Abstract
Performance improvements in DNA-modified surfaces required for microarray and biosensor applications rely on improved capabilities to accurately characterize the chemistry and structure of immobilized DNA molecules on micro-patterned surfaces. Recent innovations in imaging X-ray photoelectron spectroscopy (XPS) and time-of-flight secondary ion mass spectrometry (ToF-SIMS) now permit more detailed studies of micro-patterned surfaces. We have exploited the complementary information provided by imaging XPS and imaging ToF-SIMS to detail the chemical composition, spatial distribution and hybridization efficiency of amine-terminated single-stranded DNA (ssDNA) bound to commercial polyacrylamide-based, amine-reactive microarray slides, immobilized in both macro-spot and microarray diagnostic formats. Combinations of XPS imaging and small spot analysis were used to identify micro-patterned DNA spots within printed DNA arrays on slide surfaces and quantify DNA elements within individual microarray spots for determination of probe immobilization and hybridization efficiencies. This represents the first report of imaging XPS of DNA immobilization and hybridization efficiencies for arrays fabricated on commercial microarray slides. Imaging ToF-SIMS provided distinct analytical data on the lateral distribution of DNA within single array micro-spots before and after target hybridization. Principal component analysis (PCA) applied to ToF-SIMS imaging datasets demonstrated that the combination of these two techniques provides information not readily observable in ToF-SIMS images alone, particularly in identifying species associated with array spot non-uniformities (e.g., “halo” or “donut” effects often observed in fluorescence images). Chemically specific spot images were compared to conventional fluorescence scanned images in microarrays to provide new information on spot-spot DNA variations that affect current diagnostic reliability, assay variance and sensitivity.
Keywords: Dna, Esca, Fluorescence, Hybridization, Imaging, Multivariate Analysis, Nucleic Acid, Microarrays, Pca, Surface Characterization, Tof-sims, Xps, Diagnostics
INTRODUCTION
Patterning DNA onto surfaces has recently received considerable attention due to its applications in fundamental biology and biomedical research as genomic arrays, diagnostics and biosensors.1, 2 Several methods have been developed for fabricating micron-scale DNA patterns, including contact and non-contact printing of pre-synthesized DNA onto substrates, and in situ synthesis of microarrays using electrochemistry3 and photolithography.4–6 Micro-printing techniques are widely used for DNA microarray fabrication on commercial array slides containing hundreds to thousands of spotted features. The printing process generally involves spatially controlled delivery of nanoliter drops of DNA solutions onto reactively coated glass substrates using a robotic spotter, followed by evaporation of deposited liquid droplets within seconds. This rapid evaporative process produces increased solution ionic strength and solute concentrations in the drying DNA film, resulting in distinct differences in immobilized DNA structure, density and chemistry compared to bulk solution coupling reactions between DNA and surfaces.5, 7 A common observation is the formation of dry DNA spots with greater DNA density at the edges than in the middle as solution flows to the spot edge upon rapid evaporation.8 In addition, surface damage may also occur during the array printing process, especially with contact-based printing methods.6 The resulting immobilized DNA density and distribution within individual microarray spots have profound influences on the subsequent target capture performance.9, 10 Spot-to-spot variations in DNA surface density and distribution can therefore lead to inconsistent target capture, inaccurate data quantification and misleading results. Thus, accurate quantitative analysis of printed DNA microarray diagnostics is only possible if controlled and reliable spot uniformity (i.e., spot density, size and shape repeatability) is achieved.
Currently, fluorescence imaging (scanner-based methods and microscopy) are the most commonly used techniques to analyze and quantify fluorescently labeled DNA patterns.5 Although fluorescence imaging involves routine instrumentation with well-developed techniques widely available, it also has several limitations. For example, florescence signal generation is very sensitive to variations in surface molecular environments and does not provide chemical or structural information at each stage of the patterning process. Variable fluorescent DNA labeling and differences in quantum yields with position, label type and surface capture format all make accurate quantification difficult, and in general, chemical labeling of biological molecules with a fluorescent moiety can alter their natural activity, binding efficiency and capture kinetics. Optical imaging methods such as surface plasmon microscopy (SPR)11–13 provide quantitative images of film thickness or molecular coverage but are generally insensitive to distinct chemical species present on the surface. Atomic force microscopy provides information about nano-scale topography and phase-segregated domains on a sample surface14 but provides little information on surface chemical composition.
X-ray photoelectron spectroscopy (XPS) and time-of-flight secondary ion mass spectrometry (ToF-SIMS) are two commonly used surface analysis techniques to characterize elements and molecules present in the top 2–10 nm of a surface,15 with detection limits as low as nanograms per square centimeter.16 XPS and ToF-SIMS each has their own strengths and weaknesses with respect to generating surface chemical state information at high spatial resolution, but used together provide a powerful complementary set of techniques. The quantitative nature of XPS combined with its 2–10 nm sampling depth makes it an ideal technique for determining surface concentrations of biomolecules, including DNA.7, 17–22 We have recently demonstrated that XPS-determined phosphorus signal from the DNA backbone correlates well with 32P-radiolabeling19 and can be used to quantify both surface DNA probe and target concentrations.23 Innovations in X-ray focusing and lens/analyzer technology now permit XPS imaging at spatial resolutions less than 10 µm.24–27 Although this spatial resolution remains orders of magnitude above that obtained with other microscopy techniques, XPS has significant advantages in quantifying sample surface composition. In contrast to imaging XPS, imaging ToF-SIMS is a more surface sensitive technique (1–2 nm sampling depth), providing significantly higher spatial resolution allowing more detailed analysis of compositional variability within a biomolecular pattern on solid substrates.28–30 Previous studies have shown that static ToF-SIMS, in combination with multivariate analysis statistical methods such as principal component analysis (PCA), can provide the distribution of chemical species across a patterned surface at sub-micrometer resolution.31, 32
In this study, complementary imaging XPS, imaging ToF-SIMS and fluorescence imaging were compared, providing detailed characterization of chemical composition, spatial distribution and hybridization efficiency of amine-terminated single-stranded DNA (ssDNA) probes bound to aminereactive commercial microarray slides, immobilized in both macro-spot and microarray formats. Fluorescence scanning shows intra- and inter-spot DNA density differences and heterogeneities. Combining imaging XPS and small spot analysis, we identified DNA microarray regions on the slide surface and quantified DNA elements within individual microarray spots. Imaging ToF-SIMS was used to provide unambiguous measurements of lateral DNA distribution within micro-spots. PCA was applied to ToF-SIMS imaging datasets demonstrating that combining these two techniques improves information yield not readily observable in the ToF-SIMS images alone, particularly in identifying species causing such common non-uniformities in DNA spots such as the “halo” or “donut” effects often observed in microarray fluorescence images.
EXPERIMENTAL SECTION
Materials
Ultrapure water (UPW) was used for all solution preparation and rinsing (ASTM type I water, 18.2 MΩ·cm). High performance liquid chromatography (HPLC)-purified DNA oligomers (see Table 1) were purchased from TriLink Biotechnologies (San Diego, CA). All chemicals were used as received. Buffer salts, Tween20™, sarcosine, sodium dodecyl sulfate (SDS) and ethanolamine were ACS grade or better and purchased from Sigma-Aldrich (St. Louis, MO). 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide (EDC) and N-hydroxysuccinimide (NHS) were purchased from Acros organics (98%+ purity; Morris Plains, NJ). Polymer-coated commercial amine-reactive, polyacrylamide-based microarray slides were purchased from GE Healthcare (CodeLink™ activated slides, Piscataway, NJ) and stored according to vendor recommendations until used. Figure S1 (Supplemental Information) shows chemical structures for the various additives used in the DNA printing and hybridization.
Table 1.
Oligonucleotide sequences and terminal modifications
| DNA | Identifier | 5’-modification | Sequence | 3’-modification |
|---|---|---|---|---|
| Complementary probe1,2 | oligo1-NH2 | CTGAACGGTAGCATCTTGAC | -C6-NH2 | |
| Complementary probe1,2 | Cy3-oligo1-NH2 | Cy3-C6- | CTGAACGGTAGCATCTTGAC | -C6-NH2 |
| Non-complementary probe1 | oligo3-NH2 | GTCAAGATGCTACCGTTCAG | -C6-NH2 | |
| Target1,2 | oligo2 | GTCAAGATGCTACCGTTCAG | ||
| Target1,2 | Cy5-oligo2 | Cy5-C6- | GTCAAGATGCTACCGTTCAG | |
| Brominated target1,2,3 | Br-oligo2 | GTCAAGATGCTACCGTTCAG |
Used in microarray printing
Used in hand printing of macro-spots
The first 10 nucleotides at the 5’ end were brominated.
