Abstract
The molecular mechanisms mediating eukaryotic replication termination and pausing remain largely unknown. Here we present the molecular characterization of Rtf1 that mediates site-specific replication termination at the polar Schizosaccharomyces pombe barrier RTS1. We show that Rtf1 possesses two chimeric myb/SANT domains: one is able to interact with the repeated motifs encoded by the RTS1 element as well as the elements enhancer region, while the other shows only a weak DNA binding activity. In addition we show that the C-terminal tail of Rtf1 mediates self-interaction, and deletion of this tail has a dominant phenotype. Finally, we identify a point mutation in Rtf1 domain I that converts the RTS1 element into a replication barrier of the opposite polarity. Together our data establish that multiple protein DNA and protein–protein interactions between Rtf1 molecules and both the repeated motifs and the enhancer region of RTS1 are required for site-specific termination at the RTS1 element.
DNA replication is a highly complex process whereby genetic information and epigenetic chromatin states are duplicated, sister chromatid cohesion is established, and DNA damage repair is performed. Although there is a general understanding of the factors and mechanisms by which eukaryotic DNA replication is initiated, very little is known about the molecular processes underlying replication pausing and termination. Most replication termination occurs randomly when converging replication forks meet in termination zones between active origins (Santamaria et al. 2000). However, at special genetic elements, site-specific replication termination or pausing is deliberately induced. One class of such elements is the barriers present in the polymerase I-transcribed rDNA arrays from yeasts to metazoans (reviewed by Hyrien 2000; Codlin and Dalgaard 2003). At these replication barriers, a family of transcription termination factors mediate site-specific termination of replication forks moving in one direction while allowing replication forks moving in the other direction to pass unhindered; the factors include TTF1 (mouse and human; Gerber et al. 1997; Lopez-Estrano et al. 1998), Reb1 (Schizosaccharomyces pombe; Sanchez-Gorostiaga et al. 2004), as well as the unrelated protein Fob1 (Saccharomyces cerevisiae; Brewer and Fangman 1988; Linskens and Huberman 1988; Kobayashi and Horiuchi 1996). While the biological function(s) of the Reb1/TTF1 barriers has not been established experimentally, the Fob1 barrier has a dual function: It acts (i) to prevent collision between replication and polymerase I transcription machinery, which otherwise leads to genetic instability, by ensuring that the two types of forks move in the same direction within the polymerase I transcriptional unit (Takeuchi et al. 2003) and (ii) to induce recombination and establishment of cohesion between sister chromatids to prevent unequal crossovers and genetic instability (Kobayashi and Horiuchi 1996; Huang et al. 2006).
Interestingly, the S. pombe RTS1 element located in the mating-type region is closely related to the rDNA barriers:
RTS1 is polar, acting on replication forks moving in the cenII-distal direction, and its biological function is to optimize the replication-coupled recombination event that underlies mating-type switching (Figure 1A; Dalgaard and Klar 2001)
Replication forks stalled at RTS1 induce recombination (Ahn et al. 2005; Lambert et al. 2005).
The cis-acting sequences are related (Figure 1B). First, RTS1 region B contains four repeated ∼60-bp motifs each possessing polar barrier activity (Codlin and Dalgaard 2003). Similar rDNA motifs, which in the metazoan system are called SAL boxes, are required for barrier activity (Gerber et al. 1997; Lopez-Estrano et al. 1999; Sanchez-Gorostiaga et al. 2004). For the S. pombe Reb1 and the metazoan TTF1 factors, these rDNA barrier motifs have been shown to act as binding sites in vitro (Melekhovets et al. 1997; Zhao et al. 1997; Lopez-Estrano et al. 1998; Sanchez-Gorostiaga et al. 2004).
In addition, an ∼60-bp enhancer called region A, characterized by a purine-rich upper and a pyrimidine-rich lower strand has been defined for RTS1. Region A does not possess any independent barrier activity, but mediates in vivo a fourfold enhancement of region B activity by promoting a functional interaction between the motifs (Codlin and Dalgaard 2003). Similarly, for the metazoan rDNA elements, in vitro experiments have established the presence of a GC-rich sequence flanking one of the SAL boxes, which is required for contrahelicase activity. Like the RTS1 region A, this GC-rich sequence is characterized by an asymmetrical distribution of purines and pyrimidines on the two DNA strands (Putter and Grummt 2002).
Both the S. pombe rDNA barrier and RTS1 require Swi1 and Swi3 factors for activity, while the S. cerevisiae Fob1 rDNA barrier depends on the homologs, Tof1 and Csm3 (Mohanty et al. 2006).
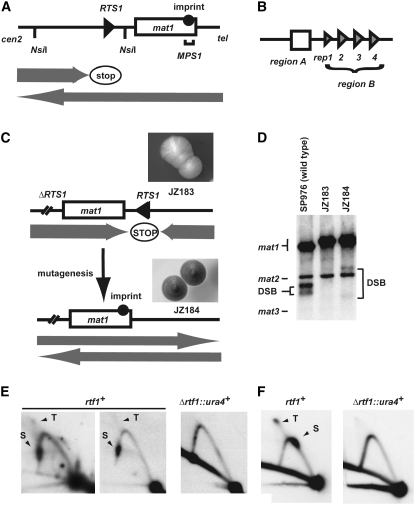
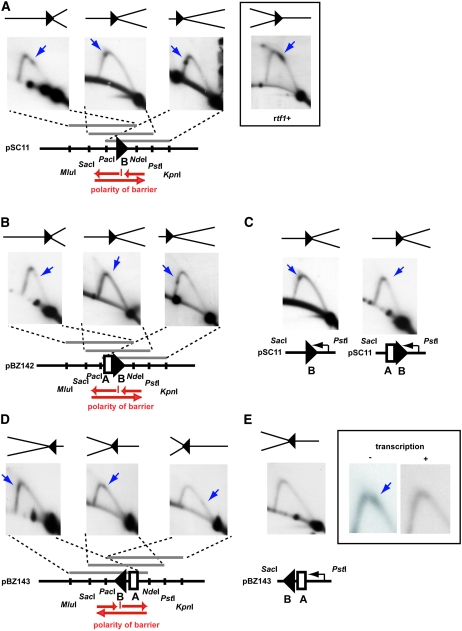
Figure 1.—
Isolation of rtf1 mutants. (A) Line drawing displaying the wild-type mat1 region on chromosome II. The positions of the imprint (solid circle), the RTS1 element (triangle) and the MPS1 (horizontal bracket) are given. Shaded arrows indicate the directions by which the replication forks are moving within the mat1 region, as well as the polarity of the RTS1 replication barrier. (B) Graphic outline of the RTS1 subelements. Region A (box) and the four repeated region B motifs (triangles; rep1, -2, -3, and -4) are shown. (C) Graphic outline of the genetic screen used for isolation of the rtf1 mutants. (Top) The rearranged mating-type region of the JZ183 strain; the site-specific terminator RTS1 has been deleted at the cen-proximal side of mat1 and inserted at the cen-distal side in the inverted orientation. (Top inset) Colonies of strain JZ183 stain yellow with iodine vapor. (Bottom) Mutagenesis (vertical arrow) of replication termination factor (rtf) genes abolishes RTS1 function and leads to a partial restoration of the wild-type direction of replication at mat1 (shaded arrows). Thus, imprinting (solid circle) and mating-type switching are partly reestablished. (Bottom inset) Colonies of rtf1 strains stain black with iodine vapor (strain JZ184, Figure 2 legend). (D) rtf1 mutations partly restore mat1 imprinting. Southern analysis of HindIII-digested chromosomal DNA (Dalgaard and Klar 1999). A probe specific to the mat1P HindIII fragment was utilized. Signals that correspond to mat1, mat2P, and mat3M fragments are indicated. The mat2P and mat3M are detected due to partial homology. mat1 imprinted DNA is fragile during purification, where hydrolysis at the imprint leads to the formation of a double-stranded break (DSB). The generated fragments are indicated within the panel. The difference in the molecular sizes of the DSB fragments from wild-type (lane 1) and mutant strain (lane 3) is due to the transposition of RTS1. (E) Rtf1 is required for RTS1 function. 2D-gel analysis of replication intermediates at the wild-type RTS1 locus in wild-type (JZ1) and Δrtf1 (SC11) strains. The genomic position of the NsiI restriction fragment analyzed is indicated in A. Stall (S) and termination (T) signals observed for wild-type replication intermediates are indicated. Note that the RTS1 element is replicated in both directions; however, we have earlier shown that while the marjority of replication forks move in the permissive direction, the small fraction of replication forks in the nonpermissive direction is stalled and terminated (Codlin and Dalgaard 2003). (F) 2D-gel analysis of plasmid-borne RTS1 from wild-type (SC1) and Δrtf1 (SC46) strains. Earlier published experiments have established that the RTS1 element at this position in the plasmid is replicated in both orientations (Codlin and Dalgaard 2003). Thus, stalling and termination signals are observed originating from forks moving in the direction where the barrier is active, while a normal Y arc is formed by replication forks moving in the other direction. Stall (S) and termination (T) signals observed for wild-type replication intermediates are indicated.
