Abstract
The blaKPC-3 and qnrB19 determinants of transferable Klebsiella pneumoniae plasmid pLRM24 reside within a complex region consisting of a Tn1331 backbone into which a Tn4401-like element and qnrB19 mobilized by an adjacent ISEcp1 insertion sequence have been inserted. This novel element represents a coalescence of genes conferring multidrug resistance in K. pneumoniae.
The activities of carbapenems and fluoroquinolones are increasingly compromised in the clinical setting by the emergence and spread of antimicrobial resistance. Resistance to the carbapenems is most frequently attributable to the expression of β-lactamases. In many cases, the expression of β-lactamases with low levels of activity against carbapenems (such as Amp-C β-lactamases) combines with permeability reductions to yield the resistant phenotype (9). In other cases, β-lactamases with specific activity against the carbapenems are encountered (5, 13). Recently, the expression of class A β-lactamases KPC-2 and KPC-3 by a range of gram-negative bacilli (but most commonly by Klebsiella pneumoniae) has become a significant problem in many medical centers (6, 8). These enzymes confer resistance to nearly all β-lactam antibiotics. The presence of blaKPC genes on transferable plasmids and the recently characterized putative KPC-2-encoding transposon Tn4401 (10) almost certainly contribute to the dissemination of these important enzymes.
High-level fluoroquinolone resistance is most commonly conferred by point mutations in cellular topoisomerases. These are essential chromosomal genes and are not transferable under normal conditions in gram-negative bacilli. Transferable low-level resistance to fluoroquinolones has been attributed to plasmid-encoded qnr genes (16), as well as to the mutant aminoglycoside-modifying enzyme gene aac-6′-Ib-cr (11). Three classes of the qnr genes (qnrA, qnrB, and qnrS, with many variants) have been reported, and their mobilization onto plasmids may involve integrons (19).
We recently reported on a K. pneumoniae strain that transferred blaKPC-3 and qnrB19 on the same plasmid (3). In the present report, we describe the genetic environment of these two resistance determinants that reveals a large composite element representing the coalescence of several smaller mobile elements.
K. pneumoniae VA367 is a clinical strain isolated from the sputum of a patient at the Louis Stokes Cleveland VA Medical Center (3). The transfer of carbapenem resistance to Escherichia coli J53 (15) was associated with the transmission of low-level resistance to ciprofloxacin (MIC, 1 μg/ml).
Genomic DNA was isolated from the transconjugant resulting from the transfer of the 80-kb plasmid from strain VA367 to E. coli J53 (Rifr), as described previously (14), followed by an alkaline lysis procedure for the isolation of large plasmids (1). Restriction fragments of interest (Fig. 1A) were identified by hybridization with probes derived from either blaKPC-3 or qnrB19, isolated from agarose gels, and ligated to prepared plasmid vector pCC1BAC (Epicenter Technologies) or pBCSK(−) (Stratagene). DNA sequencing was performed by Cogenics, Cleveland, OH. Confirmation of the distance between blaKPC-3 and qnrB19 was accomplished by PCR with primers PCR KPCoutREV (5′-CGGCCATGAGAGACAAGACAGC-3′) (within blaKPC-3) and PCR543r (5′-CCGCTCAGGTCGGCACCTG-3′) (within qnrB19) (GenAmp; Applied Biosystems).
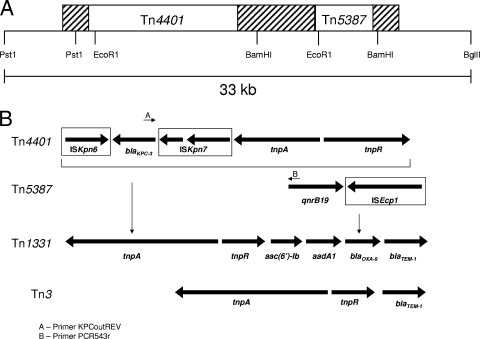
FIG. 1.
(A) Relative positions of the different putative mobile elements within the KQ element. The Tn1331 backbone is designated by the hatched areas. The positions of insertion of Tn4401 and Tn5387 are indicated. The positions of different restriction sites are also indicated. (B) Representation of the different mobile elements comprising the KQ element. The names of the individual element are indicated at the left. The left and right vertical arrows represent the insertion sites for Tn4401 and Tn5387, respectively. IS elements are boxed, and the arrows reflect the direction of transcription of their transposase genes. The locations of PCR primers KPCoutREV (arrow A) and PCR543r (arrow B) are indicated.