Macro-spot DNA Probe Immobilization
DNA oligonucleotides containing a 3’-terminal hexylamine group (see Table 1) were spotted onto activated CodeLink microarray slides. Briefly, slides were removed from vendor packaging and directly activated using standard carbodiimide chemistry with EDC/NHS according to a previously published protocol33 to ensure maximal and reliable amine-reactivity. Slides were then spotted with DNA solutions using a pipette (hand-spotting; 10 µL/spot) yielding defined macro-spotted DNA areas of approximately 5-mm diameter.7 DNA solutions (Cy3-oligo1-NH2:oligo1-NH2, 1:99) were spotted at 0, 1, 10, 20, and 40 µM in print buffer (150 mM sodium phosphate, pH 8.5 containing 0.001% Tween20 and 0.001% sarcosine). To minimize variations due to spot location, each DNA concentration was spotted in triplicate randomly across the slide. Hand-spotted samples were incubated overnight (>13 h) at 75% relative humidity, permitting drying to yield DNA film spots with immobilized density similar to microarray preparations.7
Microarray Printing
Oligonucleotides containing a 3’-terminal hexylamine group (Table 1) were printed onto activated33 CodeLink microarray slides using a non-contact Piezorray printer (Perkin-Elmer equipped with standard Perkin-Elmer piezorray pins and driven by Piezorray Microarray Printing System Version 1.1 software). Complementary (Cy3-oligo1-NH2:oligo1-NH2, 1:99) and non-complementary (oligo3-NH2) DNA probe solutions were deposited from print buffer volumes of ~333 pL +/− 33 (10% deviation) in replicates of 10 in alternating rows (layout of the printed microarray regions can be found in Figure S2, Supporting Information). Four different probe concentrations (0, 10, 20, and 40 µM) were used for microarray printing, with each array representing a single printed probe concentration. Duplicate arrays were printed on separate slides to allow comparison between probe-only and hybridized arrays (4 arrays/slide). Humidity during printing was approximately 60%. Under these print conditions, spots dry immediately to produce dried spot diameters of approximately 150 microns. Printed slides were incubated overnight (> 13 h) at room temperature under 75% relative humidity.
Post-Print Treatment and Hybridization with Target DNA
After printing and incubation, slides were treated according to the manufacturer’s recommendations. Slides were rinsed briefly with print buffer followed by UPW, and then immersed in blocking solution (50 mM ethanolamine in 0.1 M Tris, pH 9.0) at 50°C for 30 min to consume residual amine-reactive groups. Following blocking, slides were rinsed 3 times with UPW and then washed with 4X saline sodium citrate (SSC) containing 0.1% SDS at 50°C for 30 min (1X SSC: 15 mM sodium citrate and 150 mM NaCl). After washing, slides were rinsed 3 times with UPW and finally blown dry with nitrogen. Target hybridization was accomplished using either Lifterslips™ (Erie Scientific #22x50I-2-4711) or Coverwell™ perfusion chambers (Grace Bio-Labs # PC4L-A-1.0) for macro-spot and microarray samples, respectively. Briefly, 1 µM target DNA solution (1:99 Cy5-Oligo2:Br-Oligo2 or 1:99 Cy5-Oligo2:Oligo2) prepared in hybridization buffer (4X SSC/0.01% SDS) was applied to the appropriate hybridization chamber for 4 hr at room temperature and at 100% relative humidity. Following hybridization, slides were rinsed once with hybridization buffer, twice with 2X SSC/0.1% SDS (5 min each), once with 0.2X SSC (1 min), once with 4°C 0.1X SSC, and finally dried with nitrogen. All probe-only control spots were exposed to identical hybridization buffer and subsequent rinse steps. For macro-spot probe samples, the final rinse was UPW.
Fluorescence Imaging of Macro-spot and Microarray Surfaces
Modified slides were scanned using a ScanArray Express Microarray Imager (Perkin Elmer, Fremont, CA) with scanning resolutions of 10 microns for microarray samples and 50 microns for macro-spot samples. Slides were scanned using 2 channels, 543 nm and 633 nm, for Cy3- and Cy5-labeled samples, respectively.7 Laser power was set to 90% for all scans. For microarray samples, PMT sensitivity was set to 75% and 50% for probe and target measurements, respectively. For macro-spot samples, PMT sensitivity was set to 62% and 50% for probe and target measurements, respectively. High-resolution fluorescent images were acquired using an inverted Nikon TE2000U microscope equipped with a Sutter Lambda LS xenon source, a Prior scientific XYZ-stage, Chroma excitation/emission filters, and a Photometrics scientific CCD camera (Coolsnap-ES) controlled by Metamorph software (v.6.2r6, Molecular Devices, Downingtown, PA). Microarray spots were analyzed using the ScanArray software as previously described.7 Macro-spot samples were analyzed using ImageQuant software (Amersham Biosciences, v. 5.1) using grey scale image analysis. The use of separate software for analysis of the macro-spots was necessary due to the inability of the ScanArray software to handle millimeter-size spots.
XPS Analysis of DNA Macro-spot and Microarray Surfaces
All XPS measurements were performed on a Kratos Axis Ultra DLD X-ray photoelectron spectrometer employing a hemispherical analyzer for spectroscopy and a spherical mirror analyzer for imaging.24 Spectra and images were acquired with a monochromated Al-Kα X-ray source and a 0-degree take-off angle (TOA) in the “hybrid” mode. The TOA is defined as the angle between the sample surface normal and the axis of the XPS analyzer lens. A low-energy electron flood gun was used to minimize surface charging.
XPS data for macro-spot DNA samples were collected using an analysis area of 700 µm × 300 µm. For each sample, an initial compositional survey scan was acquired, followed by a detailed P2p scan using a pass energy of 80eV. High-resolution C1s spectra were also acquired for the fresh, unmodified CodeLink slides using a pass energy of 20eV and were charge-referenced to the C1s hydrocarbon peak set to 285.0eV. Values reported for the composition of fresh CodeLink slides and the DNA macro-spots were averages of values determined from three spots on two CodeLink microarray slides.
DNA spots on each microarray sample were located by taking initial XPS images of the relevant species (P2p, N1s, O1s, C1s and Si2p). Images were acquired at 400x400µm and 800x800µm fields of views at a pass energy of 80eV. Background region images were taken at a binding energy 15eV below each relevant peak. Background corrected images were obtained by subtraction of the background region image from the image at the peak of interest.26 Compositional analysis of individual microarray DNA spots was performed by collecting small-area region of interest (ROI) scans (50 µm × 50 µm) from the center of individual microarray spots. ROI spectra were acquired from a 400 µm field of view using an aperture size of 55 µm. An initial compositional survey scan was acquired at the specified location, followed by detailed (P2p) scans using a pass energy of 160eV. Four to six DNA spots from each 10×10 array were analyzed. Reported compositional data were averages of values determined from replicate spots. Data analysis was performed with Vision Processing data reduction software (Kratos Analytical Ltd.) and CasaXPS (Casa Software Ltd.).
ToF-SIMS Analysis of DNA Microarray Surfaces
ToF-SIMS data were acquired on an ION-TOF IV instrument (ION-TOF GmbH, Münster, Germany, University of Oregon) using a Bi+ primary ion source. Positive and negative ion images and spectra were acquired with a pulsed 25 keV, 1.3 pA primary ion beam in high current bunched mode from 200 µm × 200 µm areas on sample surfaces. All images obtained contained 128 × 128 pixels. These analysis conditions resulted in spatial resolution of approximately 2 µm. Data were collected using an ion dose below the static SIMS limit of 1×1012 ions/cm2. A low-energy electron beam was used for charge compensation on the DNA samples. The mass resolutions (m/Δm) for the negative spectra were typically between 6000 and 7500 for the (m/z) 25 peak in the negative spectra. Positive spectra were not used due to high sodium ion intensity from buffer salt-DNA ion exchange; this is a problem resulting from the inability to perform a final water rinse to remove counter ions and trapped Na within the Codelink matrix without also disrupting the hybridization. Principal component analysis (PCA) was performed on ToF-SIMS data as described previously31, 34 using a series of scripts written by NESAC/BIO for MATLAB (MathWorks, Inc., Natick, MA) (see supporting information for a detailed description of PCA).
RESULTS AND DISCUSSION
XPS Analysis of Surface-Immobilized and Hybridized DNA on Commercial Microarraying Substrates: Microarray vs. Macro-spot Format
Surface compositions of non-contact printed microarray spots (100–150 µm diameter) and hand-printed macro-spots (~5 mm diameter) were analyzed by XPS for direct comparison of DNA probe immobilization and hybridization efficiencies on CodeLink slides. The slide polymer chemistry has been reported elsewhere7, 8 and verified here by XPS to be consistent with polyacrylamide containing activated ester groups as attachment sites for aminated DNA probes. XPS compositional data of the fresh, as received, microarray slides indicate the presence of silicon, carbon, nitrogen, oxygen and trace ions (calcium and sodium) throughout the substrate surface (Table 2). The XPS C1s high-resolution spectrum of the unmodified microarray slide shows 70% C-C and C-H at 285 eV, 8% C-O and C-N at 286 eV and 22% N-C=O at 288 eV, similar to those published previously for Codelink slides7 and pure polyacrylamide35, but deviating slightly due to the detection of the underlying glass substrate signal (for more details see Figure S3, Supporting Information).
