Abstract
Homeotic Hox selector genes encode highly conserved transcriptional regulators involved in the differentiation of multicellular organisms. Ectopic expression of the Antennapedia (ANTP) homeodomain protein in Drosophila imaginal discs induces distinct phenotypes, including an antenna-to-leg transformation and eye reduction. We have proposed that the eye loss phenotype is a consequence of a negative posttranslational control mechanism because of direct protein–protein interactions between ANTP and Eyeless (EY). In the present work, we analyzed the effect of various ANTP homeodomain mutations for their interaction with EY and for head development. Contrasting with the eye loss phenotype, we provide evidence that the antenna-to-leg transformation involves ANTP DNA-binding activity. In a complementary genetic screen performed in yeast, we isolated mutations located in the N terminus of the ANTP homeodomain that inhibit direct interactions with EY without abolishing DNA binding in vitro and in vivo. In a bimolecular fluorescence complementation assay, we detected the ANTP–EY interaction in vivo, these interactions occurring through the paired domain and/or the homeodomain of EY. These results demonstrate that the homeodomain supports multiple molecular regulatory functions in addition to protein–DNA and protein–RNA interactions; it is also involved in protein–protein interactions.
Keywords: Drosophila, antagonism, regulation
Homeotic Hox genes are selector genes that generate morphological diversity along the antero–posterior body axis during animal development (1). They encode highly conserved transcription factors defining various cellular identities in the body segments along the antero–posterior axis of the embryo (2, 3). Hox genes share a common sequence element of 180 bp, the homeobox, first isolated in Drosophila (4, 5), encoding a 60-aa homeodomain (HD) DNA-binding domain. The HD presents a stereotypical three α-helical structure, and its mode of interaction with DNA is largely invariant. The amino acid 50, a glutamine signature for the Hox proteins, plays a fundamental role in DNA-binding specificity (6). Hox factors have very similar DNA-binding properties (7). Therefore, the limited sequence selectivity of the HD is not sufficient to explain the diversity of cell types and the batteries of downstream genes under the control of Hox proteins. Consequently, elucidating the mechanisms of Hox protein function is critical for understanding development and the diversification of serially homologous structures. Several studies have indicated that Hox factors cooperate with signaling pathways and act with cofactors that modify DNA binding and activation/repression properties (8–10). To date, the best-known Hox cofactors are the Drosophila extradenticle (EXD/PBX) and homothorax (HTH/Meis) proteins. EXD was shown to modulate the DNA-binding specificity and the activity of Hox proteins (11–13), whereas HTH promotes EXD translocation into the nucleus where they participate in a DNA-bound HOX/EXD/HTH ternary complex (14). Although EXD can raise the DNA-binding specificity of Hox proteins, some Hox gene functions appear to be EXD-independent (11, 15), suggesting that additional factors might modulate Hox activity.
To elucidate the mechanisms involved in Hox-mediated cellular transformation, we have studied the mechanism of the antenna-to-leg transformation versus the eye inhibition induced by misexpression of the Antennapedia (ANTP) protein. We have proposed that Antp inhibits the activity of the eye selector gene eyeless (ey) at the posttranslational level through direct protein–protein interactions, and we suggested that such interactions could contribute substantially to the regulation of selector genes (16).
To understand better how Antp provokes this eye loss phenotype, we first tested truncated EY proteins for their ability to antagonize the Antp-induced eye loss phenotype. Second, we analyzed the effects of mutations in key HD residues implicated in Hox DNA-binding activity for their effects on antenna-to-leg transformation and eye reduction. Third, we performed a complementary genetic screen in yeast for mutations that decrease the capacity of the ANTP HD to repress ey function without affecting DNA-binding activity. Several candidate mutants were clustered in a highly conserved motif (TLELEKEF) situated in the first α-helix of the HD. We showed by bimolecular fluorescence complementation (BiFC) in vivo that these mutants affect the direct interaction of the ANTP HD with both the paired domain (PD) and the HD of EY. In flies, this mutated ANTP protein reveals a reduced capacity to repress eye development but directs the antenna-to-leg transformation. Thus, these experiments allowed us to identify amino acids located in a conserved N-terminal region of the HD involved in protein–protein interaction and to dissociate this function from DNA-binding activity. This led us to propose that the HD is involved in protein–DNA, protein–RNA in protein–protein interactions.
Results
Truncated EY Proteins Are Able to Antagonize ANTP-Induced Eye Loss.