Finally, it should be noted that recently a Reb1-independent, but putatively Sap1-dependent barrier where replication pausing is observed was defined within the S. pombe rDNA barrier (Krings and Bastia 2005; Mejia-Ramirez et al. 2005; Krings and Bastia 2006). Interestingly, Sap1 has also been shown to bind in the mating-type region (Arcangioli and Klar 1991), but the smt-0 deletion that removes the cis-acting Sap1 binding sites does not affect the replication barriers in the mating-type region (Dalgaard and Klar 2000).
Here we characterize the trans-acting factor Rtf1 that is required for RTS1 function. Rtf1 is a paralog of the S. pombe Reb1 protein required for rDNA replication barrier activity as well as polymerase I transcription termination, and thus it is a new member of the Rtf1/Ttf1/Reb1 protein family. We address the molecular mechanism by which Rtf1 mediates site-specific replication termination at RTS1.
MATERIALS AND METHODS
UV mutagenesis:
Logarithmically growing cells (strain JZ183) were plated on either sporulation (PMA+) or rich (YEA) media-containing plates and directly irradiated with UV (24 μJ; 55% survival) using a Stratalinker (Stratagene).
PMA+ plates were incubated at 30° for 5 days and then stained with iodine vapor for identification of mutants.
YEA plates were incubated for 4 days at 30°, replicated to PMA+, followed by 2 days of incubation at 30°, and then stained with iodine vapor.
Iodine staining was performed as described by Moreno et al. (1991).
The genetic screen used for identification of the dominant rtf1 mutant was done in a similar fashion, except that rtf1+-plasmid pBZ136 had been introduced in the strain JZ183.
Strain construction and isolation:
Strains were constructed using methods described by Moreno et al. (1991). The genotypes of the strains are described in the supplemental data.
2D-gel analysis of replication intermediates:
Strains were grown either in YEA or AA −Leu (plasmid-containing strains) media. DNA from logarithmically growing cells was isolated as described by Huberman et al. (1987). Replication intermediates were enriched using BND cellulose (Sigma; Kiger and Sinsheimer 1969), digested with restriction enzymes and analyzed on two-dimensional agarose gels (Brewer and Fangman 1987). A probe specific to the 0.8-kb RTS1 fragment (Dalgaard and Klar 2001) was used for the Southern analysis. Signals were quantified using a phosphorimager and Quantity One software (Biorad). For each gel the intensity of ascending part of the Y arc was used for normalizing the pause- and termination-signal intensities. The quantification method is described in full in Codlin and Dalgaard (2003).
Protein expression, purification, and gel-shift assays:
Domains II (aa 244–466) and I + II (aa 94–466) are expressed using the Studier expression systems (Studier and Moffatt 1986). Domain I (aa 94–256) was expressed using the pMAL expression system (New England Biolabs). Partial purification was done using an amylose column or a Ni2+ column (domains I + II) followed by an amylose column (Di Guan et al. 1988; Petty 1996). Gel shifts were obtained as described by Sambrook and Russell (2001). For each figure, all lanes displayed in a given panel were run on the same gel. For a more complete description refer to the supplemental text.
Two-hybrid analysis:
Rtf1 segments were cloned into S. cerevisiae two-hybrid vectors, pGADT7 and pGBKT7 (MATCHMAKER Gal4 two-hybrid system3, BD Biosciences Clontech). The analysis was performed as described (Bartel et al. 1993) using S. cerevisiae strain AH109.
RESULTS
Identification of Rtf1:
The mating-type locus mat1 has to be replicated in a specific direction for imprinting and mating-type switching to occur (Dalgaard and Klar 1999, 2001). We have utilized the dependence of the imprinting process on the replication direction in a genetic screen for trans-acting factors involved in site-specific termination of replication at RTS1 (Dalgaard and Klar 2000). Transposition of RTS1 in the inverted orientation to the cen-distal side of mat1 changes the direction by which the mat1 locus is replicated and therefore leads to the inhibition of imprinting, mating-type switching, mating, and sporulation (Dalgaard and Klar 2001; Figure 1C, line drawing). The strain's decreased ability to sporulate can be assayed by iodine staining (Figure 1C; strain JZ183). Iodine stains the starch that is produced in the spores of this yeast. Similarly, a reduction in mat1 imprinting can be quantified by Southern analysis (Figure 1D; lanes 2 and 3). The assay utilizes the efficient conversion of the mat1 imprint into a double-stranded break (DSB) by some DNA purification methods (Arcangioli 1998; Dalgaard and Klar 1999). In our genetic screen we utilized that trans-acting mutations that abolish replication termination at RTS1 will partly restore the wild-type direction of fork progression at mat1 and, as a consequence, allow an increased number of cells to switch mating type, mate, and sporulate (Figure 1C, lower line drawing and inset). Originally, mutations in three complementation groups, named replication termination factors (rtf), were isolated in this screen (Dalgaard and Klar 2000). The majority of the mutations, 28 of 30, belong to the rtf1 complementation group described here. The sporulation levels observed for the identified rtf1 mutants varied from 31 to 61%, compared to 4.8% observed in the parental strain (JZ183) and 65% in the wild-type h90 control strain (JZ1). Importantly, haploid meiosis is not observed in these strains, establishing that derepression of the silenced donor loci, mat2P and mat3M, does not occur (data not shown). Furthermore, Southern analysis of the mat1 region of these strains detected increased levels of mat1 DSB, as expected from a partial restoration of the mat1 imprint (Figure 1D). Subsequently, subcloning and complementation studies identified rtf1 as the open reading frame SPAC22F8.07C defined in the S. pombe genome project (supplemental data). A complete rtf1 null mutation was constructed by replacing the rtf1 open reading frame with the ura4+ gene (strain SC7). Analysis of the chromosomal as well as the plasmid-borne RTS1 shows that Δrtf1 abolishes RTS1 function (Figure 1, E and F).