Table 2.
Compiled XPS compositional data for non-contact printed microarrays and hand-spotted DNA macro-spots on CodeLink slides.
| Sample | Atomic % (Std. dev) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| P 2p | N 1s | Si 2p | O 1s | C 1s | Na 1s | Ca 2p | Cl 2p | ||
| fresh, unmodified slide | 0.0 | 8.8 | 7.5 | 28.7 | 53.3 | 1.1 | 0.5 | 0.0 | |
| (0.0) | (0.1) | (0.3) | (0.3) | (0.5) | (0.1) | (0.0) | (0.0) | ||
| Microarray DNA | |||||||||
| printing buffer + blocking | 0.0 | 7.6 | 8.8 | 30.5 | 50.6 | 1.1 | 0.6 | 0.0 | |
| (0.0) | (0.5) | (0.5) | (1.5) | (1.4) | (0.3) | (0.2) | (0.0) | ||
| probe (40 µM) + blocking | 1.3 | 11.8 | 5.4 | 25.7 | 52.7 | 3.1 | 0.0 | 0.0 | |
| (0.2) | (1.1) | (0.4) | (0.8) | (1.0) | (0.7) | (0.0) | (0.0) | ||
| probe (40 µM) + blocking + target (1 µM) | 2.2 | 13.1 | 3.3 | 23.0 | 52.8 | 5.6 | 0.0 | 0.0 | |
| (0.1) | (0.9) | (0.4) | (1.7) | (2.5) | (0.6) | (0.0) | (0.0) | ||
| Macro-spot DNA | |||||||||
| printing buffer + blocking | 0.0 | 7.8 | 8.3 | 30.0 | 52.9 | 0.5 | 0.5 | 0.0 | |
| (0.0) | (0.9) | (0.5) | (0.7) | (1.3) | (0.2) | (0.1) | (0.0) | ||
| probe (40 µM) + blocking | 0.5 | 10.5 | 6.3 | 27.2 | 54.0 | 1.0 | 0.5 | 0.0 | |
| (0.1) | (0.8) | (0.5) | (0.6) | (0.6) | (0.4) | (0.1) | (0.0) | ||
| probe (40 µM) + blocking + target (1 µM) | 1.0 | 10.7 | 5.3 | 26.8 | 51.8 | 3.1 | 0.3 | 1.1 | |
| (0.1) | (0.6) | (0.7) | (1.2) | (1.6) | (0.3) | (0.2) | (0.2) | ||
XPS imaging combined with small-spot ROI analysis were used to provide quantitative surface composition for the microarrays immobilized with 0, 10, 20, and 40 µM DNA probe concentrations, and then hybridized with 1 µM DNA target. Importantly, the 0 µM (blank) concentration represents experimental controls of buffer-printed microarray slides exposed to identical incubation, post-immobilization wash and hybridization steps as the DNA-modified samples, but in the absence of DNA probes. (Figure 1 presents, to our knowledge, the first reported XPS elemental images (P, N, Na and Si) from individual spots in DNA microarrays (O and C images not shown). Background corrected P2p images (Figure 1) show higher signal intensity in the areas occupied by immobilized DNA micro-spots. Although N, Na and Si are present throughout the substrate surface, higher N1s and Na1s and lower Si2p signal intensities were also observed in the areas covered by the DNA micro-spots. Higher N and Na signal intensities correlate with nitrogen-containing DNA bases and sodium counter ions associated with the DNA polyphosphate backbone. DNA coverage in printed regions attenuates absolute signal intensity (i.e., Si) from the underlying glass substrate. In addition, XPS imaging provides the capability to distinguish between hybridized and unhybridized micro-spots on a commercial microarray slide without use of radioactive or fluorescent labels. Figure 2 shows XPS overlay images displaying P, N, Na and Si signal intensities from a hybridized (Figure 2a) and an unhybridized Figure 2b) microarray slide. Prior to exposing the microarray slide to complementary target capture, similar XPS P2p, N1s and Na1s signal intensities were detected from all micro-spots containing complementary or non-complementary probe sequences. After target hybridization, micro-spots containing complementary probe sequences exhibited higher XPS P2p, N1s and Na1s signal intensities compared to those containing non-complementary probe sequences, as expected.
Figure 1.
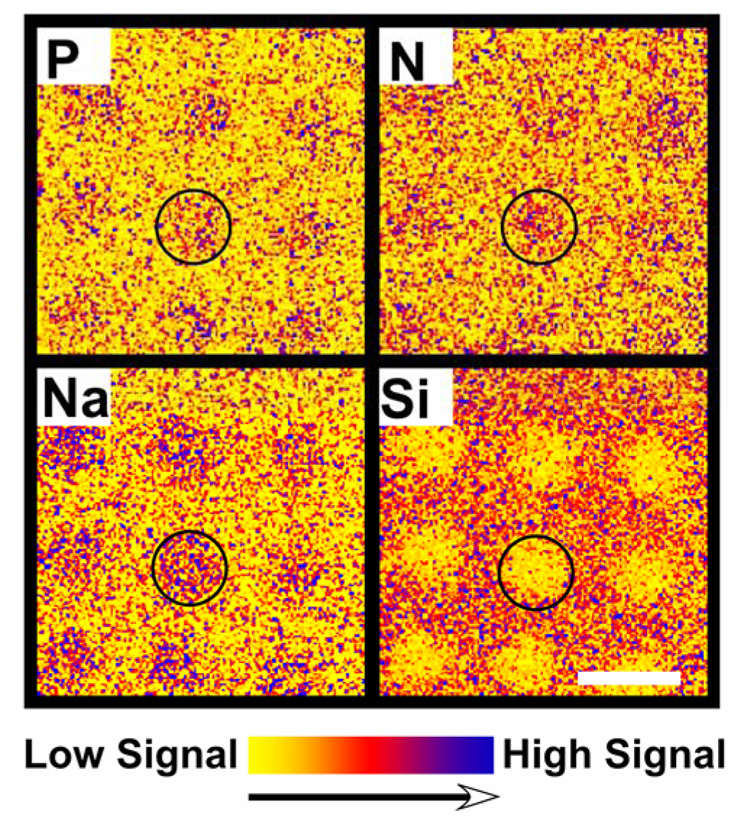
XPS elemental images (800 µm × 800 µm) of aminated DNA probes printed onto CodeLink microarray slides at 40 µM DNA concentration. While phosphorus is unique to DNA, silicon is unique to the substrate. In combination, these elemental images enable unambiguous identification of the spatial distribution of DNA for XPS region of interest (ROI) compositional analyses of the printed DNA microarray spots. The scale bar represents 200 µm.
Figure 2.
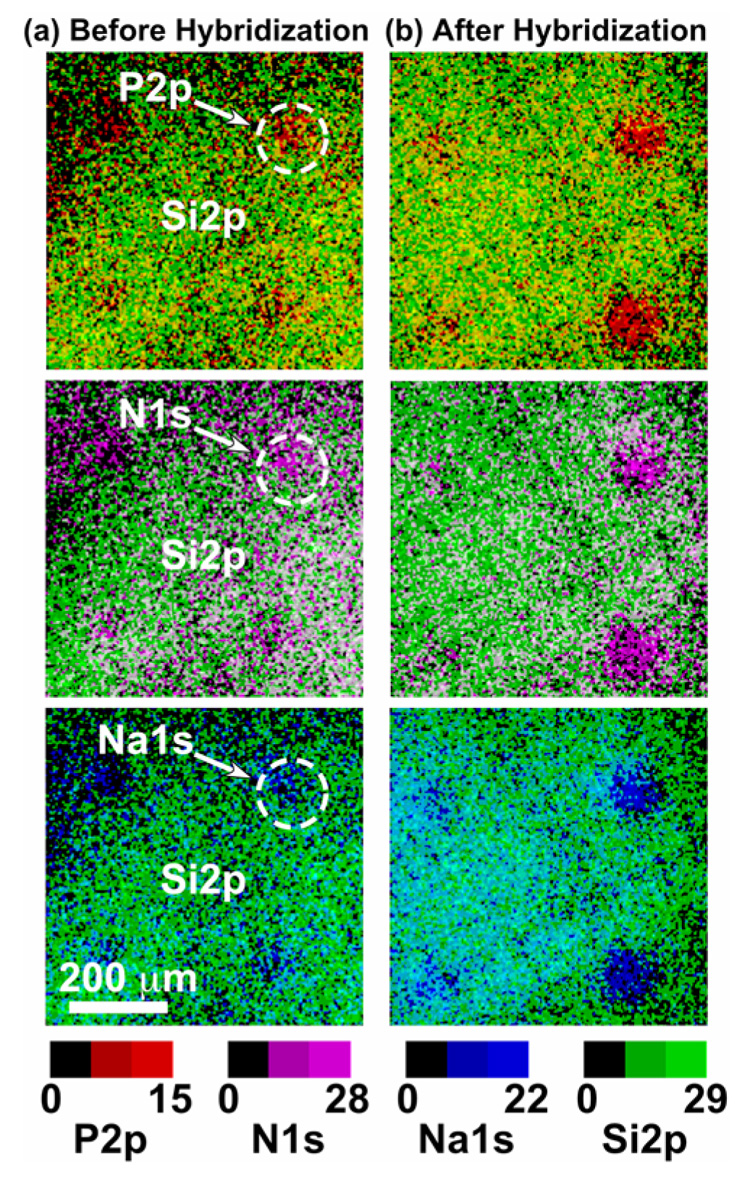
XPS overlay of phosphorous (P2p), nitrogen (N1s), sodium (Na1s) with the substrate silicon (Si2p) signal intensity images (800 × 800 µm) from printed DNA probes on CodeLink microarray slides (a) before and (b) after target hybridization. Consistent with target capture signal, the XPS P2p, N1s and Na1s signal intensities from the hybridized regions are significantly higher than from the unhybridized regions.