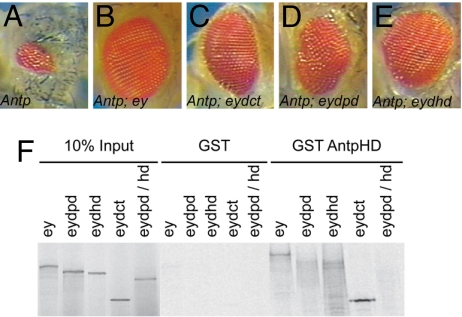
We showed that the eye loss induced by misexpression of ANTP in eye progenitor cells can be alleviated upon coexpression with transgenically supplied EY. We interpreted this effect as reflecting titration of ANTP by EY protein in vivo because parallel GST-pulldown experiments showed that EY is able to interact with ANTP through its two distinct DNA-binding domains, namely the PD and the HD (16). We therefore established a rescue assay to provide a sensitive in vivo measure for the titration of EY variants by ectopic ANTP protein to determine which EY protein domains are required to counteract Hox-induced eye loss. We generated double UAS lines coexpressing ANTP with different EY-deleted forms (17). The driver line used was OK107-Gal4 because it gives more reliable results than the widely used ey-Gal4 transgene (data not shown). However, phenotypes observed upon driving ANTP misexpression by OK107 are weaker than for ey-Gal4. Compared with misexpression of Antp alone (Fig. 1A), coexpression of EY with ANTP leads to a rescue of eye formation (Fig. 1B). EY deleted for its C terminus or its PD was still able to rescue the Antp-induced eye loss (Fig. 1 C and D). HD-deleted EY protein, with ANTP, results in lethality such that few pupae were obtained. However, the few escapers showed a partial eye rescue (Fig. 1E). All of the EY deletions able to counteract the ANTP-induced eye loss interact biochemically with the ANTP HD in vitro (Fig. 1F), albeit with less efficiency for the deleted PD and HD EY. This finding suggests that EY can interact with the ANTP HD in vivo either through its PD or its HD. Because the PD and CT EY-deleted proteins are not able to trigger eye development (17), we conclude that a functional EY protein is not required to counteract ANTP-induced eye loss.
Fig. 1.
OK107-Gal4-driven coexpression of ey deletions with Antp in the eye disc. (A) Expression of Antp. (B–D) Eye phenotype upon coexpressing Antp with ey (B), eyΔCT (C), and eyΔPD (D). (E) eyΔHD. (F) GST pulldown assay of EY deletions, reticulocyte lysate synthesis, and GST-AntpHD fusion protein.
Uncoupling of Eye Inhibition and Antenna-to-Leg Transformation.
To understand further the mechanism through which Antp mediates its inhibitory effect on eye development, we examined several critical amino acid positions potentially involved in this process. Several point mutations were introduced in the HD-changing amino acids, which are key determinants in contacting DNA. In addition to the mutations affecting the third α-helix of the HD (residues A50, A51, K50, and H53), we introduced two mutations in the N terminus changing R5 into H5 or A5. R5, which is highly conserved in evolution, was shown to be critical in stabilizing the HD on DNA by binding to the minor groove (18). Thus, mutating this amino acid should result in a protein with reduced DNA-binding activity. The H5 mutation was of interest because (i) previous evidence reported an involvement of this residue both in DNA binding and in protein–protein interaction and (ii) Benassayag et al. (19) identified EY as a direct target of protein interactions with Proboscipedia (PB) in the eye repression pathway.
To investigate whether the pupal lethality and/or eye loss induced by ANTP involves its DNA-binding activity, transgenic lines expressing similar protein expression levels were selected [supporting information (SI) Fig. S1]. We compared the antenna-to-leg transformation with eye inhibition for each mutant protein (Table 1 and Fig. S1). By using dppblk-Gal4 driver, only the H5 mutant protein was able to induce a detectable antenna-to-leg transformation, suggesting a DNA binding-dependent mechanism for the antenna-to-leg transformation. In contrast, all ANTP mutant proteins except the H53 variant were still able to inhibit eye development, albeit with different efficiencies, when drivers specifically expressed in the eye precursor cells were used (ey-Gal4 and OK107-Gal4) (Table 1). H5-induced eye loss is similar to that of the wild-type (WT) ANTP protein, whereas K50, A50, A51, and A5 were weaker. H53 neither inhibits eye development nor induces any antenna-to-leg transformation.
Table 1.
Fly phenotypes observed using different UAS-ANTP mutant lines
| Mutant lines | dpp-GAL4 |
OK107-GAL4 |
||
|---|---|---|---|---|
| Antenna to leg | Eye reduction | Pupal lethality, % | Eye reduction/WT (escapers or pupae), % | |
| UAS-Antp | + | +/++ | ∼95 | 75–100 |
| UAS-AntpH5 | + | ++ | <10 | 50–75 |
| UAS-AntpA5 | − | +/− | <10 | 25–50 |
| UAS-AntpK50 | − | − | <10 | 25–50 |
| UAS-AntpA50,51 | − | − | 0 | 10–50 |
| UAS-AntpH53 | − | − | 0 | <10 |
| UAS-AntpG15 | + | + | ∼99 | 75–100 |
| UAS-AntpG19 | + | +/− | ∼99 | 25–50 |
| UAS-AntpA27 | + | ++ | ∼99 | 75–100 |
By expressing WT ANTP in larvae using different GAL4 drivers, distinct phenotypes are distinguished: antenna-to-leg transformation, eye reduction, and pupal lethality (resulting from a headless phenotype). ANTP mutants (A5, K50, A50, 51, H53) affecting DNA binding activity are unable to induce antenna-to-leg transformation and pupal lethality. Most of these lines retain the capacity to inhibit eye development when expressed in the eye disc. By contrast, the ANTP mutant G19, retaining full DNA-binding activity in vitro, induced antenna-to-leg transformation and pupal lethality but exhibited a weaker ability to induce eye reduction. Percentage of eye reduction given was estimated by comparison with a normal WT eye size.