We have designated the 80-kb blaKPC-3 and qnrB19 plasmid in VA367 pLRM24. Within pLRM24, both blaKPC-3 and qnrB19 are located within a large composite genetic element that we have designated the KQ (KPC and QNR) element. The KQ element is built on a backbone of previously described Tn3-family transposon Tn1331 (Fig. 1A). Single-strand sequence analysis indicates 99% nucleic acid identity to the complete sequence of the 7,992-bp Tn1331 first described by Sarno and colleagues in 2002 (17), where it was reported to be integrated into K. pneumoniae plasmid pJHCMW1 (GenBank accession number AF479774). Tn1331 consists of Tn3 (tnpA, tnpR, and blaTEM-1) (4) into which three resistance genes [aac(6′)-Ib, aadA1, and blaOXA-9] are inserted at the terminus of tnpR in an insertion event that generated 520-bp direct repeats (17). We speculate that a 5 bp-duplication (underlined) of the target sequence typical of Tn3-family transposons (AGAGGTTGGAATAggggtct… agaccccGAATACAGAGG) was generated upon insertion of Tn1331 into pLRM24, although this must remain speculation because the flanking sequences (uppercase letters) have not previously been reported.
The immediate environment surrounding blaKPC-3 within the KQ element is indistinguishable from the recently reported Tn4401, as determined by partial sequencing and restriction enzyme analysis (10). Tn4401 is a ca. 10-kb putative mobile element that consists of a transposase, a resolvase, blaKPC-2 or blaKPC-3, and putative insertion sequence (IS) elements ISKpn6 and ISKpn7 (see Fig. 1B). The Tn4401-like element in pLRM24 was found to be inserted into the tnpA gene of Tn1331 at nucleotides 5502 to 5506 of the reported Tn1331 sequence, generating a 5-bp duplication of the target sequence (AGAAC), consistent with the findings described in a previous report (10).
The sequence of the pLRM24 quinolone resistance determinant confirmed that it was identical to the recently reported qnrB19 (GenBank accession number EU523120). This open reading frame was found to be inserted into the blaOXA-9 gene of Tn1331 as part of a 2,966-bp region that also included the ISEcp1 IS (Fig. 1). We have designated this IsEcp1-qnrB19 element Tn5387. Tn5387 is inserted at Tn1331 base pairs 9320 to 9324 and has a 5-bp target duplication (AATTCcctattttc… gaatctaggAATTC, where the lowercase letters represent the ends of the transposable element). Within Tn5387, qnrB19 is located immediately downstream of ISEcp1, and its open reading frame is transcribed in the direction opposite that of the transposase gene of ISEcp1 (Fig. 1B). Upstream of qnrB19 was located a sequence with similarity to the inverted repeats of ISEcp1 (CCTAGATTCTACGTCAG). Previous work has shown that such configurations contribute to the ISEcp1-mediated mobility of a variety of resistance determinants, including several blaCTX-M-type β-lactamase genes; the chromosomal β-lactamase gene from Kluyvera ascorbata; and the rmtC gene, which confers resistance to aminoglycosides (7, 12, 18). This is the first such report suggesting the ISEcp1-mediated mobility of a qnr determinant. Unlike other reports (2), the ISEcp1 IS associated with qnrB19 in pLRM24 was not positioned in a manner that would contribute an additional promoter sequence for qnrB19 expression. A distance of 14 kb between blaKPC-3 and qnrB19 within the KQ element was confirmed by a long PCR with primers specific for regions within blaKPC-3 and qnrB19 (data not shown).
Our data provide insight into the variety of mechanisms that bacteria use to create mobile resistance to multiple antimicrobial agents by using both old and newly described or created elements. Tn1331 evolved from Tn3 several years ago by the addition of two aminoglycoside-modifying enzyme genes [aadA1 and aac(6′)-Ib] and the blaOXA-9 β-lactamase gene (17). Tn1331 has now served as the backbone into which mobile elements conferring resistance to carbapenems (Tn4401) and fluoroquinolones (Tn5387) have inserted. It is not clear whether the KQ element is mobile because the insertion of Tn4401 into tnpA of Tn1331 would presumably affect transposability. The presence of the KQ element on a transferable plasmid and the likely transposability of some of the individual elements, however, confer enough mobility to predict dissemination within and perhaps beyond K. pneumoniae.
Nucleotide sequence accession number.
The complete sequence of Tn5387 was entered into GenBank under accession number EU624315.