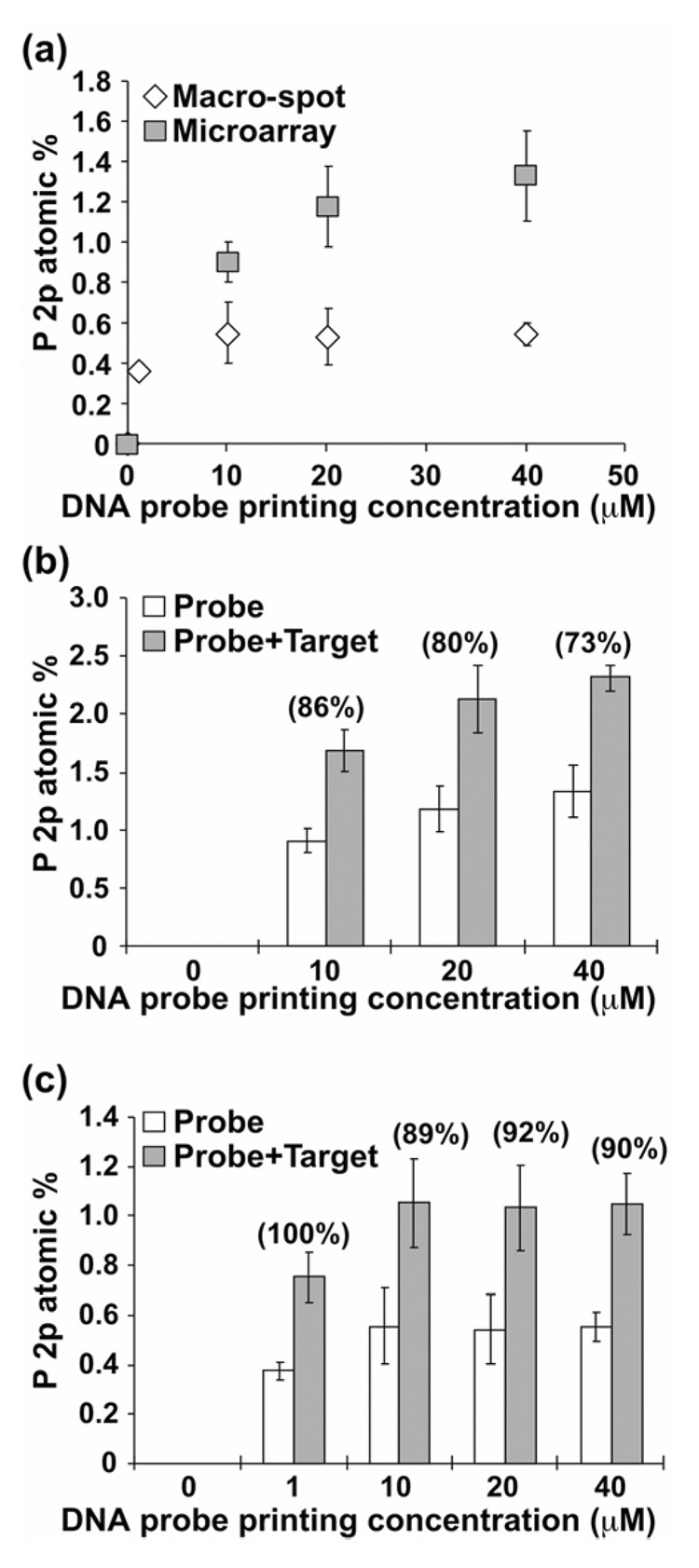
To further determine target hybridization efficiencies for microarrays printed at various printed probe concentrations (e.g., 10, 20 and 40 µM), small area ROI analyses were performed on micro-spots to obtain quantitative individual spot DNA elemental compositions both before and after hybridization. Consistent with XPS elemental images (e.g., Figure 2), regions with hybridized DNA targets show higher percentages of phosphorus (2.2 atomic percent or at%), nitrogen (13.1 at%) and sodium (5.6 at%) and lower substrate oxygen (23.0 at%) and silicon (3.3 at%) signals compared to that obtained for unhybridized non-complementary probe spots (P = 1.3, N = 11.8, Na = 3.1, O = 25.7 and Si = 5.4 at%, Table 2). Since substantial amounts of nitrogen (~8 at%) are present in the unprinted CodeLink polymer layer,7 DNA phosphorus is the only unique characteristic element useful to quantify relative amounts of surface-immobilized and hybridized DNA oligomers from the various probe printing concentrations. DNA surface coverage is proportional to phosphorus atomic concentration and has been previously reported as a quantification method for immobilized DNA on gold22, 23 as well as on polymer-modified silicon substrates.7, 17 Figures 3a and b show relative amounts of surface-immobilized probe and hybridized DNA obtained for each probe printing concentration from small area XPS analysis. As seen in Figure 3a, P2p at% from the DNA micro-spots increases with increasing spotting probe solution concentration and did not saturate for the concentration range investigated. Target hybridization efficiencies shown in parentheses above each concentration in Figure 3b were derived from P2p signal as a percentage of probe molecules hybridized [((P2p at% of hybridized spot/P2p at% of probe spot) −1) × 100%]. A hybridization efficiency of 86% was obtained for micro-spots printed at the lowest probe concentration of 10 µM. At higher probe printing concentrations (20 and 40 µM), slightly lower hybridization efficiencies (80% and 73%) were obtained. The reduction of hybridization efficiency at higher probe coverage has been reported previously,7, 9, 23 explained by steric and electrostatic crowding effects in closely packed DNA probes that hinder DNA target duplex formation on the surface.36
Figure 3.
Relative amounts of DNA probe immobilized onto CodeLink slides are compared in microarray and macro-spot formats (a). Amounts of DNA on CodeLink surfaces are proportional P2p atomic percent (at%).7 Hand-spotted macro-spot printing concentrations are 1, 10, 20 and 40 µM (150 mM sodium phosphate, pH 8.5). Microarrays were printed at three DNA probe concentrations (10, 20 and 40 µM) under the same printing conditions as the macro-spot DNA. Resulting DNA probe surface density is higher for microarrays than macro-spots at each given probe printing concentration. Target hybridization efficiencies for the microarray (b) and macro-spots (c) shown in (parentheses) above each concentration were derived as a percentage of probe molecules hybridized [((P2p at% of hybridized spot/P2p at% of probe spot) −1) × 100%]. Lower hybridization efficiencies were obtained on the microarray spots with higher surface probe densities. Hybridization efficiency slightly above 100% was rounded to 100%.