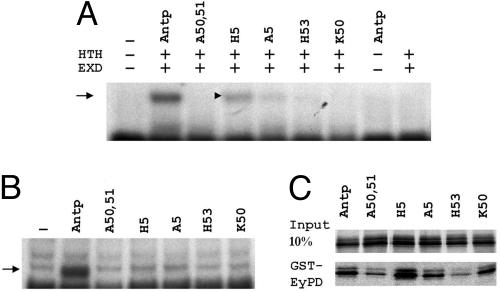
Next, we studied the effect of ANTP HD mutations on DNA-binding activity by using a bandshift assay. We used two different probes: a Hox target sequence (HB1), allowing ANTP binding in the absence of Exd, and a tripartite binding site Hox/Exd/Hth. On Hox/Exd/Hth probe, none of the mutants except H5 (and sometimes A5; note the faint signal in Fig. 2A) showed any DNA-binding activity in the presence of EXD and HTH cofactors (Fig. 2A). None of the mutated proteins tested showed any detectable DNA-binding activity on HB1 (Fig. 2B). These results suggest that ANTP-induced antenna-to-leg transformation and pupal lethality involve a DNA-binding mechanism (Table 1), whereas eye reduction does not.
Fig. 2.
Molecular characterization of the different Antp mutant proteins. (A and B) Gel shift experiments performed on a Hox/Exd/Hth site (A) or on HB1 probe (B). Arrows indicate the specific retarded complexes. (A) Only the ternary complex is shown. ANTP does not bind on its own on this site and weakly in the presence of EXD (data not shown). (C) Coretention assay between the GST-EY-PD and the mutated ANTP proteins indicated. Input lane indicates the amount of proteins used for the assay. No binding was observed with GST alone (data not shown).
Because the mutated ANTP proteins interfere differently with eye development, we compared their abilities to interact with EY by using in vitro pulldown experiments. As seen in Fig. 2C, H5 binds more strongly to EY than the WT Antp protein. A5 and K50, which are weaker in inhibiting eye development, also revealed a reduced biochemical interaction with the EY PD. Finally, the mutations that interfere least with eye development (A50, A51, and H53) also exhibited the weakest interaction with EY PD in vitro (Fig. 2C).
Search for Mutations in the ANTP HD Affecting Protein–Protein Interactions Independently of DNA-Binding Activity.
Using a derived yeast two-hybrid strategy, we showed that a GAL4AD-ANTP HD fusion protein can inhibit EY-induced transactivation of the eye-specific sine oculis enhancer (so10) in yeast (16). We therefore decided to use this experimental system to screen for mutations of the ANTP HD that reestablish EY transactivation through so10 without affecting ANTP DNA binding to a selectable HB1-HIS chimeric gene (Fig. S2A). A similar strategy was successfully used by Burz and Hanes (20) to isolate mutations of Bicoid affecting its cooperative DNA-binding activity. The ANTP HD fused to the GAL4 activation domain (GAL4AD) was mutagenized in vitro by error-prone PCR and introduced into yeast cells with a gapped-plasmid approach (21). In addition to the GAL4AD-AntpHD fusion and EY, these yeast cells carried genomic 4X HB1-HIS and so10-lacZ constructs integrated at the HIS and the URA loci, respectively. Among 400 independent clones tested, 58 were His+ and expressed lacZ. Among these 58 candidate clones, 33 contained mutations in the HD, including 16 transition mutants in its 5′ half. Although some of these mutations were unique, several hot spots were observed, including three independent mutants that replaced T7 with A (T7A), two E15G mutants, four E19G, and three T27A (Fig. 3A).
Fig. 3.
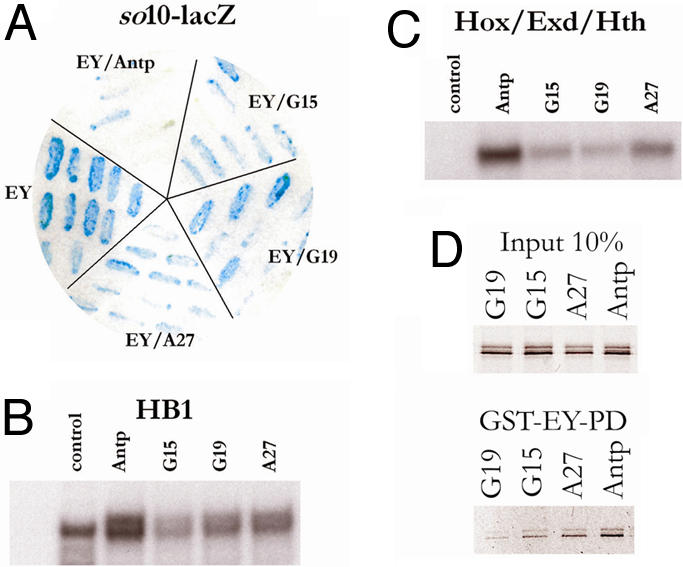
Isolation of mutations in the ANTP HD affecting EY protein interaction. (A) One-and-a-half hybrid of Antp-mediated repression of EY activity. Several independent colonies were tested independently. EY, positive control showing the activation of so10-LacZ by EY. EY/ANTP, coexpression of EY and ANTP (GAL4AD-Antp HD) results in so10-LacZ repression. EY/Antp G15 or G19 or G27, capacity of Antp mutants in repressing EY function. None of the mutants lost completely the ability to repress EY activity. (B and C) Gel shift experiments on HB1 (B) or a Hox/Exd/Hth-binding site in the presence of EXD and HTH proteins (C). (B) Upper band indicates the specific complex. (C) Only the ternary complex is shown. (D) GST pulldown experiment between the EY PD (GST-EY-PD) and full-length ANTP mutant proteins (indicated on top of each lane).