Acknowledgments
This work was supported by merit reviews from the U.S. Department of Veterans Affairs (L.B.R. and R.A.B.) and by the National Institute of Allergy and Infectious Diseases (grant AI45626 to L.B.R.).
Footnotes
Published ahead of print on 23 June 2008.
REFERENCES
- 1.Ayure, D. M. Q., Z. R. S. Moreno, F. A. A. Gutierrez, J. M. B. Morales, and D. M. Castano. 2006. Genome analysis of thirteen Colombian clostridial strains by pulsed field gel electrophoresis. Elec. J. Biotechnol. 9:541-550. http://www.ejbiotechnology.info/content/vol9/issue5/full/3/index.html. [Google Scholar]
- 2.Eckert, C., V. Gautier, and G. Arlet. 2006. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J. Antimicrob. Chemother. 57:14-23. [DOI] [PubMed] [Google Scholar]
- 3.Endimiani, A., L. L. Carias, A. M. Hujer, C. R. Bethel, K. M. Hujer, F. Perez, R. A. Hutton, W. R. Fox, G. S. Hall, M. R. Jacobs, D. L. Paterson, L. B. Rice, S. G. Jenkins, F. C. Tenover, and R. A. Bonomo. 2008. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae possessing blaKPC in the United States. Antimicrob. Agents Chemother. 52:2680-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heffron, F., B. J. McCarthy, H. Ohtsubo, and E. Ohtsubo. 1979. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell 18:1153-1163. [DOI] [PubMed] [Google Scholar]
- 5.Kim, J. Y., H. I. Jung, Y. J. An, J. H. Lee, S. J. Kim, S. H. Jeong, K. J. Lee, P. G. Suh, H. S. Lee, S. H. Lee, and S. S. Cha. 2006. Structural basis for the extended substrate spectrum of CMY-10, a plasmid-encoded class C beta-lactamase. Mol. Microbiol. 60:907-916. [DOI] [PubMed] [Google Scholar]
- 6.Landman, D., S. Bratu, S. Kochar, M. Panwar, M. Trehan, M. Doymaz, and J. Quale. 2007. Evolution of antimicrobial resistance among Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae in Brooklyn, N.Y. J. Antimicrob. Chemother. 60:78-82. [DOI] [PubMed] [Google Scholar]
- 7.Lartigue, M. F., L. Poirel, D. Aubert, and P. Nordmann. 2006. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 50:1282-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leavitt, A., S. Navon-Venezia, I. Chmelnitsky, M. J. Schwaber, and Y. Carmeli. 2007. Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob. Agents Chemother. 51:3026-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livermore, D. M. 1992. Interplay of impermeability and chromosomal β-lactamase activity in imipenem-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:2046-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naas, T., G. Cuzon, M. V. Villegas, M. F. Lartigue, J. P. Quinn, and P. Nordmann. 2008. Genetic structures at the origin of acquisition of the β-lactamase blaKPC gene. Antimicrob. Agents Chemother. 52:1257-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park, C. H., A. Robicsek, G. A. Jacoby, D. Sahm, and D. C. Hooper. 2006. Prevalence in the United States of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme. Antimicrob. Agents Chemother. 50:3953-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel, L., J. W. Decousser, and P. Nordmann. 2003. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M beta-lactamase gene. Antimicrob. Agents Chemother. 47:2938-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Queenan, A. M., and K. Bush. 2007. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20:440-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice, L. B., S. H. Marshall, and L. L. Carias. 1992. Tn5381, a conjugative transposon identifiable as a circular form in Enterococcus faecalis. J. Bacteriol. 174:7308-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice, L. B., S. H. Willey, G. A. Papanicolaou, A. A. Medeiros, G. M. Eliopoulos, R. C. Moellering, Jr., and G. A. Jacoby. 1990. Outbreak of ceftazidime resistance caused by extended-spectrum β-lactamases at a Massachusetts chronic care facility. Antimicrob. Agents Chemother. 34:2193-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 17.Sarno, R., G. McGillivary, D. J. Sherratt, L. A. Actis, and M. E. Tolmasky. 2002. Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 46:3422-3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wachino, J., K. Yamane, K. Kimura, N. Shibata, S. Suzuki, Y. Ike, and Y. Arakawa. 2006. Mode of transposition and expression of 16S rRNA methyltransferase gene rmtC accompanied by ISEcp1. Antimicrob. Agents Chemother. 50:3212-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu, J. J., W. C. Ko, S. H. Tsai, and J. J. Yan. 2007. Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob. Agents Chemother. 51:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]