One key difference between robotic microarray spotting and bulk media surface immobilization (i.e., large area hand-spotting or solution phase capture) is the effect of probe spot drying. Microarray printing delivers nanoliter drops of DNA solution to the assay surface that evaporate within seconds, minimizing opportunities for equilibrium DNA covalent reactions with the surface and yielding a dried DNA aggregated film. This rapid evaporative process therefore produces substantially different DNA immobilization states in spots compared to slow-or non-drying bulk media reactions.7
To compare immobilized probe and hybridized target densities obtained from these two different printing methods, DNA probes were hand-printed in macro-spot formats (~5 mm diameter) using the same printing conditions as the microarrays and analyzed by XPS. XPS compositional data for the macro-spots are summarized in Table 2 and Figures 3a and c. Reduced DNA (P and N) and increased substrate (Si and O) signals were detected from macro-spots versus microarrays for the same probe printing concentrations (Table 2). Figure 3a shows that lower percentages of phosphorus, indicative of DNA surface density, were observed for macro-spots over the entire probe printing concentration range (0–40 µM). Amounts of immobilized probe in macro-spots saturate at 10 µM probe solution concentration. Observed differences in amounts of immobilized DNA between macro- and micro-spot formats can be explained by different spot drying rates.7 Rapid evaporation during microarray printing significantly increases ionic strength of the spotting buffer as well as DNA concentration, promoting higher DNA probe surface immobilization density.7 Target hybridization increases the percentage of phosphorus and nitrogen on macro-spots while decreasing the oxygen and silicon substrate signals, as observed with DNA microarrays (Table 2). Lower probe density on the macro-spots reduces steric and electrostatic problems on the surface, yielding higher hybridization efficiencies (Figure 3c). A hybridization efficiency of 100% was obtained on the lowest probe concentration regions (DNA spotting concentration of 1 µM). At the higher probe spotting concentrations (10–40 µM), a small reduction in target hybridization efficiency (~90%) is again observed.
Fluorescence Analysis of Surface-Immobilized and Hybridized DNA
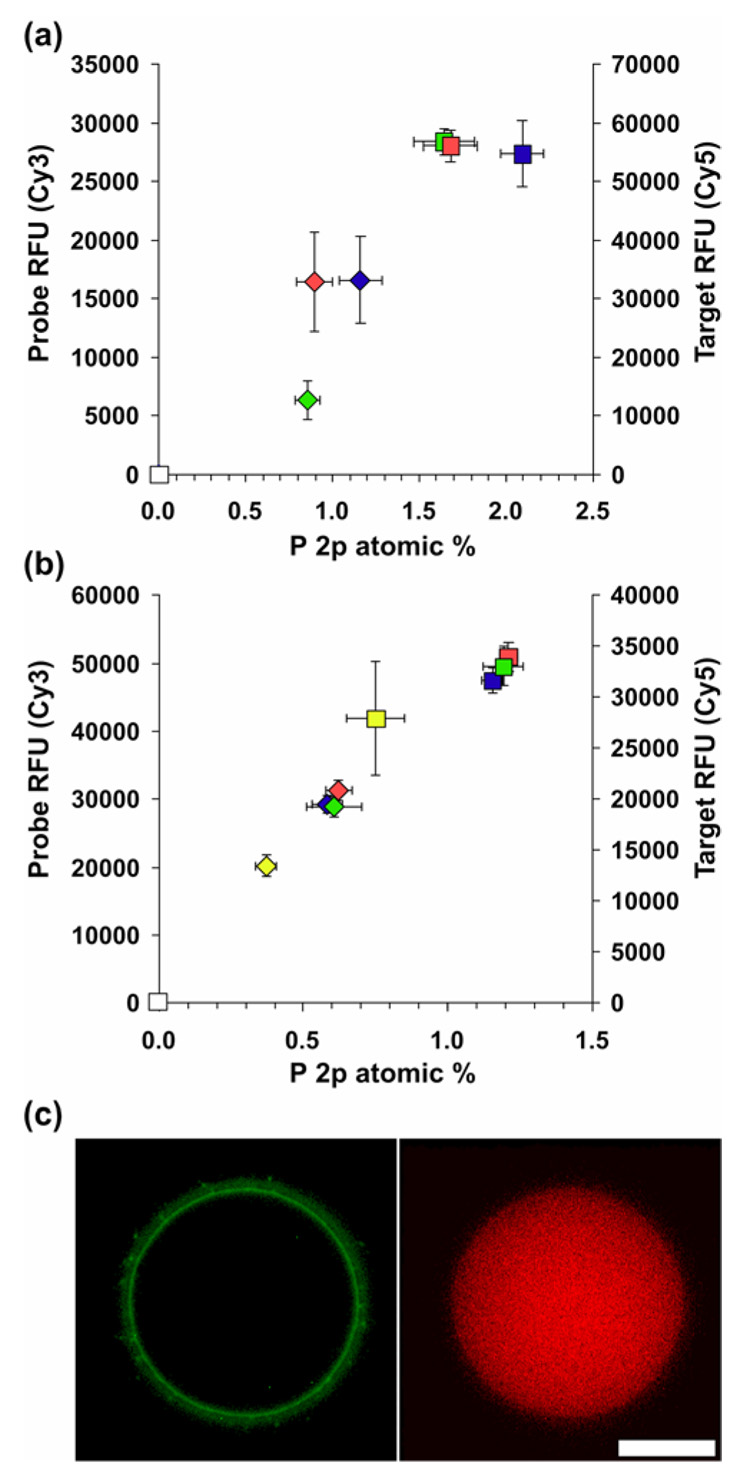
Printed slides were compared using a fluorescence scanner to provide complementary, more conventional information regarding relative density and homogeneity of probes and targets in microarray and macro-spots. Figures 4a and b show correlations between relative fluorescence signal (Cy3 probe and Cy5 target) and XPS P2p at% (shown in Figure 3) for microarray and macro-spot samples. Data show reasonable correlations between RFUs and P2p at%, both representative of immobilized or hybridized oligo densities. Under current experimental conditions (e.g., standard 75% humidity), large variations in fluorescence intensity in the 20–40 µM range (hand-spotted) and 10–40 µM range (printed array) were absent due to spot solution evaporation as discussed above. At 1 µM coupling concentration (hand-spotting, Fig. 4b) macro-spot RFUs were different compared to those for higher coupling concentrations. Although XPS results for hand-spotted macrospot data agree well with fluorescence measurements, XPS proved to be more sensitive to variations in probe density for microspots compared to fluorescence measurements. Additionally, fluorescence measurements provide relative, qualitative assessment of hybridization efficiencies, while XPS provides quantitative % efficiencies. Hand-spotted samples incubated overnight at 100% humidity to better duplicate bulk immobilzations where evaporation effects are absent produced a much greater dependence of resulting RFU on coupling concentration as previously shown7 (data not shown). This is likely due to a combination of bulk phase aqueous hydrolysis and neutralization of surface reactive ester groups, and diffusion-limiting DNA-surface coupling reactions.7, 33
Figure 4.
Correlation between P 2p atomic % from XPS and relative fluorescence units (RFU) data collected from microarray printed (a) and hand-spotted (b) samples. (c) fluorescent image of printed probe (left) and hybridized target (right) collected using a fluorescent microscope showing a “halo effect”; scale bar represents 50 micrometers. For this analysis, all samples were incubated at 75% humidity overnight to facilitate probe covalent attachment. Symbol key: diamonds, probe; squares, target. Probe concentration key: white, 0 µM; yellow, 1 µM; green, 10 µM; red, 20 µM; blue, 40 µM.
Unlike XPS, fluorescence measurements can provide high-resolution area information about the homogeneity of array spots as seen in Fig. 4c. The halo effect shown in Figure 4c is much more prevalent when using higher resolution fluorescence microscopy compared with standard fluorescent scanning or imaging XPS. XPS is able to detect and quantify total probe surface presence, but is unable to resolve the 1% Cy3-labeled probe doped into the probe print solution. High resolution fluorescence images shows that this 1% Cy3-labeled probe has segregated to the spot periphery producing the observed fluorescent halo effect. However, from the uniform target hybridization fluorescent image, it is clear that although the 1% labeled probe resides at the periphery, the printed spot has active, unlabeled probe immobilized throughout its area. At this time we do not have a satisfactory explanation for the observed effect. However, we will show in a related study that the amount of Cy3-labeled probe doped into the probe print solution (0 to 100%) dramatically affects spot homogeneity as examined using fluorescence microscopy. Given these results, the practice of doping fluorescent probe into unlabeled probe at low concentrations to reduce the amount of the more expensive fluorescently labeled oligo, may produce unrepresentative images of arrays when scanned, thus making interpretation difficult. Observation of halo or donut features in many printed microarrays,8, 33 has confounded microarray fluorescence analysis and data interpretation.37, 38 Reasons for these defective printed features are not elucidated. Therefore, to determine if the observed fluorescence halo was correlated to specific molecules in the print solution, imaging ToF-SIMS was utilized to examine individual micro-spots.