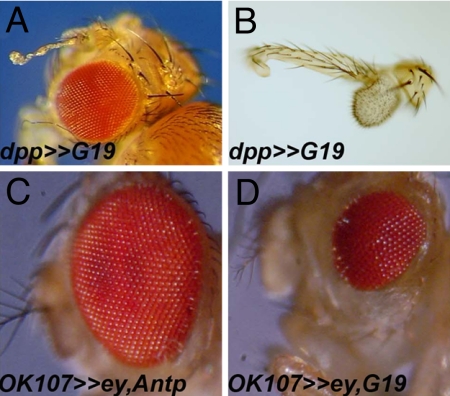
The E to G substitutions concentrated at positions 15 and 19 reside in a highly conserved motif (TLELEKEF) within the first α-helix of the HD (Fig. S2B). We decided to analyze the highly represented E15G, E19G, and T27A mutants further. We first studied the DNA-binding activity of these mutants. One-hybrid experiments were conducted with the HB1-LacZ yeast line to verify that all of the Antp mutants could bind and activate the HB1 motif in yeast to the same extent as the WT (Fig. S2C). In addition, we confirmed by bandshift assays the ability of the mutants to bind to HB1 monomer or dimer DNA in vitro (Fig. 3B) and to a Hox/Exd/Hth motif in the presence of EXD/HTH (Fig. 3C). From these experiments, we conclude that the point mutations found in HD helices 1 or 2 differ from the others in that they slightly decrease but do not disrupt DNA binding, alone or in combination with EXD and HTH, this decrease not being detectable in the yeast one-hybrid experiment. Because these mutants are less efficient for EY repression in yeast (Fig. 3A), we investigated the capacity of mutant ANTP proteins to interact physically with the EY PD in pulldown experiments. As shown in Fig. 3D, all of the mutants retained the capacity to interact with the EY-PD, albeit less than for WT and are consistent with the results obtained in yeast. To study the effect of these mutations in vivo, we generated UAS-ANTP transgenic fly lines for the G15, G19, and A27 mutations. Phenotypes were analyzed with regard to eye and antennal development after crossing those newly generated Antp lines with ey-Gal4, OK107-Gal4, or dppblk-Gal4. The G15 and A27 mutant proteins that only partially repress EY in yeast (Fig. 3A) and show mildly diminished EYPD-binding compared with the WT protein in vitro (Fig. 3D) induce phenotypes in flies close to that seen with the WT ANTP protein itself (Table 1). In contrast, the G19 mutation, which shows the weakest EY inhibition in yeast and the weakest interaction with EY in vitro (Fig. 3 A and D), only weakly interferes with eye development when expressed during larval stages using dppblk-Gal4 (Fig. 4A). Crosses to ey-Gal4 and OK107-Gal4 both resulted in a dominant headless pupal phenotype. The few escaper flies exhibit a moderate eye loss (Table 1). To overcome the lethality to measure G19 activity better, we applied the “eye rescue assay” used to provide a more sensitive in vivo measure for the titration of EY by the WT and G19 ANTP proteins. Coexpressing EY with WT ANTP protein under OK107-Gal4 control resulted in a rescue of the induced lethality (head loss) and eye loss-induced phenotype (Fig. 4C). Coexpressing EY with ANTP G19, lethality (head loss) is rescued, but a moderate eye reduction is observed, likely reflecting the EY inability to counteract ANTP efficiently (Fig. 4D). Using the dppblk-Gal4 driver, we observed an antenna-to-leg transformation, confirming the in vivo DNA-binding activity of the G19 protein (Fig. 4 A and B). Thus, in contrast to the mutations disrupting DNA-binding activity, the G19 mutant retains the capacity to transform the antenna into leg (Fig. 4B) but only weakly interferes with eye development (Table 1).
Fig. 4.
Functional analysis in flies of the ANTP G19 mutation. (A–D) Adult phenotype of transgenic flies harboring the UAS-Antp-G19 driven with the dppblink-Gal4 (A). (A) Adult head with a weak eye reduction and antenna transformed into leg. (B) Dissected G19 transformed antennae. (C and D) Coexpression of ey and Antp proteins (WT or G19) in the eye disc driven by OK107-Gal4 resulted in a complete (C) or partial rescue (D) of the Antp-induced eye loss, illustrating the inability of ey to interact in vivo with the Antp G19 mutant.
