Abstract
On the basis of multiple complete mitochondrial DNA genome sequences, we describe the temporal phylogeography of Atlantic cod (Gadus morhua), a lineage that has undergone a complex pattern of vicariant evolution, postglacial demographic shifts, and historic sharp population declines due to fishing and/or environmental shifts. Each of 32 fish from four spawning aggregations from the northwest Atlantic and Norway has a unique mtDNA sequence, which differs by 6–60 substitutions. Phylogenetic analysis identifies six major haplogroups that range in age from 37 to 75 KYA. The widespread haplotype identified by previous single-locus analyses at the center of a “star phylogeny” is shown to be a paraphyletic assemblage of genome lineages. The coalescent that includes all cod occurs 162 KYA. The most basal clade comprises two fish from the western Atlantic. The most recent superclade that includes all fish examined from Norway, and which includes 84% of all fish examined, dates to 128 KYA at the Sangamon/Würm interglacial, when ocean depths on continental shelves would have favored transcontinental movement. The pairwise mismatch distribution dates population expansion of this superclade to the middle of the Wisconsinan/Weichsel glaciation 59 KYA, rather than to a postglacial emergence from a marine refugium 12 KYA, or to more recent historic events. We discuss alternative scenarios for the expansion and distribution of the descendants of the “codmother” in the North Atlantic. Mitochondrial phylogenomic analyses generate highly resolved trees that enable fine-scale tests of temporal hypotheses with an accuracy not possible with single-locus methods.
EVOLUTIONARY analysis of mitochondrial DNA (mtDNA) has graduated from RFLP mapping (Brown et al. 1979; Wilson et al. 1985) to direct sequencing of single-loci (Kocher et al. 1989; Carr and Marshall 1991) to comparisons of complete genome sequences among species (Horai et al. 1995; Inoue et al. 2001; Coulson et al. 2006). Recent intraspecific analyses of complete human mtDNA genomes have supported the “mitochondrial Eve” hypothesis and clarified the historical emergence of her daughters “out of Africa” (Ingman et al. 2000; Torroni et al. 2006).
Atlantic cod (Gadus morhua L. 1758) is another lineage that has undergone a complex pattern of phylogeographic evolution, including vicariant events and population fluctuations attributable to long-term geological events, short-term ecological history, and contemporary anthropogenic fishing and/or environmental shifts (Myers et al. 1995; Hutchings 1996; Rose et al. 2001; Rose 2004, 2007; Coulson et al. 2006; International Council for the Exploration of the Sea 2006).
We have shown (Carr et al. 1999; Coulson et al. 2006) that the basal gadine genera are endemic to the northeast Atlantic (Melanogrammus and Merluccius). The sister genera to Gadus L. 1758 are the Polar basin Arctogadus and Boreogadus. The genus Gadus comprises three nominal species, including Atlantic cod (G. morhua), its sister species walleye pollock [G. (= Theragra) chalcogrammus], and Pacific cod (G. macrocephalus) (cf. Figure 4 of Coulson et al. 2006). The latter two species are found on both Pacific coasts and north of the Bering Strait. Greenland cod, found in the Davis Strait west of Greenland and previously considered a separate species G. ogac, is a subspecies of G. macrocephalus and apparently represents a tertiary invasion of the western Atlantic via the Canadian arctic archipelago. We presented a model in which Gadus is of North Atlantic origin, and the two Pacific species derive from separate but simultaneous invaders of the Pacific through the Bering Strait 3.5 MYA. Pogson and Mesa (2004) suggested instead that the genus was of Pacific origin, with morhua reentering the North Atlantic via a polar route. Although this latter model requires only a single event and Pacific to Atlantic vicariance is more common (Vermeij 1991), we suggested that it is difficult to understand how speciation of macrocephalus and chalcogrammus and their current distribution could have arisen in sympatry.
Given our model, current patterns would have carried the ancestor of morhua through the polar basin east of Greenland into the northeastern Atlantic. Then, morhua spread westward via Iceland and Greenland to the coast of North America. Pleistocene glaciations of the Grand Banks and the rest of the Newfoundland and Labrador continental shelf may then have restricted suitable cod habitat to southerly marine refugia, such as the Flemish Cap, an offshore seamount and putative marine refugium during the Wisconsinan glaciation and especially the last glacial maximum 8–13 KYA (Shaw 2006).
Population genetic analysis of cod goes back to the roots of experimental population genetics, including first identification of the Wahlund effect (Wahlund 1928), an observed deficiency of heterozygotes in a geographically structured population vs. the expectation of panmixis. Subsequent investigations by protein allozymes (Cross and Payne 1978) and DNA microsatellites (Ruzzante et al. 1999, 2001) have typically adopted phenetic methods of analysis, in which fish are aggregated a priori, for example, as samples from geographically delimited management zones. Analysis may assume that aggregations drawn for a particular zone are necessarily representative of the entire zone, rather than, for example, of a latitudinal cline. Peculiarities of particular loci may bias results (Nielsen et al. 2006).