ToF-SIMS Analysis of Microarray Spot Uniformity
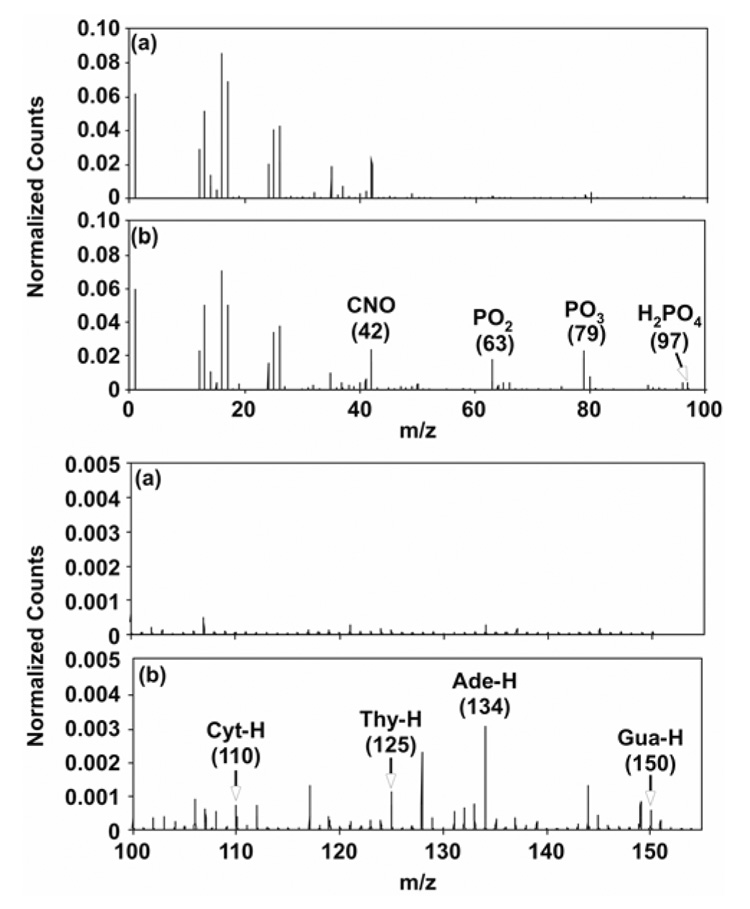
ToF-SIMS was used to characterize the distribution of DNA molecules within individual DNA micro-spots via detection of the characteristic DNA ion fragments (i.e., DNA bases and phosphate backbone).18, 21, 39, 40 Negative ion ToF-SIMS ROI spectra from the substrate and from DNA micro-spotted regions are shown in Figures 5a and b. The negative ion spectra from the DNA region (Figure 5b) show DNA phosphate fragments (PO−, PO2−, PO3− and H2PO4−) at m/z 47, 63, 79 and 97, respectively.18, 21, 39, 40 DNA bases, including adenine (Ade-H, m/z = 134), thymine (Thy-H, m/z = 125), guanine (Gua-H, m/z = 150) and cytosine (Cyt-H, m/z = 110), were also detected by ToF-SIMS in negative ion mode.18, 21 Absence of these peaks in the negative spectra from the unspotted substrate region (Figure 5a) confirm their origin from surface-immobilized DNA molecules. Individual negative ion ToF-SIMS images from selected masses reveal the distribution of printed DNA within an unhybridized micro-spot containing non-complementary DNA probes (Figure 6a), and a hybridized micro-spot containing complementary probes (Figure 6b) from the microarray region printed with 40 µM DNA solution. Detectable spot diameter from the ToF-SIMS POx- images was approximately 150 µm, comparable to that observed using fluorescence microscopy after printing (Figure 4c). These ToF-SIMS images, acquired at a spatial resolution of approximately 2 µm, indicate that printed DNA molecules are distributed non-uniformly within individual microarray spots upon drying. In addition, ToF-SIMS Si images of the unhybridized micro-spots reveal “halo” features around the probe spot consistent with those seen in fluorescent images (Figure 4c). (PCA analysis of these “halos” is presented later in this paper.) Also consistent with XPS data, images for characteristic DNA fragments (m/z 47, 63, 79, 97, 110, 125, 134, and 150) show higher signal intensities for the hybridized DNA micro-spot (Figure 6b).
Figure 5.
Negative ion ToF-SIMS ROI spectra from the (a) substrate and the (b) DNA regions of the microarray surface. The DNA region shows characteristic nucleic acid peaks at m/z 42 (CNO−), 63 (PO2−), 79 (PO3−), 97 (H2PO4−), 110 (C4H4N3O−, Cyt-H), 125 (C5H5N2O2−, Thy-H), 134 (C5H4N5−, Ade-H), and 150 (C5H4N5O− Gua-H) that were absent or present at much lower intensities in the background substrate region.
Figure 6.
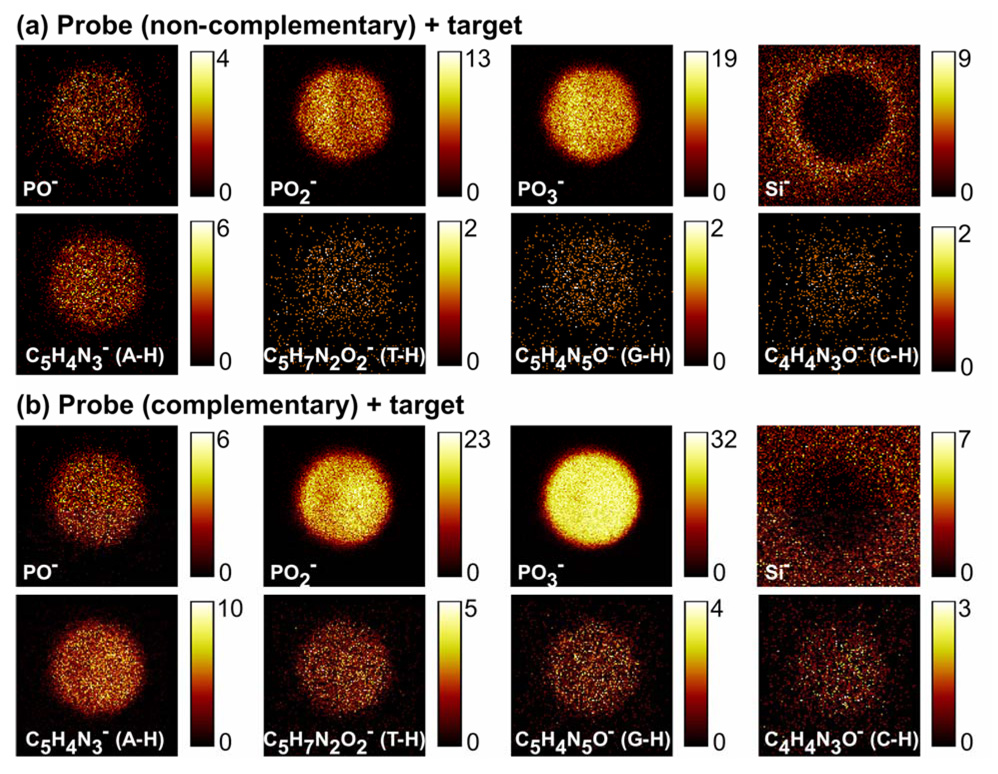
Representative negative ion ToF-SIMS images showing the distribution of DNA and substrate fragments within single (a) unhybridized and (b) hybridized microarray spot. The DNA fragments are localized to the non-contact printed regions but distributed inhomogeneously within the micro-spot. The Si image from the unhybridized probe spot (a) showed a “halo” feature around the spot. Brighter pixel intensity corresponds to higher DNA or substrate signals (counts per pixel). Images are 200 µm ×200 µm.
ToF-SIMS was also used to identify DNA hybridization signal by hybridizing a complementary target sequence having 50% of the DNA bases each modified with one Br atom (Table 1 and Figure 7a). Printed probe microarrays exposed to Br-modified DNA complementary targets produced strong Br signals from hybridized probe spots (Figure 7b and c) compared to non-complementary Br-containing targets (controls). ToF-SIMS’ intrinsic high sensitivity in detecting brominated species, as well as the ability to acquire images with submicron spatial resolution, opens the possibility to exploit this analytical method to determine hybridization uniformity across single microarray spots.
Figure 7.