Definition of functional Rtf1 domains:
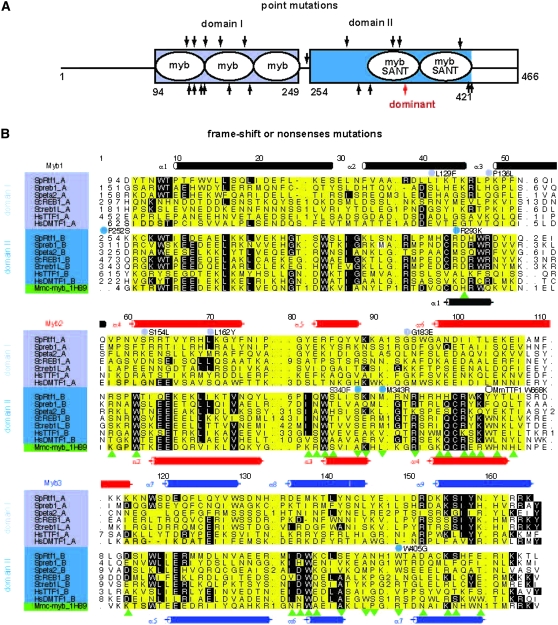
The large number of isolated rtf1 alleles allowed us to define the functional domains of the Rtf1 protein. The alleles include 10 single amino acid (aa) substitutions, six frameshifts (one in an intron splice junction), and four nonsense mutations (Figure 2, A and B; supplemental Figure S1). All the mutants isolated in the initial screen were recessive (data not shown). The distribution of point mutations suggested that in addition to the known myb motif, an additional functional domain might be present, thus, we employed bioinformatics for its identification. The Rtf1 sequence (CAF31329; SpRtf1) and related sequences were used to search a nonredundant protein sequence database through the World Wide Web interface to the PSI-BLAST program (default parameter settings). An ∼400-aa Rtf1 segment showed statistically significant similarity to proteins from a variety of species (E-value ≪ 0.05) and was retained for further analysis. Previously, an ∼200-aa conserved segment (here domain II) encompassing the two myb/SANT motifs was identified in Mus musculus TTF1 (MmTTF1), S. cerevisiae Reb1, and M. musculus c-myb (Evers et al. 1995, which refers to the two myb/SANT motifs as domains I and II). The myb motif is an ∼50-aa sequence which folds into a domain consisting of three helices characterized by tryptophan (Trp) residues essential for DNA binding. In the case of this protein family, mutation of Trp668 to Lys (W668K) in MmTTF1 was found to abolish binding of the dsDNA recognition sequence (Evers et al. 1995). In addition, a subclass of the myb motifs called the SANT motif has been shown to interact with histone tails (Boyer et al. 2004). Interestingly, the two domain II c-myb motifs of Rtf1 are identified on the sequence level to belong to this subclass. A more careful examination of the PSI-BLAST output revealed that the conserved domain II, present in the second half of the protein, displayed similarity to a putatively related domain in the first half, i.e., the ∼400-aa Rtf1/Reb1 conserved segment can be divided into two structurally related regions both predicted to interact with DNA via myb-like folds (Figure 2, A and B; domains I and II). A careful computational analysis, using a hidden Markov model, the Conserved Domain Database, and the PhD structural predictions establishes that this family of proteins possesses two chimeric putative DNA-binding domains, both displaying an overall similarity to metazoan c-myb. These two domains potentially contain in total five structural myb motifs, two of which might also be SANT motifs (supplemental text and Figure 2, A and B).
Figure 2.—
Bioinformatics analysis of the Rtf1 amino acid sequence. (A) Graphic outline of the position of the two c-myb-like domains (blue boxes) and their structural motifs (white ellipses) as well as identified mutations. In the gel-shift experiment presented below domain II encompasses the C-terminal tail. The positions of missense and frameshifts/nonsense mutations are given at the top and bottom, respectively. The position of the dominant mutation (strain ES8, R346*) is highlighted (red arrow). The strain names and identified mutations are JZ184, W405G; JZ185, S340F; JZ221, Q300*; JZ223, R293K; JZ226, L129F; JZ229, S154L; JZ231, T131*frame-1; JZ232, L162Y; JZ237, Q175*; JZ238, P136L; JZ240, Q145*; JZ241, L318*; JZ242, K200*frame-1; JZ243, G183E; JZ245, T420*frame+1; JZ249, M343R; JZ250, Q147*splice junction; JZ251, P252S; JZ252, T420*frame-1; JZ253, L129*frame-1; and JZ254, P252S. (B) Alignment of the two conserved c-myb-like domains of the Reb1/Rtf1/TTF1 protein family to the human c-myb protein sequence. Domains I and II are highlighted in light and dark blue, respectively. Proteins aligned are S. pombe Reb1 (Q9P6H9), Eta2 (BAC54905), S. cerevisiae Reb1 (CAA84992), Reb1L (NP_010309), Homo sapiens DMTF1 (AAH07447), and H. sapiens TTF1 (AAI04640). Residues that display >40% conservation are highlighted. The α-helices shown above the alignment are those predicted for the Rtf1 domain using the PhD program package. The last sequence shown is that of M. musculus c-myb (Mmc-myb_1H89). The positions of the α-helices in the three-dimensional structure of mouse c-myb (Weinstein et al. 1986; Ogata et al. 1993; Tahirov et al. 2002) are displayed below the alignment together with residues that interact with DNA (green triangles) and metal ions (green inverted triangles). The three structural myb motifs (Myb1, Myb2, and Myb3) are highlighted in yellow and the corresponding helices are in black, red, and blue, respectively. Substitutions in Rtf1 domains I and II identified as leading to loss of function are given at the top of the alignment and within the spacer line, respectively. Open circles indicate the nonsynonymous coding single nucleotide polymorphism HsTTF1 473K,E (rs12336746) and the location of the 668W-to-K mutation in M. musculus TTF1, which abolishes sequence-specific DNA binding (Evers et al. 1995).
Rtf1 domain I can bind to RTS1 regions A and B:
To characterize the DNA-binding specificities of the two domains, fusion proteins between a 6× His-tagged maltose binding protein (MBP) and Rtf1 segments encompassing domain I, domain II, and the chimeric domains (domains I + II) were purified (supplemental Figure S2A). Using the domain I, gel-shift assays were performed with a labeled dsDNA oligonucleotide corresponding to motif 4 from region B (Codlin and Dalgaard 2003; Figure 3A). The analysis detected several sharply defined mobility shifts characteristic of protein binding, and potentially of more than one molecule. It should be noted that Western analysis of shifted material verifies that the shift is due to binding of domain I (supplemental Figure S2B), and that binding can be outcompeted with excess cold-specific competitor (supplemental Figure S2C). Furthermore, gel shifts with dsDNA oligonucleotides resembling three shorter segments of motif 4 establish that domain I binds to the middle third of the motif (Figure 3B; left). A linker scanning mutagenesis of motif 4 has earlier defined two linker substitutions that abolish motif 4 barrier activity in vivo (Codlin and Dalgaard 2003). We used the five dsDNA oligonucleotides synthesized for that study to further identify sequences within motif 4 required for Rtf1 domain I binding. Interestingly, none of the substitutions completely abolished binding (data not shown; Codlin and Dalgaard 2003). However, gel-shift assays using the rep4-mut3 substitution, which in vivo abolishes barrier activity, leads to a marked reduction in the amount of shifted material (Figure 3B; right). Together these experiments establish that the main domain I binding site is located in the middle third of motif 4.
