Abstract
Clinical isolates of Klebsiella pneumoniae were tested for a correlation between tigecycline MIC and expression of ramA by using real-time PCR. At MICs of 4 and 8 μg/ml, the expression of ramA was statistically significantly different from MICs of 2 μg/ml or less, supporting the tigecycline susceptibility breakpoint of ≤2 μg/ml for K. pneumoniae.
Tigecycline is a novel expanded-broad-spectrum glycylcycline antibiotic that is not affected by classical tetracycline resistance mechanisms, including ribosomal protection and efflux by tetracycline-specific pumps (8). Decreased tigecycline susceptibility in gram-negative bacteria is associated with constitutive overexpression of multidrug efflux pumps such as MexXY, AcrAB, and AdeABC (3-5, 9, 10, 12).
Although Klebsiella pneumoniae is generally susceptible to tigecycline, a few clinical strains with decreased tigecycline susceptibility have been isolated. Decreased susceptibility to tigecycline in K. pneumoniae is associated with RamA, a transcriptional activator that is involved in the upregulation of the AcrAB multidrug efflux pump (11). The additional possibility that RamA might affect tigecycline susceptibility by regulating efflux pumps other than AcrAB was not ruled out (11). The aim of this study was to further investigate the role of RamA and AcrAB in decreased susceptibility to tigecycline in K. pneumoniae by studying the relationship between acrAB and ramA expression and tigecycline MIC in a large collection of K. pneumoniae clinical isolates in order to assess the appropriateness of MIC breakpoints for distinguishing between susceptible and resistant organisms.
The study included a total of 72 strains collected from a diverse group of patients that represented various regions and infection sites and were enrolled in phase 3 clinical trials for tigecycline. Tigecycline MICs for this set of isolates ranged from 0.25 μg/ml to 8 μg/ml; the distribution of MICs is shown in Table 1. All available K. pneumoniae isolates with tigecycline MICs of 4 and 8 μg/ml were included in the study, and multisusceptible strains were included for comparison. The strains were propagated at 37°C in Luria-Bertani broth or agar. The MIC of tigecycline was determined by a standard broth microdilution test (1, 2). Tests for tigecycline susceptibility were performed using fresh Mueller-Hinton broth (<12 h old).
TABLE 1.
Distribution of tigecycline MICs for K. pneumoniae isolates used in this study
| MIC (μg/ml) | na |
|---|---|
| 0.25 | 9 |
| 0.5 | 14 |
| 1 | 16 |
| 2 | 14 |
| 4 | 10 |
| 8 | 8 |
Number of strains.
Preparation of RNA templates and TaqMan quantitative real-time PCR (RT-PCR) analysis of gene expression were done as described previously (10). Oligonucleotide primers and probes used for RT-PCR are shown in Table 2. RNA templates were used at final concentrations of 4 μg/ml for acrA and ramA and 0.0004 μg/ml for rrsE expression analyses; these concentrations produced the optimal amplification efficiencies. Each sample was run in duplicate. Cycle threshold (CT) values were generated by iCycler iQ5 software. In the previous studies, which involved only a few strains, relative quantification of the target gene expression was performed by using a normalized expression analysis method, where the 16S rRNA gene served as a reference gene and one of the susceptible strains served as a reference condition (5, 10, 11). As this study involved a large population of clinical isolates, relative quantification of gene expression was performed by calculating delta CT values for each strain and each target gene, implying that no single strain was used as a reference condition in order to avoid biases. To adjust for the differences in concentration between the target genes (acrA and ramA) and the housekeeper gene (rrsE), a value of log2 10,000 (10,000 is the ratio between 4 μg/ml and 0.0004 μg/ml) was subtracted from each rrsE CT value. Adjusted rrsE CT values were then subtracted from the corresponding CT values for the target genes, resulting in the delta CT values, which were used for the statistical analyses. Because there is an inverse correlation between delta CT and gene expression level, the lower delta CT value implies that gene expression is increased.
TABLE 2.
Primers and fluorescent probes used for RT-PCR
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Fluorescent probea (5′-3′) |
|---|---|---|---|
| acrA | GTCCGGCACGCTGGA | ATAGCGCGTAGCGTGATTGA | CGGATGTCACCGTCGATCAGACCAC |
| ramA | GCATCAACCGCTGCGTATT | CGTTGCAGATGCCATTTCG | ATCGCTCGCCATGCCGGGTAT |
| rrsE | TTGACGTTACCCGCAGAAGAA | GCTTGCACCCTCCGTATTACC | TAACTCCGTGCCAGCAGCCG |
Labeled with 6-carboxyfluorescein at the 5′ end and with 6-carboxytetramethylrhodamine at the 3′ end.
The association between MIC and expression level was addressed by two statistical methods, analysis of variance (ANOVA) and linear regression. For ANOVA, mean delta CT values were calculated for each MIC and pairwise comparisons of mean expression levels were made between MICs. The pairwise comparisons were summarized using the least-significant difference approach and also using the Waller-Duncan approach, which adjusts for multiple comparisons. Pairwise comparisons were based on t statistics, using the error term from the ANOVA. The error term was also used to calculate appropriate standard errors and 95% confidence limits for the mean expression levels. In the ANOVA, delta CT was a dependent variable and MIC was an independent, categorical variable. In the regression analysis, MIC was an independent continuous variable and was log2 transformed for the analysis. Statistical analyses were performed using SAS for Windows, version 9.1, with SAS procedure GLM. Statistical significance was established by using a conventional P level of 0.05.