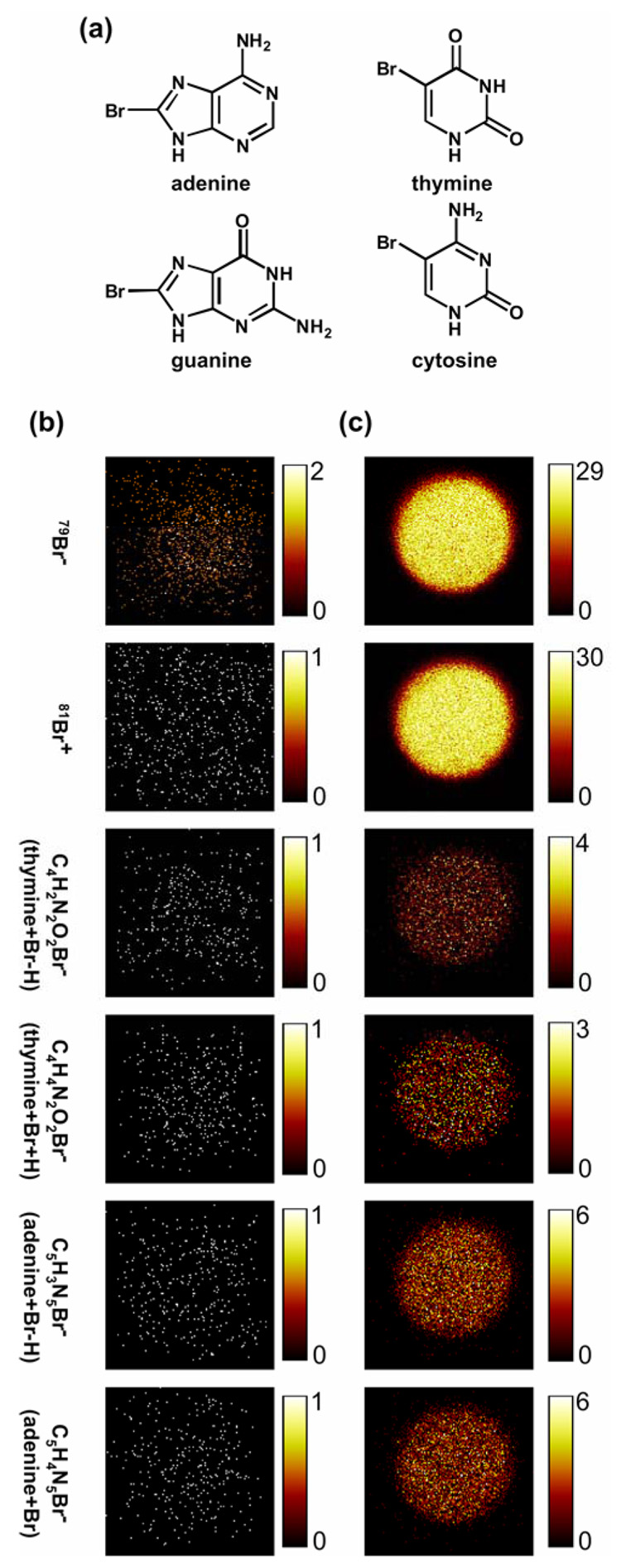
(a) Bromine modification of DNA target (50% brominated DNA bases comprise the DNA target sequence). Representative negative ion ToF-SIMS images showing no Br fragments detected from the non-complementary (unhybridized) micro-spots (b). Microarrays exposed to Br-modified DNA targets produce strong Br signals for complementary (hybridized) micro-spots after target hybridization (c). Brighter pixel intensity corresponds to higher DNA or substrate signals (counts per pixel). Images are 200 µm × 200 µm.
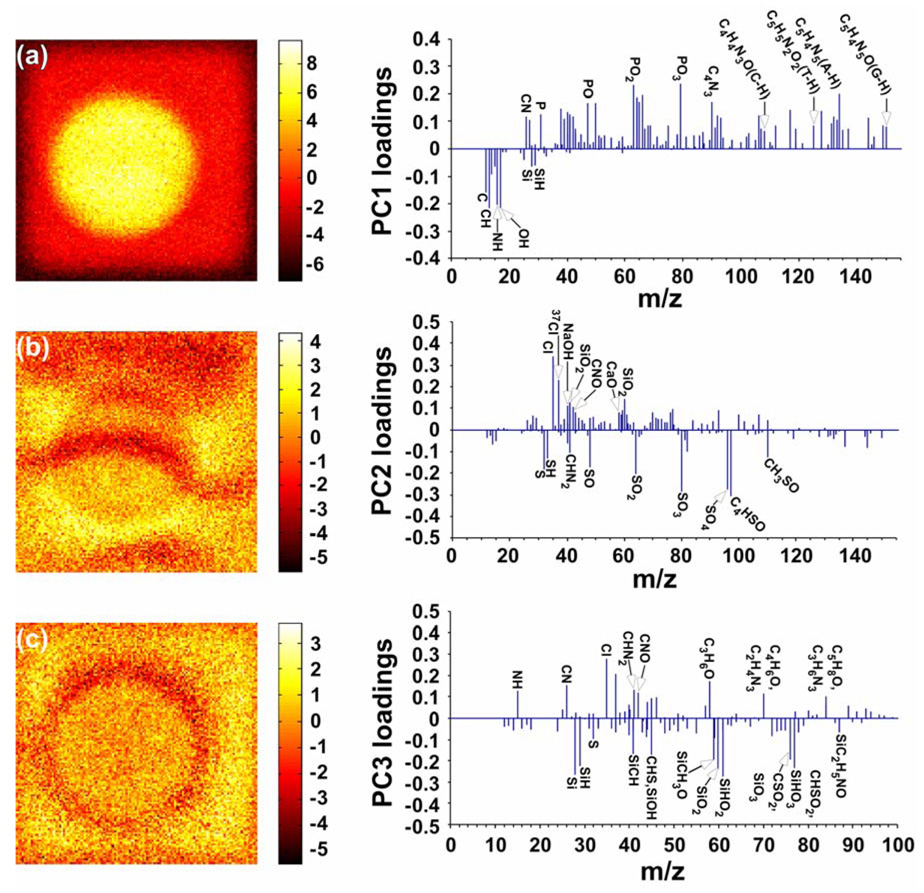
In addition to the DNA molecular fragments identified above, the energetic SIMS process yields hundreds of peaks in the 0–200 m/z range, making the interpretation of ToF-SIMS data difficult. To simplify data interpretation and identify image features related to other chemical species (e.g., salt ions, detergent molecules, polymer layer, etc. For more details see Figure S1, Supporting Information), a multivariate analysis technique, PCA, was used for more detailed analyses of the ToF-SIMS images as described in the Supporting Information section.31, 34 PCA was performed on the ToF-SIMS negative ion image of the unhybridized DNA micro-spot shown in Figure 6a to gain a better understanding of the chemical species related to the halo feature. The first three image scores and loadings from PCA are shown in Figures 8a, b, and c. Principal component 1 (PC 1, Figure 8a) clearly distinguishes the image features that correspond to the DNA micro-spot (bright regions) and the substrate (dark regions). From the PC 1 loadings plot (Figure 8a) we confirmed that most major peaks with positive PC 1 loadings are associated with the micro-spot region in the ToF-SIMS images and are phosphate- and nitrogen-containing DNA fragments, while most major peaks with negative PC1 loadings are hydrocarbon fragments and silicon-containing species from the substrate polymer layer. PC 2 (Figure 8b) reflects the image features that correspond to the salt ions, including Cl−, NaOH−, CaO−, etc, (bright regions) and SDS fragments (dark regions). PC 3 (Figure 8c) captures the image feature that corresponds to the halo around the probe spot (dark regions). PC 3 loadings plot (Figure 8c) indicate that most major peaks with negative PC 3 loadings associated with the halo in the ToF-SIMS image are silicon- and sulfur-containing fragments possibly from the polymer-coated glass substrate exposed as a result of polymer layer damage from the microarray printing process (for more details see Figures S4 and S5, Supporting Information) or from silicon-containing contaminants wicking to the outside of the spot upon spotting. Capillary phenomena during drying have been suggested as the reason for the appearance of ring-like halo features in other contexts (e.g., drying of nanoparticle solutions).41, 42 Other factors such as surface tension, droplet shape and droplet impact on the substrate could also play a role. Further study is required to address this issue in further detail.
Figure 8.
Image scores and loadings for PC 1 (a), PC 2 (b) and PC 3 (c) for an unhybridized micro-spot (negative ion images). PC 3 loadings (c) from the negative ToF-SIMS image data matrix indicate that the “halo” feature detected in spots by ToF-SIMS imaging is characterized by Si-containing fragments from the polymer-coated glass substrate. Images are 200 µm × 200 µm.