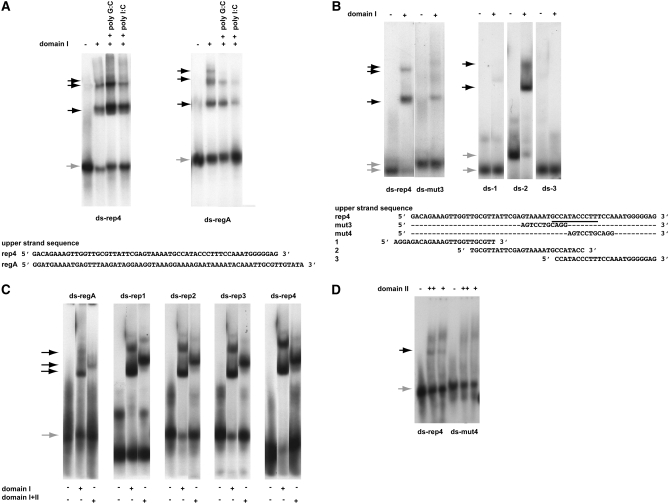
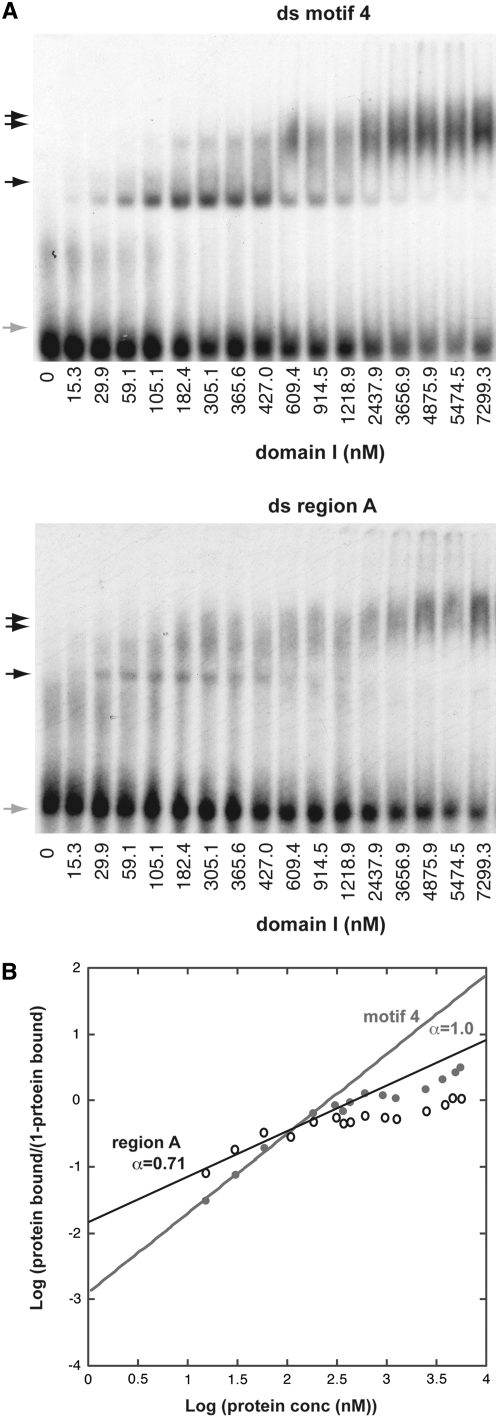
Figure 3.—
DNA-binding specificities of the two Rtf1 c-myb-like domains. A key above the panels defines the experimental conditions used. Unbound (shaded arrows) and shifted material (solid arrows) are indicated for each panel. Unless otherwise stated, the experiments were done in the presence of 100 μg/ml poly G:C. The DNA olignucleotides utilized are given at the bottom of each panel. Western analysis of shifted material is provided for A and B as supplemental data (Figure S2F). (A) Gel-shift assays using purified domain I protein and a dsDNA oligonucleotide resembling motif 4 (ds-rep4; left) and region A (ds-regA; right). The sequences of the “upper” strands of the dsDNA oligonucleotides are displayed. In both panels, DNA binding challenged by the addition of 50 μg/ml nonspecific competitors (given) does not abolish the observed shifts. However, binding can be efficiently outcompeted by the addition of specific competitors constituted by unlabeled substrates (supplemental Figure S2G). (B) Definition of the domain I's binding site within the motif 4 sequence. (Left) Three gel-shift assays using three different segments of motif 4. The sequences of the upper strands of the three dsDNA oligonucleotides, 1, 2, and 3, are displayed. (Right) Gel-shift assay utilizing dsDNA oligonucleotides ds-rep4 or ds-mut3. The different mobilities observed for unbound wild-type and mutant dsDNA oligonucleotides are due to the presence of a GATC overhang on the ds-mut3 oligonucleotide. (C) Comparison of domain I's and the chimeric domain's affinities to the five different dsDNA oligonucleotides constituting region A (ds-regA) and each of the four repeated region B motifs (ds-rep1, 2, 3, and 4). Unbound and shifted material is indicated to the left of the panels with shaded and solid arrows, respectively. The names of the utilized oligonucleotides are shown above the panels. It should be noted, that the retardation observed using the chimeric domains is greater than that observed for the individual domains. Since the increased retardation reflects the increased molecular size the observation establishes independently that the gel-shifts are due to binding of the purified domain(s). (D) Domain II interacts weakly with motif 4. Gel-shift assay using purified domain II protein and dsDNA oligonucleotides ds-rep4 and ds-mut4. A weak gel-shift is only observed with the wild-type sequence (ds-rep4) but not the mutant (ds-mut4). Importantly, we do not see this shift in the presence of unspecific poly I:C competitor DNA (data not shown), suggesting that while the result obtained using the ds-mut4 oligo indicates that the interaction is sequence specific the interaction must be weak as it can be outcompeted with an unspecific competitor. Also, the smear observed in the top section of lanes 2, 3, 5, and 6 is due to the domain II interacting with single-stranded oligo DNA (see below).
Interestingly, in this part of motif 4, purines and pyrimidines are distributed asymmetrically between the two strands. As mentioned in the introduction the RTS1 element possesses an enhancer region characterized by an asymmetric distribution of pyrimidines and purines. We decided to investigate if domain I also displays an affinity for region A dsDNA (Figure 3A; right). Again, gel-shift assays detected DNA binding. The binding could somewhat be outcompeted with poly I:C but not poly G:C, thus displaying some specificity. Western analysis of shifted material verifies that the shift is due to binding of domain I (supplemental Figure S2B), and that binding can be outcompeted with excess cold-specific competitor (supplemental Figure S2C). However, the domain I displays a lower affinity for region A (Kd = 3467 nm) than for motif 4 dsDNA (Kd = 549 nm; Figure 4). Importantly, assays with the segment containing the chimeric domains detected similar binding specificities as observed for domain I only; shifts of a slightly reduced intensity are observed for all four region B motifs as well as for the enhancer region A (Figure 3C, domains I + II).