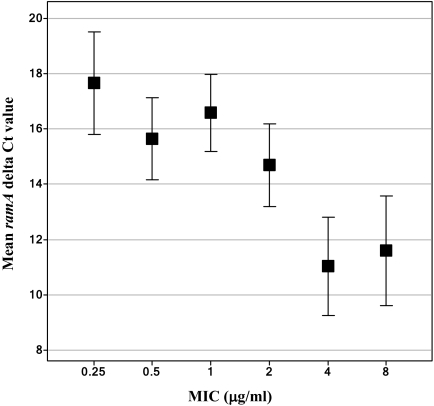
The ANOVA indicated that there was a statistically significant association of ramA delta CT values with the tigecycline MICs (P < 0.001). Mean ramA expression levels, along with their 95% confidence limits, are shown in Fig. 1. According to the Waller-Duncan method, mean delta CT values for the MICs of 4 and 8 μg/ml were statistically significantly different from those for MICs of 2 μg/ml or less, which is in agreement with the currently established tigecycline-susceptible breakpoint of ≤ 2 μg/ml for K. pneumoniae (Tygacil package insert, available at http://www.fda.gov/cder/foi/label/2005/021821lbl.pdf; Wyeth Pharmaceuticals Inc., Collegeville, PA). The linear regression of ramA delta CT on log2 MIC was statistically significant, with a P value of <0.0001 (data not shown). There was a statistically significant linear trend for a lower ramA delta CT as the tigecycline MIC increased, with a predicted decrease of 6.55 in mean delta CT (corresponding to a 94-fold increase in expression) between MICs of 0.25 and 8 μg/ml. These results confirm the previously established role of transcriptional activator RamA in decreased tigecycline susceptibility in K. pneumoniae (11).
FIG. 1.
ANOVA of ramA expression on the tigecycline MIC. The expression of ramA was analyzed by TaqMan RT-PCR. Mean ramA expression levels (delta CT values) and 95% confidence limits (based on ANOVA) are shown for each MIC from 0.25 to 8 μg/ml.
In contrast to ramA expression, although there was a statistically significant linear trend for lower acrA delta CT values as the tigecycline MIC increased, neither ANOVA nor linear regression analysis provided sufficient statistical evidence that mean acrA expression levels differ over the range of MICs. As suggested previously, RamA might have AcrAB-independent functions, e.g., regulation of efflux pumps other than AcrA (11). Further experiments are required to identify those additional functions of RamA in K. pneumoniae.
The most important implication from this study is an agreement between the results of quantitative analyses and the currently established tigecycline susceptibility breakpoint. An MIC breakpoint can be defined as a discriminating concentration used in the interpretation of results of susceptibility testing to define isolates as susceptible (will probably respond to antibiotic treatment), intermediate (the response is indeterminate or uncertain), or resistant (will probably not respond to antibiotic treatment) (6). According to the Clinical and Laboratory Standards Institute (CLSI, formerly NCCLS), several features of both antibiotic and bacterial pathogens must be considered when determining susceptibility breakpoints, including in vitro characteristics of the drug, distribution of susceptibilities for at least 500 isolates, pharmacokinetic/pharmacodynamic parameters, and clinical outcome statistics (7). The results of this study indicate that an understanding of the resistance mechanisms coupled with the quantitative methods, such as RT-PCR and statistical analyses, for monitoring the expression of resistance determinants may be used as an additional factor to facilitate the determination or to assess the appropriateness of MIC breakpoints.
Footnotes
Published ahead of print on 14 July 2008.
REFERENCES
- 1.CLSI. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard M7-A7, 7th ed., vol. 26. Clinical and Laboratory Standards Institute, Wayne, PA.
- 2.CLSI. 2007. Performance standards for antimicrobial susceptibility testing: M100-S17, 17th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Dean, C. R., M. A. Visalli, S. J. Projan, P.-E. Sum, and P. A. Bradford. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47:972-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keeney, D., A. Ruzin, and P. A. Bradford. 2007. RamA, a transcriptional regulator, and AcrAB, a RND-type efflux pump, are associated with decreased susceptibility to tigecycline in Enterobacter cloacae. Microb. Drug Resist. 13:1-6. [DOI] [PubMed] [Google Scholar]
- 5.Keeney, D., A. Ruzin, F. McAleese, E. Murphy, and P. A. Bradford. 2008. MarA-mediated overexpression of the AcrAB efflux pump results in decreased susceptibility to tigecycline in Escherichia coli. J. Antimicrob. Chemother. 61:46-53. [DOI] [PubMed] [Google Scholar]
- 6.MacGowan, A. P., and R. Wise. 2001. Establishing MIC breakpoints and the interpretation of in vitro susceptibility tests. J. Antimicrob. Chemother. 48(Suppl. 1):17-28. [DOI] [PubMed] [Google Scholar]
- 7.NCCLS. 2003. Development of in vitro susceptibility testing criteria and quality control parameters; approved guideline: M23-A2, 2nd ed., vol. 18. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 8.Petersen, P. J., N. V. Jacobus, W. J. Weiss, P. E. Sum, and R. T. Testa. 1999. In vitro and in vivo antimicrobial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob. Agents Chemother. 43:738-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruzin, A., D. Keeney, and P. A. Bradford. 2005. AcrAB efflux pump plays a role in decreased susceptibility to tigecycline in Morganella morganii. Antimicrob. Agents Chemother. 49:791-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruzin, A., D. Keeney, and P. A. Bradford. 2007. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J. Antimicrob. Chemother. 59:1001-1004. [DOI] [PubMed] [Google Scholar]
- 11.Ruzin, A., M. A. Visalli, D. Keeney, and P. A. Bradford. 2005. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 49:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visalli, M. A., E. Murphy, S. J. Projan, and P. A. Bradford. 2003. AcrAB multidrug efflux pump is associated with reduced levels of susceptibility to tigecycline (GAR-936) in Proteus mirabilis. Antimicrob. Agents Chemother. 47:665-669. [DOI] [PMC free article] [PubMed] [Google Scholar]