BiFC.
To test whether a direct physical interaction between Ey PD and HD and ANTP HD occurred in cells, we used BiFC analysis (22, 23). This method is based on the reconstitution of yellow fluorescent protein (YFP) fluorescence when two nonfluorescent YFP fragments are brought together in the cell. The two nonfluorescent fragments of Venus YFP (VN and VC; 23), an improved variant of YFP (24), were covalently linked to the ANTP HD, and EY PD or HD, respectively (Fig. S3A). The resulting plasmids were cotransfected into human embryonic kidney (HEK) 293 cells in combination with a control plasmid encoding a cytoplasmic enhanced cyan fluorescent protein (ECFP).
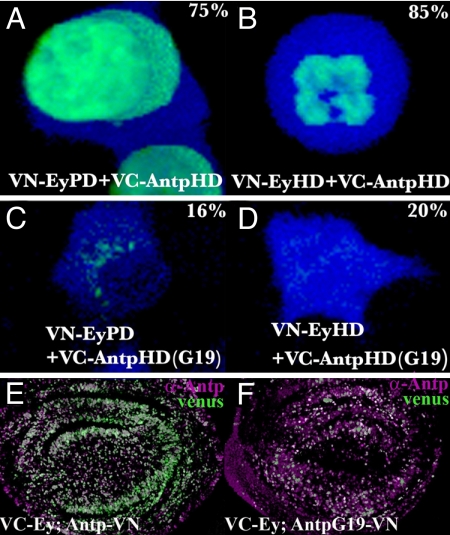
No fluorescence is detected in control experiments when only one fusion protein was cotransfected with the complementary YFP fragment alone (Fig. S3B). By contrast, strong interactions (80% of the cells) are observed between the ANTP HD and the EY PD (Fig. 5A), and between the ANTP HD and the EY HD (Fig. 5B). This interaction is severely reduced with the G19 ANTP HD for which only 20% of cells exhibited a detectable YFP signal (Fig. 5 C and D). No homodimer formation between ANTP HD and EY HD by BiFC was observed (data not shown), suggesting a strict specificity in the interaction between different HD classes. These results also preclude a nonspecific fluorescence when the two protein partners come into contact upon random binding to neighboring DNA sites.
Fig. 5.
BiFC analysis of protein–protein interactions. (A–D) Interaction between EyPD and AntpHD (A) or between EyHD and AntpHD (B) resulted in a strong Venus fluorescence signal. Percentage positive cells are indicated. The G19 mutation dramatically reduces the interactions to the EyPD (C) or EyHD (D). CFP counterstains the cytoplasm. (E and F) Full-length EY/ANTP WT and G19 interactions were detected in wing discs (green). Level of ANTP expression is monitored by immunostaining (magenta).
We next applied the BiFC method in Drosophila larvae by using the full-length ANTP and EY proteins fused to VN and/or VC, respectively (Fig. 5E). When ANTP and EY fused to VN or VC are coexpressed by the heat shock Gal4/UAS system, a strong fluorescence is observed in imaginal discs (Fig. 5E). In contrast, when G19 ANTP fused to VC is coexpressed with VN EY, the interaction is reduced as illustrated by a much fainter fluorescence signal detected in wing imaginal discs (Fig. 5F). Taken together, these results provide compelling evidence for direct, functionally relevant protein–protein interactions involving the EY PD and HD and the first α-helix of the ANTP HD.
Discussion
Hox proteins are transcription factors harboring a conserved HD involved in sequence-specific DNA binding. Such binding capacity, occurs via surprisingly simple DNA “TAAT” core motifs. How can the binding of these sequences by a conserved DNA-binding protein lead to the highly refined outputs required to specify a body segment? The discovery of Exd/Hth cofactors conferring additional specificity provided one new element of response (9). Also, the spacing specific residues located in the N-terminal arm of the HD also contribute to Hox specificity (25). Other nuclear partners, generating different combinations of HOX transcription factor complexes integrating a variety of positional cues, generate the specificity and versatility required for context-dependent Hox function (8, 25–28). Thus, the activity of the Hox protein involves joint inputs of bound target DNA sequences and cooperating protein partners.
Inhibition of the Eye Developmental Pathway Involves Competitive Protein–Protein Interactions.
For Antp, ectopically expressed protein is capable of inducing a variety of dominant phenotypes. One function is its homeotic role, transforming antennae to mesothoracic (T2) legs, the head capsule into notum, and the eyes into wings (29). Other effects of ectopic Antp-, such as the eye loss examined here, are less clearly related to its normal selector function. To understand how a Hox gene can function in normal development, it is important to understand the parameters that contribute to its pleiotropic function. We identified a functional antagonism between hox and eyeless (16, 19). In both cases, the Hox proteins interact primarily via their conserved HD with the EY PD, leading to apoptosis and to epistatic reduction of the compound eyes. The relationship of ANTP with EY thus offers a useful model for examining how protein interactions can modify cell fate. As shown, an EY protein deleted for its HD retains the capacity to rescue ey loss-of-function mutants. In contrast, PD or C-terminal deleted EY proteins do not rescue (17). Testing these same proteins for their capacity to reverse Antp-induced eye reduction revealed important roles for both the PD and HD and supported the notion that the EY PD and/or HD domain plays the primary role in counteracting ANTP by a direct competitive interaction.