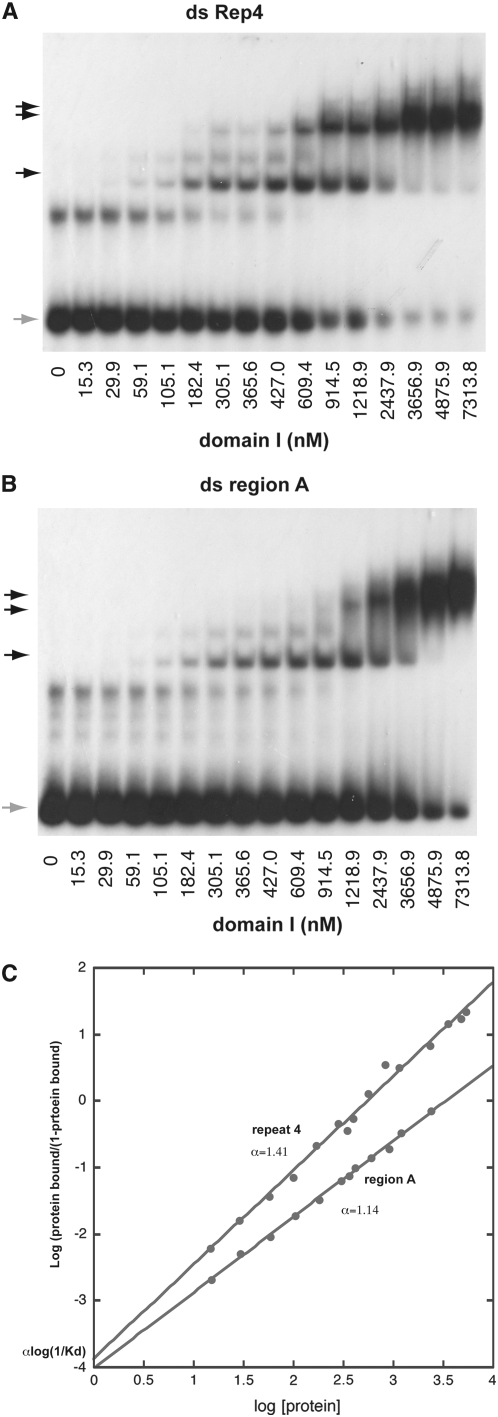
Figure 4.—
Characterization of domain I binding to double-stranded region A and repeat 4. (A) Gel shift of ds repeat 4 DNA while titrating domain I. (B) Gel shift of ds region A DNA while titrating domain I. (C) Hill plot of the data points obtained above. The binding to repeat 4 and region A DNA fit a simple model with a Hill coefficient of 1.45 and 1.16, respectively, characteristic of low or no synergistic binding. The dissociation constant (Kd) for domain I binding to repeat 4 DNA is determined as 549 nm. Binding to region A is slightly weaker than to repeat 4 with a Kd of 3467 nm.
Rtf1 domain II binds region B dsDNA:
As mentioned above, a TTF1 domain II mutation which abolishes dsDNA binding has been identified (Evers et al. 1995). We therefore tested if the purified domain II displays an affinity for region A or motif 4 dsDNA oligonucleotide. No domain II binding was detected using the region A dsDNA oligonucleotide (data not shown), however, a weak shift is observed for motif 4 dsDNA oligonucleotide (Figure 3D). Importantly, the shift is only observed in the absence of unspecific competitor poly I:C DNA, suggesting that the interaction either is sequence unspecific or that the domain also can interact in a sequence-unspecific manner (data not shown). We therefore proceeded to test whether the motif 4 linker substitutions described above affected binding and found that when the rep4-mut4 mutation is introduced, the shift is abolished, showing that the detected interaction is sequence specific (data not shown; Figure 3C). This substitution, which also abolishes motif 4 barrier function in vivo (Codlin and Dalgaard 2003), affects the sequence which shows similarity to the binding sequence defined for S. pombe Reb1 (Melekhovets et al. 1997). Thus, the observations are consistent with Rtf1 domain II interacting with the motif's Reb1-like recognition sequence (Codlin and Dalgaard 2003).
Importantly, we have previously established that a single motif can act as a weak replication barrier, and that in the absence of region A, the introduction of additional motifs has an additive effect on the overall barrier activity (Codlin and Dalgaard 2003). The datasets are therefore consistent with Rtf1 molecules binding each of the four repeats present in region B in vivo. We also establish that domain I, but not domain II, can interact specifically but with a lesser affinity with region A dsDNA. This potentially allows at least five Rtf1 molecules to act at RTS1 (see discussion).
Domain I is involved in establishment of the polarity of the RTS1 barrier activity:
To gain further insight into the mechanism of Rtf1-mediated replication termination at RTS1, we decided to investigate the in vivo activity of mutant rtf1 alleles, containing aa substitutions. The analyzed domain II point mutations either strongly reduced or abolished barrier activity (mutations rtf1-S340F, rtf1-R293K, and rtf1-M343R; data not shown). However, while abolishment of the wild-type barrier activity is observed in the six mutant domain I alleles, a novel barrier signal could be observed in some; the signal is the strongest in the rtf1-S154L genetic background (Figure 5A), is detectable in the rtf1-L162Y strain (supplemental Figure S2D), barely detectable in the rtf1-P136L strain, and is absent for rtf1-L129F and rtf1-G183E (data not shown). When the SacI–PstI fragment is analyzed, the wild-type signal is located close to the apex on the ascending part of the Y arc (Figure 5A, inset), however, the novel signal is located on the descending part (Figure 5A, middle). This novel barrier signal is strongest when only the cis-acting region B is present; for unknown reasons the presence of region A causes a reduction of the signal intensity (compare Figure 5A and 5B). There are two possible explanations for the appearance of this novel barrier signal: either the forks replicate through the RTS1 sequence and pause at a de novo site outside the element or the RTS1 barrier activity has inverted its polarity now pausing replication forks moving in the opposite direction (we conclude that only replication pausing occurs as we do not observe any termination signal). To discriminate between the two possibilities, we first excluded that replication forks were stalling at a different position within the plasmid DNA. An analysis of an empty plasmid detected no barrier signal (supplemental Figure S2E), thus, the RTS1 cis-acting sequence is still required for Rtf1-S154L-mediated pausing. We also verified that the novel Rtf1-S154L barrier is dependent on swi1+ and swi3+ activities (supplemental Figure S2F), and that the novel signal could be observed when the element was cloned in both orientations within the plasmid (Figure 5, A, B, and D). Again in the presence of region A, the barrier intensity of the signal is lower and only clearly visible when located close to the middle of the fragment (Figure 5D; also a relative difference in intensity is observed for the wild-type barrier in the two orientations, supplemental Figure S2G; left). Finally, we excluded that the novel barrier is due to “collisions” with polymerase II transcription initiated at the flanking nmt1 promoter, similar to the collisions recently observed between transcription forks initiated by polymerase III and replication forks (Krings and Bastia 2006). Changes between repressed “low-level” and induced “high-level” nmt1-promoter mediated polymerase II transcription has no effect on the wild-type RTS1 activity (supplemental Figure S2G). However, while we observed no effect of polymerase II transcription on Rtf1-S154L barrier activity, when the transcription forks move in the same direction as the paused replication forks (Figure 5C), a reduction of the barrier activity is observed when the transcription occurs in the opposite direction (Figure 5E). A possible explanation is that transcription displaces Rtf1-S154L molecules bound to the DNA. We then investigated the second possibility that the polarity of the RTS1 barrier has changed in the Rtf1-S154L genetic background. We utilized the method where the polarity of a replication barrier can be established by analyzing overlapping restriction fragments of replication intermediates such that the position of the barrier is moved from one end of the DNA fragment to the other. This analysis was done for plasmids containing RTS1-derived elements in both orientations, and it verified that the polarity of the Rtf1-S154L barrier is inverted (Figure 5, A and D). To investigate the possibility that the change in polarity was due to the S154L mutation affecting domain I DNA binding, we purified the mutant domain and analyzed its binding to motif 4 and region A dsDNA. We observed gel-shift signals using the S154L-domain I at lower concentrations than observed with the wild-type domain I (Kd = 264 nm and 343 nm for motif 4 and region A, respectively; Figure 6), establishing that the mutant domain is binding with a greater affinity than the wild-type domain. However, at the lower protein concentrations we also observed a smaller Hill coefficient in both cases: 1.0 and 0.71 vs. 1.41 and 1.14 for motif 4 and region A, respectively (Figures 4 and 6). At higher protein concentrations there is no linear fit but a stronger negative cooperativity. Thus, the mutation affects the domain's ability to form multimeric complexes with both region A and motif 4.