CONCLUSIONS
Imaging XPS and imaging TOF-SIMS are complementary, sensitive tools for analysis of elemental composition, chemical structure, relative density and spatial distributions of micro-patterned DNA arrays on glass substrates. Combined with routine fluorescence imaging, a more complete assessment of the chemistry and physical disposition of DNA array spotting can be realized. Individual DNA microarray spots were analyzed at high-resolution for the first time. A combination of XPS imaging and small spot analysis allowed identification of the micro-patterned DNA arrays and quantification of DNA elements within individual microarray spots to determine probe immobilization and hybridization efficiencies. XPS comparisons of DNA immobilized in both macro-spot and microarray formats demonstrated distinct differences in probe densities and hybridization efficiencies resulting from the two different printing processes (non-equilibrium drying microarray printing process vs. macro-spot reactions). Imaging ToF-SIMS provided different information on DNA spatial distribution and relative density, even against a complex organic matrix background. Bromine modification of the DNA bases provides unique target DNA fragments in ToF-SIMS data and high quality images of DNA micro-spots with little interference from the other organic species present in the surface region. Finally, application of PCA to ToF-SIMS imaging datasets provided new, unique information not readily observable in the univariate ToF-SIMS images alone, allowing identification of species involved in spot non-uniformities (e.g., “halo” often observed in fluorescence images). The ability to accurately quantify surface-immobilized DNA molecules with these methods is more convenient than radiolabeling and more informative than fluorescence scanning alone. The approach is likely to prove extremely useful in the future development and optimization of micro-patterned DNA surfaces to improve the performance and accuracy of genomic arrays and biosensor applications. Obtaining detailed information about distribution of chemical species within a DNA microspot is the first step in developing correlations between the DNA surface properties (i.e., structure and composition) and hybridization properties at the microscopic level. This information is also needed to determine the influence of experimental conditions (print additives, non-equilibrium drying, buffer, etc.) on the DNA surface structure and composition.
Supplementary Material
Experimental data is available as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGMENTS
The authors gratefully acknowledge support from NESAC/BIO (NIH Grant No. EB-002027) and NIH Grant EB-001473. Dan Graham is thanked for assistance with PCA analysis. We also thank Steve Golledge for expert technical assistance with the ToF-SIMS experiments performed at the Center for Advanced Materials Characterization in Oregon. Ping Gong is thanked for supplying patterned arrays used for exploratory XPS imaging experiments done prior to experiments reported in this study.
REFERENCES
- 1.Bejjani BA, Shaffer LG. J. Mol. Diagn. 2006;8:528–533. doi: 10.2353/jmoldx.2006.060029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamartine J. Mater. Sci. Eng. C-Biomimetic Supramol. Syst. 2006;26:354–359. doi: 10.1016/j.msec.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egeland RD, Southern EM. Nucleic Acids Res. 2005;33:e125. doi: 10.1093/nar/gni117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbulovic-Nad I, Lucente M, Sun Y, Zhang MJ, Wheeler AR, Bussmann M. Crit. Rev. Biotechnol. 2006;26:237–259. doi: 10.1080/07388550600978358. [DOI] [PubMed] [Google Scholar]
- 5.Dufva M. Biomol. Eng. 2005;22:173–184. doi: 10.1016/j.bioeng.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Pirrung MC. Angew. Chem.-Int. Edit. 2002;41:1276–1289. doi: 10.1002/1521-3773(20020415)41:8<1276::aid-anie1276>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 7.Gong P, Harbers GM, Grainger DW. Anal. Chem. 2006;78:2342–2351. doi: 10.1021/ac051812m. [DOI] [PubMed] [Google Scholar]
- 8.Ramakrishnan R, Dorris D, Lublinsky A, Nguyen A, Domanus M, Prokhorova A, Gieser L, Touma E, Lockner R, Tata M, Zhu XM, Patterson M, Shippy R, Sendera TJ, Mazumder A. Nucleic Acids Res. 2002;30:e30. doi: 10.1093/nar/30.7.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson AW, Heaton RJ, Georgiadis RM. Nucleic Acids Res. 2001;29:5163–5168. doi: 10.1093/nar/29.24.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herne TM, Tarlov MJ. J. Am. Chem. Soc. 1997;119:8916–8920. [Google Scholar]
- 11.Piliarik M, Vaisocherova H, Homola J. Biosens. Bioelectron. 2005;20:2104–2110. doi: 10.1016/j.bios.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 12.Shumaker-Parry JS, Aebersold R, Campbell CT. Anal. Chem. 2004;76:2071–2082. doi: 10.1021/ac035159j. [DOI] [PubMed] [Google Scholar]
- 13.Wolf LK, Fullenkamp DE, Georgiadis RM. J. Am. Chem. Soc. 2005;127:17453–17459. doi: 10.1021/ja056422w. [DOI] [PubMed] [Google Scholar]
- 14.Wadu-Mesthrige K, Xu S, Amro NA, Liu GY. Langmuir. 1999;15:8580–8583. [Google Scholar]
- 15.Castner DG, Ratner BD. Surf. Sci. 2002;500:28–60. [Google Scholar]
- 16.Wagner MS, McArthur SL, Shen MC, Horbett TA, Castner DG. J. Biomater. Sci.-Polym. Ed. 2002;13:407–428. doi: 10.1163/156856202320253938. [DOI] [PubMed] [Google Scholar]
- 17.Shen G, Anand MFG, Levicky R. Nucleic Acids Res. 2004;32:5973–5980. doi: 10.1093/nar/gkh932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee CY, Canavan HE, Gamble LJ, Castner DG. Langmuir. 2005;21:5134–5141. doi: 10.1021/la0472302. [DOI] [PubMed] [Google Scholar]
- 19.Lee C-Y, Gong P, Harbers GM, Grainger DW, Castner DG, Gamble LJ. Anal. Chem. 2006;78:3316–3325. doi: 10.1021/ac052137j. [DOI] [PubMed] [Google Scholar]
- 20.Lee C-Y, Gamble LJ, Grainger DW, Castner DG. Biointerphases. 2006;1:82–92. doi: 10.1116/1.2219110. [DOI] [PubMed] [Google Scholar]
- 21.May CJ, Canavan HE, Castner DG. Anal. Chem. 2004;76:1114–1122. doi: 10.1021/ac034874q. [DOI] [PubMed] [Google Scholar]
- 22.Petrovykh DY, Kimura-Suda H, Whitman LJ, Tarlov MJ. J. Am. Chem. Soc. 2003;125:5219–5226. doi: 10.1021/ja029450c. [DOI] [PubMed] [Google Scholar]
- 23.Gong P, Lee C-Y, Gamble LJ, Castner DG, Grainger DW. Anal. Chem. 2006;78:3326–3334. doi: 10.1021/ac052137j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walton J, Fairley N. Surf. Interface Anal. 2006;38:1230–1235. [Google Scholar]
- 25.Walton J, Fairley N. Surf. Interface Anal. 2006;38:388–391. [Google Scholar]
- 26.Vohrer U, Blomfield C, Page S, Roberts A. Appl. Surf. Sci. 2005;252:61–65. [Google Scholar]
- 27.Walton J, Fairley N. Surf. Interface Anal. 2004;36:89–91. [Google Scholar]
- 28.Belu AM, Yang ZP, Aslami R, Chilkoti A. Anal. Chem. 2001;73:143–150. doi: 10.1021/ac000771l. [DOI] [PubMed] [Google Scholar]
- 29.Yang ZP, Belu AM, Liebmann-Vinson A, Sugg H, Chilkoti A. Langmuir. 2000;16:7482–7492. [Google Scholar]
- 30.Belu AM, Graham DJ, Castner DG. Biomaterials. 2003;24:3635–3653. doi: 10.1016/s0142-9612(03)00159-5. [DOI] [PubMed] [Google Scholar]
- 31.Wagner MS, Graham DJ, Castner DG. Appl. Surf. Sci. 2006;252:6575–6581. [Google Scholar]
- 32.Tyler BJ. Appl. Surf. Sci. 2006;252:6875–6882. [Google Scholar]
- 33.Gong P, Grainger DW. Surf. Sci. 2004;570:67–77. [Google Scholar]
- 34.Wickes BT, Kim Y, Castner DG. Surf. Interface Anal. 2003;35:640–648. [Google Scholar]
- 35.Garg DH, Lenk W, Berwald S, Lunkwitz K, Simon F, Eichhorn KJ. J. Appl. Polym. Sci. 1996;60:2087–2104. [Google Scholar]
- 36.Heaton RJ, Peterson AW, Georgiadis RM. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3701–3704. doi: 10.1073/pnas.071623998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li QH, Fraley C, Bumgarner RE, Yeung KY, Raftery AE. Bioinformatics. 2005;21:2875–2882. doi: 10.1093/bioinformatics/bti447. [DOI] [PubMed] [Google Scholar]
- 38.Tran PH, Peiffer DA, Shin Y, Meek LM, Brody JP, Cho KWY. Nucleic Acids Res. 2002;30 doi: 10.1093/nar/gnf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hellweg S, Jacob A, Hoheisel JD, Grehl T, Arlinghaus HF. Appl. Surf. Sci. 2006;252:6742–6745. [Google Scholar]
- 40.Hashimoto H, Nakamura K, Takase H, Okamoto T, Yamamoto N. Appl. Surf. Sci. 2004;231–2:385–391. [Google Scholar]
- 41.Maillard M, Motte L, Ngo AT, Pileni MP. J. Phys. Chem. B. 2000;104:11871–11877. [Google Scholar]
- 42.Ohara PC, Heath JR, Gelbart WM. Angew. Chem.-Int. Edit. Engl. 1997;36:1078–1080. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental data is available as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.