DNA Binding Is Required for ANTP-Induced Antenna-to-Leg Transformation.
We have also examined the reciprocal contributions of ANTP to the interaction by expressing normal or mutant protein forms in eye cells that contain endogenous EY protein. This work suggests that both the homeotic transformation and head loss require intact DNA-binding activity. However, the two mutations H5 and G19 retained our attention because both are able to transform the antenna into a leg. Whereas H5 does not bind HB1 in vitro, H5 and G19 bind the tripartite Hox/Exd/Hth-binding site albeit less efficiently than the WT ANTP protein. This result might suggest that ANTP-driven antenna-to-leg transformation requires a tripartite ANTP/Exd/Hth complex on the DNA, a finding that is in contradiction with the current model that ANTP represses Hth and, consequently, nuclear Exd (30). Because it is not clear whether an ANTP/Exd/Hth complex forms in the antenna cells, characterization of in vivo ANTP target sequences is a necessary step to understand the mechanisms controlling this process. Through the characterization of a ss antenna-specific enhancer, Emmons et al. (31) suggested that antennal identity is not determined solely by hth, but is specified by the combined action of Hth, Dll, and Ss. Because ANTP is able to repress this element, ANTP might modify these combinatorial protein complexes or compete at the DNA level with antenna-specific proteins complexes.
In contrast, we show here that eye loss can be uncoupled from DNA binding because DNA-binding-defective variants of ANTP can still induce eye loss. Although these mutant proteins (A5, A50, A51, and K50) interfere with eye development when they are expressed in the eye primordium using the robust ey-Gal4 driver, little if any effect is observed on expressing these proteins at larval stages (probably at lower levels) when using the dpp-GAL4 driver. We inferred that the ability of the ANTP protein to induce eye reduction depends on its affinity for EY. In vitro tests indicated a clear correlation between eye reduction and ANTP protein binding to EY, independent of its DNA-binding capacity. These observations supporting our titration model of specific protein–protein interactions are corroborated by the direct interactions of these proteins detected by BiFC in cell culture assays and in vivo in imaginal discs.
Dual Functions of the HD: Regulating Transcription Through DNA or Protein Partners.
The ensemble of in vivo and in vitro results suggested that eye reduction and homeotic functions of ANTP protein are at least in part separable, the former being because of protein–protein interactions, the latter depending on DNA-binding activity. A forward genetic screen in yeast performed to identify ANTP sequences involved in EY repression but not DNA-binding activity identified several mutations. The G19 (E19G) mutant was particularly instructive. In vivo, gain-of-function experiments revealed that G19 retains the ability to induce an antenna-to-leg transformation and a strong headless phenotype, as for WT ANTP. As expected based on the selection strategy, this protein retains DNA-binding activity. In contrast, G19 is clearly diminished in its ability to interfere with eye development, in agreement with its impaired binding to EY protein in vitro and in cultured cells. This mutation clearly illustrates the multiplicity of HD functions and the possibility of separating, or modulating, these functions.
Cross-Regulatory Protein–Protein Interactions Among Transcription Factors.
Direct competitive interactions among different Hox proteins and between Hox and Pax transcription factors have been confirmed in cultured cells by BiFC, using the G19 mutant isolated for its reduced inhibition of EY in yeast cells as a negative control. These Hox and Pax interactions may account for a more general mechanism, resulting not only in gene repression but also leading to gene activation as recently suggested by Gong et al. (32). These findings have wide implications for gene regulation in different developmental pathways, in particular for understanding the epistatic relationships among different transcription factor genes. In one example designated as “phenotypic suppression,” a posterior Hox gene suppresses the action of a more anteriorly expressed one (33). Earlier findings led to the conclusion that cross-regulatory interactions between homeotic genes at the transcriptional level cannot explain their epistatic relationship (34). Subsequently, it was shown that the phenotypic suppression of the homeobox gene empty spiracles (ems) is prevented by buttonhead (btd), a gene encoding a zinc finger class transcription factor, correlated with protein interactions between EMS and BTD in vitro (35). Although the structural details of this interaction have to be clarified, it appears that a Hox–zinc finger protein interactions may also be systemic (36). These converging observations may be indicate an entire network of protein–protein interactions among transcription factors, superimposed on the transcriptional networks controlled by these genes and essential for the specification of the different developmental pathways. It remains to be seen whether the transcription factors form large protein complexes or whether they interact as heterodimers or oligomers in solution and/or bound to DNA.
Materials and Methods
Fly Strains.
All stocks and crosses were reared at 22–25°C on standard yeast–agar–cornmeal medium. Fly lines or alleles used are described in refs. 16 and 17). Fly lines generated by standard P germ-line transformation in pUAST were UAS-Antp A5, UAS-Antp H5, UAS-Antp H53, UAS-Antp G15, UAS-Antp G19, UAS-Antp A27 and UAS-eyΔCT, UAS-Antp-VN, UAS-Antp-VC, UAS-Antp G19-VN, UAS-Antp G19-VC, UAS-VN-ey, and UAS-VC-ey.