Figure 5.—
(A) The domain I point mutation, S154L, changes the polarity of the RTS1 replication barrier activity. Pause signals are indicated by blue arrows. Determination of the polarity of the rtf1-S154L region B replication barrier using 2D-gel analysis of replication intermediates. The polarity is determined by analyzing overlapping fragments where the position of region B is shifted from one end of the analyzed fragment to the other. The polarity of the barrier activity can be determined by comparing the position of the barrier signal on the arc constituted by Y structures between the three panels. The position on the Y arc relative to the 1N and 2N signals shows how far the replication fork has traveled into the analyzed fragment before it was paused. The polarity of the replication barrier determined by the analysis is given below (red arrows). The polarity and position of the stalled fork is displayed above each panel. Inset, 2D-gel analysis of the wild-type region B, SacI–PstI fragment. A 2D-gel analysis of the genomic RTS1 element in the rtf1-S154L genetic background, verifies the loss of the wild-type barrier activity, but fails to detect an activity with inverted polarity suggesting that the novel barrier is only observed when RTS1 is located on a plasmid (supplemental Figure S2I). (B) The rtf1-S154L barrier activity is not enhanced by region A. Analysis of regions A and B, cloned in the same orientation as region B shown in B. The pause signal is indicated by a blue arrow. The barrier signal is only clearly visible on the analyzed PacI–KpnI fragment. (C) Transcription initiated at the flanking nmt-promoter does not affect the rtf1-S154L barrier activity. The pause signal is indicated by a blue arrow. (D) Analysis of the polarity of the Rtf1-S154L RTS1 barrier, cloned in inverted orientation. See A for the description of symbols. Pause signals are indicated by blue arrows. (E) Transcription initiated at the nmt1-promoter reduces Rtf1-S154L RTS1 barrier activity when transcription moves in the opposite direction of that of the stalled replication forks. The line drawing at the bottom displays the relative orientation of the nmt1 promoter and the RTS1 element within plasmid pBZ143. Inset displays an enlargement of the apex of the Y arcs observed when analyzing the Pst1–SacI fragment in the presence and absence (middle autoradiograph in D) of transcription. The pause signals are indicated by blue arrows.
Figure 6.—
Characterization of domain I-S154L binding to double-stranded motif 4 and region A DNA. (A) Gel shift of ds motif 4 and region A DNA while titrating domain I-S154L. (B) Hill plot of the data points obtained above, only data points for the five lowest concentrations were used for the linear fit. The Hill coefficient of 1.0 and 0.71 were obtained for motif 4 and region A, respectively. The dissociation constant Kd for domain I-S154L binding to repeat 4 and region A DNA is estimated at 265 nm and 343 nm, respectively.
The Rtf1 C-terminal region is required for function and can mediate dimerization/polymerization:
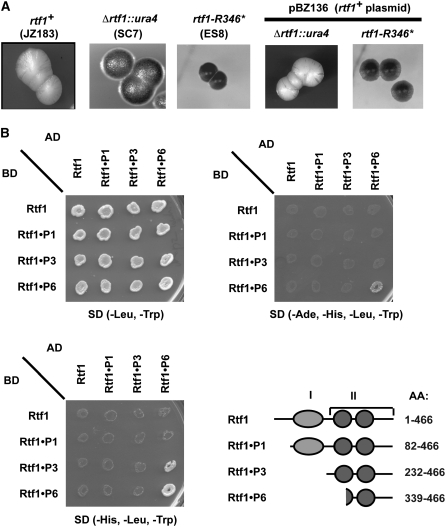
Finally, a genetic screen for dominant mutants was conducted. A multicopy plasmid carrying the rtf1 gene was transformed into the JZ183 strain, and the obtained strain was mutagenized. One mutant with increased iodine staining was isolated. Analysis of RTS1 replication intermediates verified that there is a complete loss of replication barrier activity in this mutant (supplemental data; supplemental Figure S2J). By crossing the isolated mutant strain with the Δrtf1 strain (SC8), and observing that no crossovers occurred in 27 tetrads analyzed, it was established that the mutation is closely linked to Rtf1 (data not shown). Sequence analysis of the rtf1 gene detected a mutation introducing a nonsense codon at position 346, leading to a 120-aa truncation of the Rtf1 protein. Transformation of the strain with an rtf1+ plasmid (pBZ136) verified that the isolated strain carried a partially rtf1+-dominant mutation (Figure 7A; strain ES8). One possible model for the partially dominant effect of this truncation is that it inhibits a functionally important dimerization or oligomerization of the Rtf1 molecules. To test this hypothesis, we employed a two-hybrid analysis. A self-interaction could be detected with the 127-aa C-terminal region of Rtf1 that includes one of the myb-sant domains (Figure 7B). However, this interaction was masked by the presence of DNA-binding domains, probably because the fusion proteins could bind at other positions in the S. cerevisiae genome with greater affinity than at the reporter genes used for the assay. Thus, our genetic analysis shows that the Rtf1 C-terminal region is required for RTS1 function, and the two-hybrid results establish that this is through a role in Rtf1 dimerization or polymerization.
Figure 7.—
The Rtf1 C-terminal tail is required for function. (A) Characterization of the dominant rtf1-R346* mutation. The three left sections display the iodine staining phenotypes of sporulating wild-type, Δrtf1, and the rtf1-R346* colonies, respectively. The strains carry the RTS1 allele that allows quantification of in vivo barrier activity by iodine staining of sporulating colonies (Figure 1, C and D). In this genetic background wild-type rtf1+ strains stain yellow, while rtf1 mutants stain black. The two right sections show that the introduction of the rtf1+ plasmid (pBZ136) complements the sporulation phenotype of the recessive Δrtf1 mutant but not the dominant rtf1-R346* mutation (right). Strain names are given in parentheses. (B) Two-hybrid analysis of Rtf1 amino acid segments' ability to interact. Different Rtf1 segments (graphic outline) were fused to the GAL4 activation domain (AD) and GAL4 DNA-binding domain (BD). Interactions between two fusion proteins are detected by increased expression of the two reporter genes GAL1-HIS3 and GAL2-ADE2. Expression allows the ade2 his3 S. cerevisiae strain to survive in the absence of histidine and adenine supplements in the media. Only when the Rtf1 C-terminal tail (P6) fused to both the AD- and the BD-domain combined, cells become histidine and adenine prototrophs.
DISCUSSION
The analysis presented here allows us to propose a model for Rtf1-mediated impediment of replication fork progression at RTS1 (Figure 8A). In summary, the presented data suggest that at least five Rtf1 molecules can bind to the double-stranded RTS1 element through interactions involving both of the protein's myb domains but mainly promoted by domain I (Figures 2, 3, 4, and 8A, top). Importantly, Rtf1 is able to interact both with the repeated region B motifs and the enhancer region A.