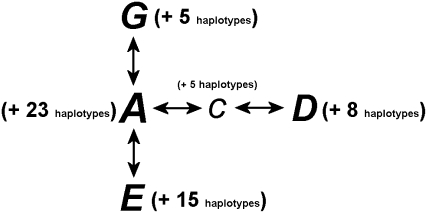
Phylogenetic approaches based on DNA sequences of individual fish have the potential to identify reciprocally monophyletic population lineages a posteriori (Slatkin and Maddison 1989; Slatkin and Hudson 1991; Avise 2000). Previous sequence analysis of a 0.3–0.4 kbp portion of the mitochondrial Cytochrome b locus of almost 1300 fish from the western Atlantic, Greenland, Iceland, and Norway identified 60 haplotypes, of which 4 accounted for ∼90% of the observed variation (Figure 1). One haplotype (“A”) occurs in 54% of all fish examined, and only three others occur at overall frequencies of >3%: “D” (10%), “E” (15%), and “G” (9%) (Árnason 2004). Haplotype A accounts for >70% of cod in the northwest Atlantic (Carr and Marshall 1991; Pepin and Carr 1993; Carr et al. 1995; Carr and Crutcher 1998). Eastern and mid-Atlantic cod populations have much greater single-locus mtDNA diversity as compared with the northwest Atlantic (Árnason 2004). The single-locus picture of cod in the northwest Atlantic is thus a “star phylogeny” centered on haplotype A, which might suggest a relatively recent population expansion (Árnason 2004). For example, protein (Cross and Payne 1978), microsatellite (Bentzen et al. 1996; Ruzzante et al. 1999, 2001), and single-locus mtDNA studies (Carr and Crutcher 1998) all suggest that the most genetically distinctive population in the offshore northwest Atlantic occurs at Flemish Cap, in accordance with the refugial model. Unfortunately, the shallow depth of the star phylogram does not provide sufficient temporal resolution to make a rigorous test of the hypothesis that the distribution of continental fish populations on the continental shelves reflects postglacial expansion <13 KYA.
Figure 1.—
Single-locus mtDNA cytochrome B haplotypes (after Árnason 2004). Analysis of almost 1300 individual cod from the North Atlantic has identified 60 haplotypes in a 0.2–0.4 kbp region of the mitochondrial cytochrome b locus. One of these (A) occurs in 54% of all fish examined; only 3 others occur at overall frequencies of >3%: D (10%), E (15%), and G (9%). Most of the remaining haplotypes have been observed in only 1 or a few fish each and differ from 1 of the common types by a single substitution. Of these, 23 are derived from A, 8 from D, 15 from E, and 5 from G. Haplotype C (2%) is intermediate between A and D and comprises 5 related haplotypes.
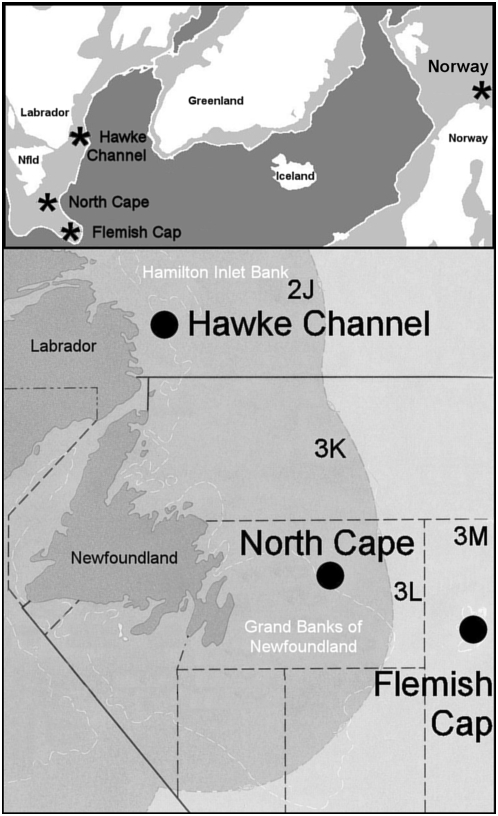
During historic times, populations of Atlantic cod in the northwest Atlantic have been observed to spawn in a variety of inshore and offshore areas along the continental shelf of Newfoundland and Labrador, including populations in the “northern cod” stock complex on the Grand Banks [Northwest Atlantic Fisheries Organization (NAFO) Divisions 3K and 3L], the adjacent Hamilton Bank (NAFO 2J), and at Flemish Cap, an offshore seamount outside the Canadian economic zone (NAFO 3M) (Figure 2). Although it sustained the world's richest fishery for >500 years (Rose 2007), the estimated biomass of northern cod declined >98% from an historic high of 3 × 106 metric tons in the early 1960s to <0.1 × 106 metric tons by the early 1990s (International Council for the Exploration of the Sea 2006). Despite closure of the fishery in 1992, numbers have not recovered (Shelton and Healey 1999; Department of Fisheries and Oceans 2005), and northern cod have been assessed by the Committee on the Status of Endangered Wildlife in Canada (COSEWIC) as “Endangered” (Anonymous 2006).