Antibody Staining and Phenotypic Analysis.
Antibody staining was performed according to ref. 16. Mouse α-Antp 4C3 or 8C11 (1/1,000) was from Hybridoma Bank. Secondary antibodies (anti-mouse Cy3, anti-rabbit DTAF, anti-rat AMAC; 1/500) were from Jackson Laboratory. Discs were mounted in Vectashield (Vector Laboratories). Adult fly heads were photographed directly or dissected and mounted in Hoyer's medium. Detailed antenna phenotypes were analyzed by light microscopy (Zeiss Axiophot) after mounting dissected samples.
Pulldown, Western Blotting, and Gel Shift Experiments.
GST fusion proteins were produced and purified according to the manufacturer's specifications (Amersham Pharmacia). Pulldown assay were performed as described in ref. 16. Western blotting experiments were performed essentially as described by Punzo et al. (17). Gel shift assays were performed as described by Plaza et al. (16) with the HB1 double-stranded oligonucleotide (37) and a modified HOX/EXD element with an additional HTH site (5′-GGGTGATTTATGGGCGACTGTCACCGCTC and its complementary fragment).
“One-and-a-Half Hybrid” Screen.
One-hybrid experiments were performed by using the MATCHMAKER one-hybrid system according to the manufacturer's specifications (Clontech). ey cDNA was subcloned in-frame with the Gal4DB in the pAS vector. To perform the mutagenesis, the Antp expression plasmid pACTII-Antp 287–378 (16) was used as the template for mutagenic PCR (21). Primers of 70 nt corresponding to the 5′ and 3′ part of the Antp insert also overlap the plasmid sequences to ensure a correct length necessary for homologous recombination in yeast and will be given upon request. The AntpHD-containing region was mutagenized according to Muhlrad et al. (21), and the amplification products were cotransformed into the host with 40 ng of linearized pACTII. The transformation mixture was plated on ura−his−trp−leu− medium supplemented by 15 mM 3-amino-1,2,4-triazole. Colonies growing (HIS+) were picked up onto new plates to produce replicates onto Whatman filters to detect their β-galactosidase activity. Clones presenting a strong β-galactosidase activity were selected. The ANTP HD region was further PCR-amplified and sequenced.
Plasmid Constructions.
To generate the eyΔCT construct, the embryonic ey cDNA was cut by NruI, and a stop codon was inserted just after the last amino acid of the HD. In vitro transcription/translation was performed by using Eyeless deletion cDNAs (17) or Antp mutants cloned into pBSK. Antp cDNA mutations were generated by PCR, and the mutated fragments were cloned into the pBS Antp (16) and further cloned into pUAST. HD and PD domains were Pfu PCR-amplified with restriction site-containing primers and inserted into the BiFC plasmids (28). To generate the WT and G19 pUAST-Antp-VN, pUAST-Antp-VC, the C-terminal part of the Antp WT and G19 cDNAs, was replaced by the VN and VC sequences. The pUAST-VN-ey and pUAST-VC-ey were generated by inserting VN or VC in-frame at the ey N terminus. All constructions were confirmed by sequencing. Detailed cloning procedures will be given upon request.
BiFC.
Venus BiFC plasmids and enhanced cyanofluorescent protein (pECFP-N1) (Clontech) were cotransfected into HEK293 cells by using FuGENE 6 reagent (Roche). Fluorescence signals were observed 24–48 h after transfection by using a Leica TCS confocal microscope. BiFC flies were crossed with a hs-Gal4 line. Third-instar larvae were heat-shocked for 20 min, fixed, dissected, and analyzed after a 10- to 12-h recovery. Protein expression of Antp was monitored by antibody staining.
Supplementary Material
Acknowledgments.
We are grateful to Yasushi Saka and Jim Smith (Gurdon Institute, University of Cambridge) for Venus BiFC plasmids. We also thank Tom Kerppola (University of Michigan, Ann Arbor, MI) for BiFC plasmids and information on his web site (http://sitemaker.umich.edu/kerppola.lab/kerppola_lab_bifc). This work was supported by the Kantons of Basel-Stadt and Basel-Land, a grant from the Swiss National Science Foundation, and the European Network of Excellence “Cells into Organs.” S.P. was supported by the Centre National de la Recherche Scientifique and an EMBO fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806106105/DCSupplemental.