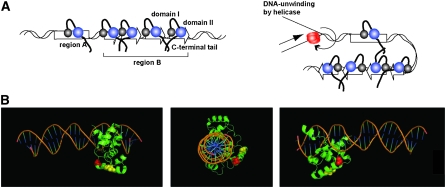
Figure 8.—
(A) Model of Rtf1's mediated termination of replication at RTS1. (B) Model of c-myb in complex with its target DNA. c-myb residues (Ogata et al. 1993) that align with the Rtf-S154L and -L162Y residues are highlighted in red and yellow, respectively (Figure 2B).
The Rtf1 binding to the cis-acting sequences might be stabilized through protein–protein interactions between Rtf1 molecules involving the Rtf1 C-terminal domain (Figure 6). One possibility is that Rtf1 DNA binding at multiple sites within the RTS1 in combination with interactions between Rtf1 molecules acts as a topological constraint for DNA unwinding by the replicative helicase (Figure 7A). Such a constraint could be augmented by DNA looping, a property which already has been observed for c-myb; the c-myb and c/EBP transcription factor complex together mediate DNA looping required for transcriptional activation (Tahirov et al. 2002). In addition, binding of multiple Rtf1 molecules within region B combined with the interaction between Rtf1 molecules could act to recruit Rtf1 to the lower affinity site within the region A dsDNA. Indeed, the dominant phenotype of the Rtf1 allele lacking the C-terminal region (Figure 7) combined with the observation that region A has no intrinsic barrier activity but mediates a cooperative enhancement of the region B activity (Codlin and Dalgaard 2003) strongly support a role of C-terminal domain's self-interaction in recruitment of Rtf1 to the enhancer region A. One possibility we are investigating is that the protein interacts with single-stranded DNA formed at region A when the DNA is unwound by the replicative helicase (T. Eydmann and J. Z. Dalgaard, unpublished observation).
We identify a domain I mutation that changes the polarity of RTS1 (Figure 5). When the Rtf1 domain I mutations that cause this inversion of the barrier's polarity are superimposed on the known structure of c-myb in complex with its dsDNA-binding site, it is evident that the mutation is not located on the DNA-binding surface (Figure 8B). When the initial Kd is estimated for this domain, we find that it is lower than that of the wild-type domain, suggesting a stronger DNA affinity; however, at the higher protein concentrations we observe a decreased affinity and a Hill coefficient <1. Thus while the mutation does not significantly affect the initial complex formation, the mutant protein does display a decreased ability to form a multimeric complex. The characteristics of this rtf1 allele add some support to the model that unknown protein–protein interaction(s) involving domain I and replication protein(s) are affected by the mutation. Among the replication proteins, the replicative helicase [mini-chromosome maintenance proteins (MCMs), reviewed by Takahashi et al. 2005], as well as Rtf2 (Codlin and Dalgaard 2003), Swi1, and Swi3 factors are likely candidates. Swi1 and Swi3 travel with the replication fork (Katou et al. 2003; Noguchi et al. 2004) and act at MPS1 to coordinate pausing of leading-strand replication in response to a lagging-strand signal (Figure 1A; Vengrova and Dalgaard 2004). The identification of a swi1-rtf mutation, which only affects termination of replication at RTS1 but not at other replication barriers establishes that such RTS1-specific interactions involving replication fork proteins do occur (Codlin and Dalgaard 2003; Krings and Bastia 2004). This parallels the situation in Escherichia coli, where the transacting factor Tus is thought to mediate replication termination through direct interactions with the replicative helicase DnaB (Mulugu et al. 2001). Importantly, the observation that in the Rtf1-S154L genetic background there is a loss of replication termination activity affecting the forks moving in one direction, but a gain of replication pausing activity acting on forks moving in the other, shows that the proposed Rtf1 domain I interactions are of importance when the element is replicated in both directions: Wild-type Rtf1 domain I interactions are required for efficient replication termination of the forks moving in one direction, but also must act to prevent pausing of the forks moving in the opposite.
Finally it should be noted that the identification of two DNA binding domains within the Rtf1 protein could have implications for understanding the molecular mechanisms underlying a wide range of activities attributed to the Reb1/TTF1/Rtf1 protein family; polymerase II transcription activation (Carmen and Holland 1994; Graham and Chambers 1994; Packham et al. 1996; Wang and Warner 1998), polymerase I transcription activation/repression (Wang et al. 1990), and termination (Lang and Reeder 1993; Lang et al. 1994; Mason et al. 1997; Melekhovets et al. 1997; Zhao et al. 1997), as well as chromatin insulator function (Fourel et al. 2001). Interactions with double-stranded DNA as well as dynamic changes in these interactions could play an important role for all these molecular processes.
Acknowledgments
We thank our colleagues at the Marie Curie Research Institute for helpful suggestions and interactions. A special thanks to Rob Cross, Natalie Mansfield, S. Jack Carlisle, Doug Drummond, Sonya Vengrova, and Michael Bonaduce for technical assistance. This work was supported by the Intramural Research Program of the National Cancer Institute of the National Institutes of Health (A.J.S.K.), the Marie Curie Cancer Care (J.Z.D.) and the Association of International Cancer Research (J.Z.D.).
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. DS:[57973].
References
- Ahn, J. S., F. Osman and M. C. Whitby, 2005. Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast. EMBO J. 24 2011–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli, B., and A. J. Klar, 1991. A novel switch-activating site (SAS1) and its cognate binding factor (SAP1) required for efficient mat1 switching in Schizosaccharomyces pombe. EMBO J. 10 3025–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangioli, B., 1998. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 17 4503–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, P. L., C.-T. Chien, R. Sternglanx and S. Fields, 1993. Using the two-hybrid system to detect protein-protein interactions, pp. 153–179 in Cellular Interaction in Development: A Practical Approach, edited by D. A. Hartley. Oxford University Press, Oxford.
