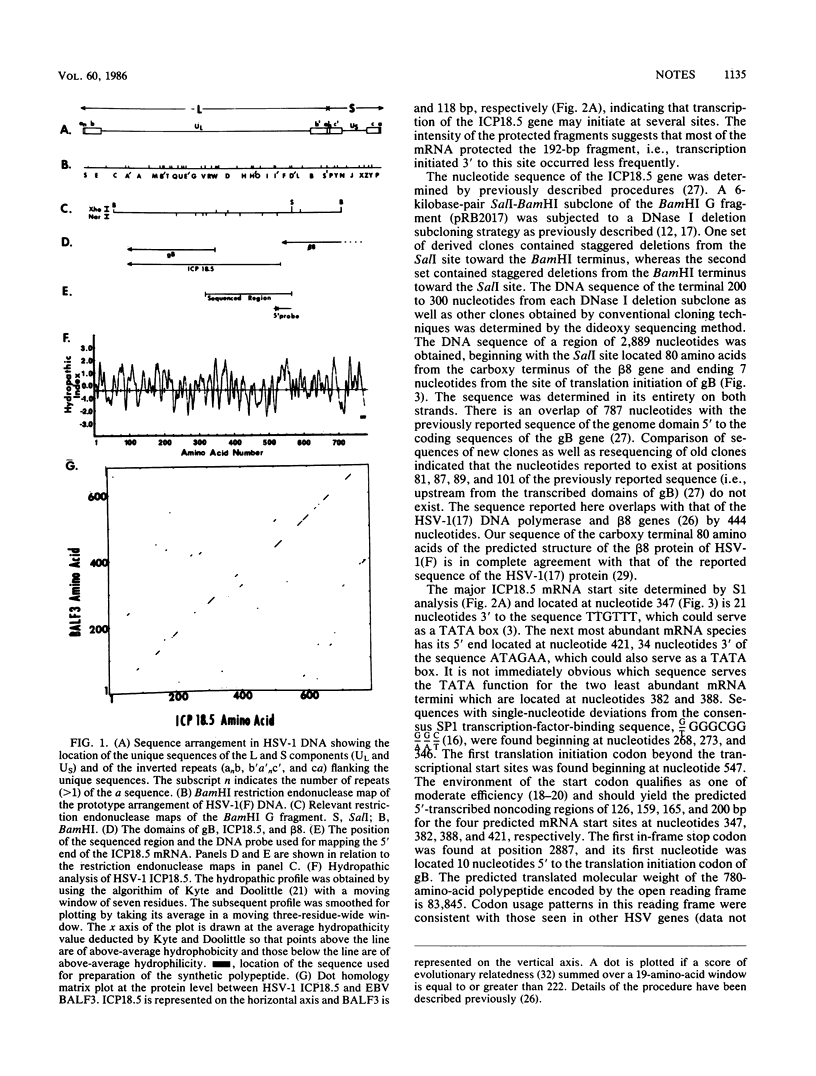
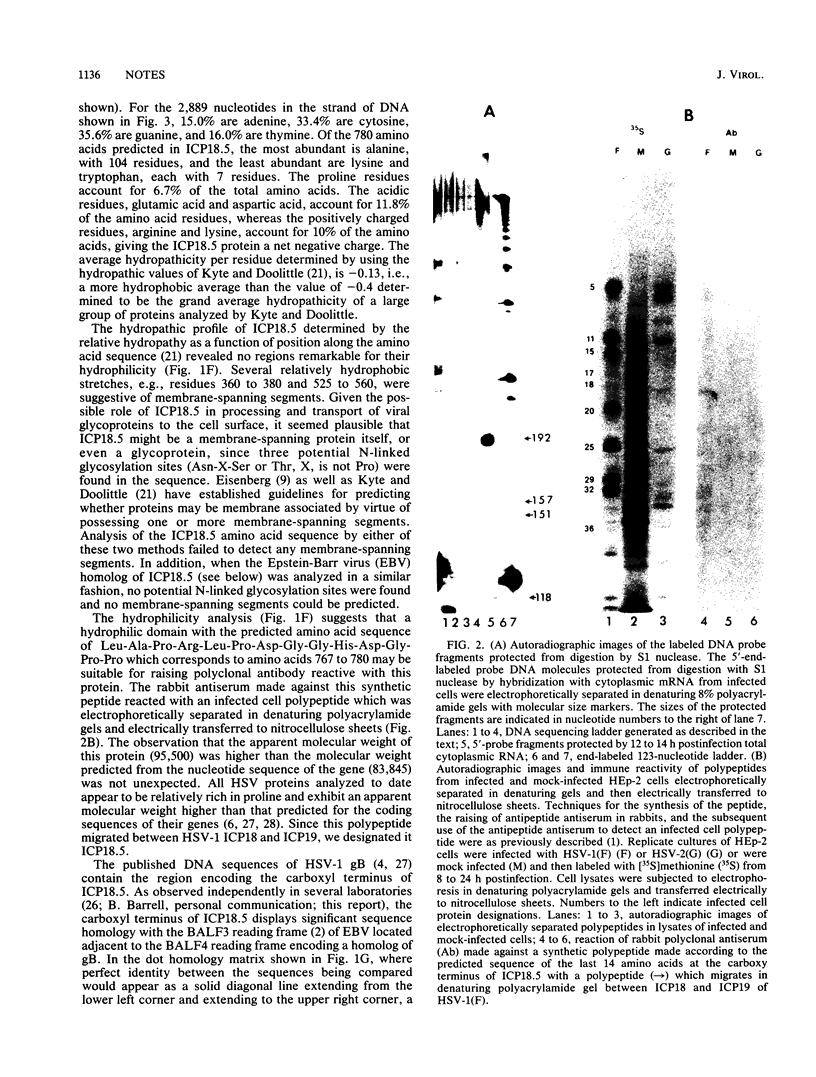
Abstract
Earlier reports have localized mutations which affect the processing and transport of herpes simplex virus 1 glycoproteins to a region located between the genes specifying glycoprotein B and the major viral DNA-binding protein (beta 8). The nucleotide sequence of this region contains a single long open reading frame encoding a 780-amino-acid protein with a predicted molecular weight of 83,845. To confirm the existence of this protein, rabbit polyclonal antibody was made against a synthetic peptide made according to the predicted sequence of a hydrophilic domain near the carboxy terminal of the protein. This antibody reacted with an infected cell protein of an apparent molecular weight of 95,500. We designated this protein infected cell protein 18.5 (ICP18.5). S1 nuclease analysis suggested that the 5.6-kilobase mRNA encoding ICP18.5 is initiated predominantly from one site, but three weaker initiation sites also seemed to occur within a 74-base-pair stretch of DNA. This gene appears to be conserved in the Epstein-Barr virus (EBV) genome, inasmuch as 174 of the 780 amino acids of ICP18.5 align with corresponding amino acids predicted by the EBV open reading frame BALF3. The EBV gene is located adjacent to the gene specifying a homolog of the herpes simplex virus 1 glycoprotein B.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackermann M., Sarmiento M., Roizman B. Application of antibody to synthetic peptides for characterization of the intact and truncated alpha 22 protein specified by herpes simplex virus 1 and the R325 alpha 22- deletion mutant. J Virol. 1985 Oct;56(1):207–215. doi: 10.1128/jvi.56.1.207-215.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. Nucleotide sequence specifying the glycoprotein gene, gB, of herpes simplex virus type 1. Virology. 1984 Mar;133(2):301–314. doi: 10.1016/0042-6822(84)90397-0. [DOI] [PubMed] [Google Scholar]
- Campbell M. E., Palfreyman J. W., Preston C. M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J Mol Biol. 1984 Nov 25;180(1):1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- Chou J., Roizman B. The terminal a sequence of the herpes simplex virus genome contains the promoter of a gene located in the repeat sequences of the L component. J Virol. 1986 Feb;57(2):629–637. doi: 10.1128/jvi.57.2.629-637.