References
- 1.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence PA, Morata G. Homeobox genes: Their function in Drosophila segmentation and pattern formation. Cell. 1994;78:181–189. doi: 10.1016/0092-8674(94)90289-5. [DOI] [PubMed] [Google Scholar]
- 3.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 4.McGinnis W, Garber RL, Wirz J, Kuroiwa A, Gehring WJ. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984;37:403–408. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- 5.Scott MP, Weiner AJ. Structural relationships among genes that control development: Sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci USA. 1984;81:4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA-binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 7.Biggin MD, McGinnis W. Regulation of segmentation and segmental identity by Drosophila homeoproteins: The role of DNA binding in functional activity and specificity. Development. 1997;124:4425–4433. doi: 10.1242/dev.124.22.4425. [DOI] [PubMed] [Google Scholar]
- 8.Affolter M, Mann R. Development. Legs, eyes, or wings: Selectors and signals make the difference. Science. 2001;292:1080–1081. doi: 10.1126/science.1060856. [DOI] [PubMed] [Google Scholar]
- 9.Mann RS, Chan SK. Extra specificity from extradenticle: The partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet. 1996;12:258–262. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 10.Morata G, Sanchez-Herrero E. Patterning mechanisms in the body trunk and the appendages of Drosophila. Development. 1999;126:2823–2828. doi: 10.1242/dev.126.13.2823. [DOI] [PubMed] [Google Scholar]
- 11.Peifer M, Wieschaus E. Mutations in the Drosophila gene extradenticle affect the way specific homeodomain proteins regulate segmental identity. Genes Dev. 1990;4:1209–1223. doi: 10.1101/gad.4.7.1209. [DOI] [PubMed] [Google Scholar]
- 12.Chan SK, Jaffe L, Capovilla M, Botas J, Mann RS. The DNA-binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 13.van Dijk MA, Murre C. Extradenticle raises the DNA-binding specificity of homeotic selector gene products. Cell. 1994;78:617–624. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]
- 14.Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- 15.Pinsonneault J, Florence B, Vaessin H, McGinnis W. A model for extradenticle function as a switch that changes HOX proteins from repressors to activators. EMBO J. 1997;16:2032–2042. doi: 10.1093/emboj/16.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plaza S, et al. Molecular basis for the inhibition of Drosophila eye development by Antennapedia. EMBO J. 2001;20:802–811. doi: 10.1093/emboj/20.4.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Punzo C, Kurata S, Gehring WJ. The eyeless homeodomain is dispensable for eye development in Drosophila. Genes Dev. 2001;15:1716–1723. doi: 10.1101/gad.196401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otting G, et al. Protein–DNA contacts in the structure of a homeodomain–DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990;9:3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benassayag C, et al. Evidence for a direct functional antagonism of the selector genes proboscipedia and eyeless in Drosophila head. Development. 2003;130:575–586. doi: 10.1242/dev.00226. [DOI] [PubMed] [Google Scholar]
- 20.Burz DS, Hanes SD. Isolation of mutations that disrupt cooperative DNA binding by the Drosophila Bicoid protein. J Mol Biol. 2001;305:219–230. doi: 10.1006/jmbi.2000.4287. [DOI] [PubMed] [Google Scholar]
- 21.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 22.Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 23.Saka Y, Hagemann A, Piepenburg O, Smith JC. Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus. Development. 2007;134:4209–4218. doi: 10.1242/dev.010645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 25.Joshi R, et al. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bondos SE, Tan XX, Matthews KS. Physical and genetic interactions link Hox function with diverse transcription factors and cell signaling proteins. Mol Cell Proteomics. 2006;5:824–834. doi: 10.1074/mcp.M500256-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Luo L, Yang X, Takihara Y, Knoetgen H, Kessel M. The cell cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature. 2004;427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- 28.Wang N, et al. TGFβ/BMP inhibits the bone marrow transformation capability of Hoxa9 by repressing its DNA-binding ability. EMBO J. 2006;25:1469–1480. doi: 10.1038/sj.emboj.7601037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prince F, et al. The YPWM motif links Antennapedia to the basal transcriptional machinery. Development. 2008;135:1669–1679. doi: 10.1242/dev.018028. [DOI] [PubMed] [Google Scholar]
- 30.Casares F, Mann RS. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- 31.Emmons RB, Duncan D, Duncan I. Regulation of the Drosophila distal antennal determinant spineless. Dev Biol. 2007;302:412–426. doi: 10.1016/j.ydbio.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong KQ, Yallowitz AR, Sun H, Dressler GR, Wellik DM. A Hox–Eya–Pax complex regulates early kidney developmental gene expression. Mol Cell Biol. 2007;21:7661–7668. doi: 10.1128/MCB.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macias A, Morata G. Functional hierarchy and phenotypic suppression among Drosophila homeotic genes: The labial and empty spiracles genes. EMBO J. 1996;15:334–343. [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalez-Reyes A, Urquia N, Gehring WJ, Struhl G, Morata G. Are cross-regulatory interactions between homoeotic genes functionally significant? Nature. 1990;344:78–80. doi: 10.1038/344078a0. [DOI] [PubMed] [Google Scholar]
- 35.Schock F, et al. Phenotypic suppression of empty spiracles is prevented by buttonhead. Nature. 2000;405:351–354. doi: 10.1038/35012620. [DOI] [PubMed] [Google Scholar]
- 36.Mahaffey JW. Assisting Hox proteins in controlling body form: Are there new lessons from flies (and mammals)? Curr Opin Genet Dev. 2005;15:422–429. doi: 10.1016/j.gde.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Haerry TE, Gehring WJ. Intron of the mouse Hoxa-7 gene contains conserved homeodomain-binding sites that can function as an enhancer element in Drosophila. Proc Natl Acad Sci USA. 1996;93:13884–13889. doi: 10.1073/pnas.93.24.13884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.