- Boyer, L. A., R. R. Latek and C. L. Peterson, 2004. The SANT domain: A unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5 158–163. [DOI] [PubMed] [Google Scholar]
- Brewer, B. J., and W. L. Fangman, 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51 463–471. [DOI] [PubMed] [Google Scholar]
- Brewer, B. J., and W. L. Fangman, 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55 637–643. [DOI] [PubMed] [Google Scholar]
- Carmen, A. A., and M. J. Holland, 1994. The upstream repression sequence from the yeast enolase gene ENO1 is a complex regulatory element that binds multiple trans-acting factors including REB1. J. Biol. Chem. 269 9790–9797. [PubMed] [Google Scholar]
- Codlin, S., and J. Z. Dalgaard, 2003. Complex mechanism of site-specific DNA replication termination in fission yeast. EMBO J. 22 3431–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard, J. Z., and A. J. Klar, 1999. Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400 181–184. [DOI] [PubMed] [Google Scholar]
- Dalgaard, J. Z., and A. J. Klar, 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102 745–751. [DOI] [PubMed] [Google Scholar]
- Dalgaard, J. Z., and A. J. Klar, 2001. A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev. 15 2060–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Guan, C., P. Li, P. D. Riggs and H. Inouye, 1988. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene 67 21–30. [DOI] [PubMed] [Google Scholar]
- Evers, R., A. Smid, U. Rudloff, F. Lottspeich and I. Grummt, 1995. Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J. 14 1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel, G., C. Boscheron, E. Revardel, E. Lebrun, Y. F. Hu et al., 2001. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, J. K., E. Gogel, C. Berger, M. Wallisch, F. Muller et al., 1997. Termination of mammalian rDNA replication: polar arrest of replication fork movement by transcription termination factor TTF-I. Cell 90 559–567. [DOI] [PubMed] [Google Scholar]
- Graham, I. R., and A. Chambers, 1994. A Reb1p-binding site is required for efficient activation of the yeast RAP1 gene, but multiple binding sites for Rap1p are not essential. Mol. Microbiol. 12 931–940. [DOI] [PubMed] [Google Scholar]
- Huang, J., I. L. Brito, J. Villen, S. P. Gygi, A. Amon et al., 2006. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 20 2887–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman, J. A., L. D. Spotila, K. A. Nawotka, S. M. el-Assouli and L. R. Davis, 1987. The in vivo replication origin of the yeast 2 microns plasmid. Cell 51 473–481. [DOI] [PubMed] [Google Scholar]
- Hyrien, O., 2000. Mechanisms and consequences of replication fork arrest. Biochimie 82 5–17. [DOI] [PubMed] [Google Scholar]
- Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka et al., 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424 1078–1083. [DOI] [PubMed] [Google Scholar]
- Kiger, Jr., J. A., and R. L. Sinsheimer, 1969. Vegetative lambda DNA. IV. Fractionation of replicating lambda DNA on benzoylated-naphthoylated DEAE cellulose. J. Mol. Biol. 40 467–490. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., and T. Horiuchi, 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1 465–474. [DOI] [PubMed] [Google Scholar]
- Krings, G., and D. Bastia, 2004. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 101 14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings, G., and D. Bastia, 2005. Sap1p binds to Ter1 at the ribosomal DNA of Schizosaccharomyces pombe and causes polar replication fork arrest. J. Biol. Chem. 280 39135–39142. [DOI] [PubMed] [Google Scholar]
- Krings, G., and D. Bastia, 2006. Molecular architecture of a eukaryotic DNA replication terminus-terminator protein complex. Mol. Cell. Biol. 26 8061–8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, S., A. Watson, D. M. Sheedy, B. Martin and A. M. Carr, 2005. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121 689–702. [DOI] [PubMed] [Google Scholar]
- Lang, W. H., B. E. Morrow, Q. Ju, J. R. Warner and R. H. Reeder, 1994. A model for transcription termination by RNA polymerase I. Cell 79 527–534. [DOI] [PubMed] [Google Scholar]
- Lang, W. H., and R. H. Reeder, 1993. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol. Cell. Biol. 13 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens, M. H., and J. A. Huberman, 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8 4927–4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Estrano, C., J. B. Schvartzman, D. B. Krimer and P. Hernandez, 1998. Co-localization of polar replication fork barriers and rRNA transcription terminators in mouse rDNA. J. Mol. Biol. 277 249–256. [DOI] [PubMed] [Google Scholar]
- Lopez-Estrano, C., J. B. Schvartzman, D. B. Krimer and P. Hernandez, 1999. Characterization of the pea rDNA replication fork barrier: putative cis-acting and trans-acting factors. Plant. Mol. Biol. 40 99–110. [DOI] [PubMed] [Google Scholar]
- Mason, S. W., M. Wallisch and I. Grummt, 1997. RNA polymerase I transcription termination: similar mechanisms are employed by yeast and mammals. J. Mol. Biol. 268 229–234. [DOI] [PubMed] [Google Scholar]
- Mejia-Ramirez, E., A. Sanchez-Gorostiaga, D. B. Krimer, J. B. Schvartzman and P. Hernandez, 2005. The mating type switch-activating protein Sap1 Is required for replication fork arrest at the rRNA genes of fission yeast. Mol. Cell. Biol. 25 8755–8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melekhovets, Y. F., P. S. Shwed and R. N. Nazar, 1997. In vivo analyses of RNA polymerase I termination in Schizosaccharomyces pombe. Nucleic Acids Res. 25 5103–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty, B. K., N. K. Bairwa and D. Bastia, 2006. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, S., A. Klar and P. Nurse, 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194 795–823. [DOI] [PubMed] [Google Scholar]
- Mulugu, S., A. Potnis, J. Shamsuzzaman, K. Taylor, K. Alexander et al., 2001. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. Proc. Natl. Acad. Sci. USA 98 9569–9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi, E., C. Noguchi, W. H. McDonald, J. R. Yates III and P. Russell 2004. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol. Cell. Biol. 24 8342–8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, K., H. Kanai, T. Inoue, A. Sekikawa, M. Sasaki et al., 1993. Solution structures of Myb DNA-binding domain and its complex with DNA. Nucleic Acids Symp. Ser. 29 201–202. [PubMed] [Google Scholar]
- Packham, E. A., I. R. Graham and A. Chambers, 1996. The multifunctional transcription factors Abf1p, Rap1p and Reb1p are required for full transcriptional activation of the chromosomal PGK gene in Saccharomyces cerevisiae. Mol. Gen. Genet. 250 348–356. [DOI] [PubMed] [Google Scholar]
- Petty, K. J., 1996. Metal-chelate affinity chromatography, unit 10.11B in Current Protocols in Molecular Biology, edited by F. M. Ausubel. John Wiley & Sons, Malden, MA. [DOI] [PubMed]
- Putter, V., and F. Grummt, 2002. Transcription termination factor TTF-I exhibits contrahelicase activity during DNA replication. EMBO Rep. 3 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D. W. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanchez-Gorostiaga, A., C. Lopez-Estrano, D. B. Krimer, J. B. Schvartzman and P. Hernandez, 2004. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol. Cell. Biol. 24 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria, D., E. Viguera, M. L. Martinez-Robles, O. Hyrien, P. Hernandez et al., 2000. Bi-directional replication and random termination. Nucleic Acids Res. 28 2099–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier, F. W., and B. A. Moffatt, 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189 113–130. [DOI] [PubMed] [Google Scholar]
- Tahirov, T. H., K. Sato, E. Ichikawa-Iwata, M. Sasaki, T. Inoue-Bungo et al., 2002. Mechanism of c-Myb-C/EBP beta cooperation from separated sites on a promoter. Cell 108 57–70. [DOI] [PubMed] [Google Scholar]
- Takahashi, T. S., D. B. Wigley and J. C. Walter, 2005. Pumps, paradoxes and ploughshares: mechanism of the MCM2–7 DNA helicase. Trends Biochem. Sci. 30 437–444. [DOI] [PubMed] [Google Scholar]
- Takeuchi, Y., T. Horiuchi and T. Kobayashi, 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengrova, S., and J. Z. Dalgaard, 2004. RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genes Dev. 18 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., P. R. Nicholson and D. J. Stillman, 1990. Identification of a Saccharomyces cerevisiae DNA-binding protein involved in transcriptional regulation. Mol. Cell. Biol. 10 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K. L., and J. R. Warner, 1998. Positive and negative autoregulation of REB1 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 18 4368–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein, Y., J. N. Ihle, S. Lavu and E. P. Reddy, 1986. Truncation of the c-myb gene by a retroviral integration in an interleukin 3-dependent myeloid leukemia cell line. Proc. Natl. Acad. Sci. USA 83 5010–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, A., A. Guo, Z. Liu and L. Pape, 1997. Molecular cloning and analysis of Schizosaccharomyces pombe Reb1p: sequence-specific recognition of two sites in the far upstream rDNA intergenic spacer. Nucleic Acids Res. 25 904–910. [DOI] [PMC free article] [PubMed] [Google Scholar]