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A., Sabourin D. J., Schaffer P. A. Genetic analysis of temperature-sensitive mutants which define the genes for the major herpes simplex virus type 2 DNA-binding protein and a new late function. J Virol. 1983 Jan;45(1):343–353. doi: 10.1128/jvi.45.1.343-353.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper K. G., Frink R. J., Wagner E. K. Detailed characterization of an apparently unspliced beta herpes simplex virus type 1 gene mapping in the interior of another. J Virol. 1982 Sep;43(3):1123–1128. doi: 10.1128/jvi.43.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D. Three-dimensional structure of membrane and surface proteins. Annu Rev Biochem. 1984;53:595–623. doi: 10.1146/annurev.bi.53.070184.003115. [DOI] [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Frame M. C., Marsden H. S., Dutia B. M. The ribonucleotide reductase induced by herpes simplex virus type 1 involves minimally a complex of two polypeptides (136K and 38K). J Gen Virol. 1985 Jul;66(Pt 7):1581–1587. doi: 10.1099/0022-1317-66-7-1581. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Garoff H., Lehrach H. A subcloning strategy for DNA sequence analysis. Nucleic Acids Res. 1980 Dec 11;8(23):5541–5549. doi: 10.1093/nar/8.23.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorioso J. C., Levine M., Holland T. C., Szczesiul M. S. Mutant analysis of herpes simplex virus-induced cell surface antigens: resistance to complement-mediated immune cytolysis. J Virol. 1980 Sep;35(3):672–681. doi: 10.1128/jvi.35.3.672-681.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Draper K. G., Frink R. J., Costa R. H., Wagner E. K. Herpes simplex virus mRNA species mapping in EcoRI fragment I. J Virol. 1982 Aug;43(2):594–607. doi: 10.1128/jvi.43.2.594-607.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L. E., Sandri-Goldin R. M., Goldin A. L., Glorioso J. C., Levine M. Transcriptional and genetic analyses of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J Virol. 1984 Mar;49(3):947–959. doi: 10.1128/jvi.49.3.947-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousoulas K. G., Pellett P. E., Pereira L., Roizman B. Mutations affecting conformation or sequence of neutralizing epitopes identified by reactivity of viable plaques segregate from syn and ts domains of HSV-1(F) gB gene. Virology. 1984 Jun;135(2):379–394. doi: 10.1016/0042-6822(84)90194-6. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 1984 Mar 15;308(5956):241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Little S. P., Jofre J. T., Courtney R. J., Schaffer P. A. A virion-associated glycoprotein essential for infectivity of herpes simplex virus type 1. Virology. 1981 Nov;115(1):149–160. doi: 10.1016/0042-6822(81)90097-0. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Clements J. B. Organization of the herpes simplex virus type 1 transcription unit encoding two early proteins with molecular weights of 140000 and 40000. J Gen Virol. 1983 May;64(Pt 5):997–1006. doi: 10.1099/0022-1317-64-5-997. [DOI] [PubMed] [Google Scholar]
- Pancake B. A., Aschman D. P., Schaffer P. A. Genetic and phenotypic analysis of herpes simplex virus type 1 mutants conditionally resistant to immune cytolysis. J Virol. 1983 Sep;47(3):568–585. doi: 10.1128/jvi.47.3.568-585.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P. E., Biggin M. D., Barrell B., Roizman B. Epstein-Barr virus genome may encode a protein showing significant amino acid and predicted secondary structure homology with glycoprotein B of herpes simplex virus 1. J Virol. 1985 Dec;56(3):807–813. doi: 10.1128/jvi.56.3.807-813.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P. E., Kousoulas K. G., Pereira L., Roizman B. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J Virol. 1985 Jan;53(1):243–253. doi: 10.1128/jvi.53.1.243-253.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P. E., McKnight J. L., Jenkins F. J., Roizman B. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5870–5874. doi: 10.1073/pnas.82.17.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. P., McGeoch D. J. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the genes for DNA polymerase and the major DNA binding protein. Nucleic Acids Res. 1985 Nov 25;13(22):8143–8163. doi: 10.1093/nar/13.22.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon F. J., McGeoch D. J. Detailed analysis of the mRNAs mapping in the short unique region of herpes simplex virus type 1. Nucleic Acids Res. 1985 Feb 11;13(3):953–973. doi: 10.1093/nar/13.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento M., Haffey M., Spear P. G. Membrane proteins specified by herpes simplex viruses. III. Role of glycoprotein VP7(B2) in virion infectivity. J Virol. 1979 Mar;29(3):1149–1158. doi: 10.1128/jvi.29.3.1149-1158.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]