar]
- 39.Liu, E., Li, X., Yan, F., Zhao, Q., and Wu, X. (2004) J. Biol. Chem. 279 17283-17288 [DOI] [PubMed] [Google Scholar]
- 40.Lin, J. J., and Dutta, A. (2007) J. Biol. Chem. 282 30357-30362 [DOI] [PubMed] [Google Scholar]
- 41.Hochegger, H., Dejsuphong, D., Sonoda, E., Saberi, A., Rajendra, E., Kirk, J., Hunt, T., and Takeda, S. (2007) J. Cell Biol. 178 257-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen, V. Q., Co, C., and Li, J. J. (2001) Nature 411 1068-1073 [DOI] [PubMed] [Google Scholar]
- 43.Thomer, M., May, N. R., Aggarwal, B. D., Kwok, G., and Calvi, B. R. (2004) Development 131 4807-4818 [DOI] [PubMed] [Google Scholar]
- 44.Kondo, T., Kobayashi, M., Tanaka, J., Yokoyama, A., Suzuki, S., Kato, N., Onozawa, M., Chiba, K., Hashino, S., Imamura, M., Minami, Y., Minamino, N., and Asaka, M. (2004) J. Biol. Chem. 279 27315-27319 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.