Figure 2.—
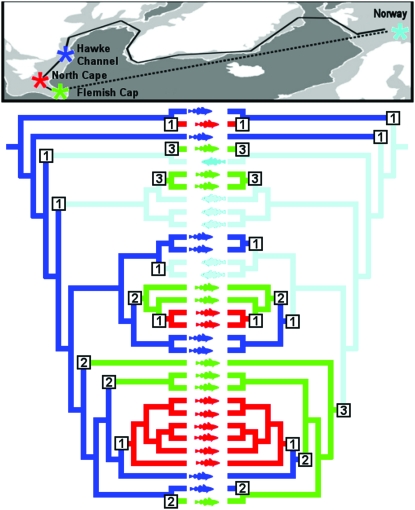
Map of the North Atlantic and sources of codfish samples. The northern cod complex comprises NAFO Divisions 2J, 3K, and 3L: Hawke Channel is a deepwater channel associated with Hamilton Inlet Bank off Labrador and the North Cape is a northward projection of the Grand Banks. Flemish Cap is an off-shore seamount in 3M, outside the Canadian economic zone. The Norwegian coastal sample is from a population near Tromsö, Norway.
We can thus distinguish at least four a priori phenomena over six orders of annual magnitude to explain observed biogeographic patterns of genetic variability in cod: geographic separation from their closest ancestor several 103 KYA, transatlantic vicariance during glacial cycles 101–2 KYA, restriction to and recovery from glacial refugia several tens of thousands of years ago, and fishing pressure and stock collapse within historic times, tens to hundreds of years ago. Just as whole-mtDNA genome data sets lead to statistically robust interspecific phylogenies (Inoue et al. 2001; Miya et al. 2004; Coulson et al. 2006), so too can they provide highly resolved trees to investigate temporal and geographic patterns in intraspecific phylogeography (Ingman et al. 2000; Achilli et al. 2004, 2008). We show here that analysis of a highly corroborated tree provides opportunities for tests of hypotheses, with a precision of temporal discrimination not previously possible.
MATERIALS AND METHODS
We studied 32 fish from four spawning aggregations (Figure 2), two from the northern cod complex at Hawke Channel (Northwest Atlantic Fishery Organization Division 2J, n = 8) and the North Cape of the Grand Banks (3L, n = 9), one from Flemish Cap (3M, n = 9), and one from a Norwegian coastal population near Tromsö, Norway (n = 6). The consensus length was 16,576 base pairs (bp), with a length variant of 1 bp in one individual. The control region (CR) includes a variable number of 40 bp tandem repeats; we excluded 921 bp of this region. The cod CR is otherwise not particularly variable (Johnstone et al. 2007). Genome sequences have been submitted to the NCBI GenBank database and assigned the accession numbers EU877710–EU877741.
A set of 20 primer pairs was identified that amplifies the mitochondrial genome in fragments of 750–1400 bp, with overlaps between adjacent fragments of 80–300 bp (Coulson et al. 2006). Most genomes were sequenced with the BigDye chemistry v. 2.0 (Applied Biosystems) on the ABI377 Prism automated sequencer. Both DNA strands were sequenced. Sequence assemblies were done with Sequencher 4.5 (Gene Codes). Four of the Flemish Cap genomes were sequenced on a custom iterative resequencing microarray (Affymetrix) (Carr et al. 2008), including a quality-control algorithm (Flynn and Carr 2007).
A neighbor-joining tree was constructed with PAUP* 4.0 (Swofford 2002], on the basis of two-parameter maximum likelihood distances (Ts/Tv = 8.5, γ = 0.95) and 10,000 bootstrap replications, with the molecular clock constraint enforced for calculations of the most recent common ancestor (MRCA). Pairwise mismatches and tests of the mismatch distribution were calculated with DnaSP 4.1.10 (Rozas and Rozas 1995). The alternative hypotheses in Figure 3 were evaluated with the help of MacClade (Maddison and Maddison 2000).
Figure 3.—
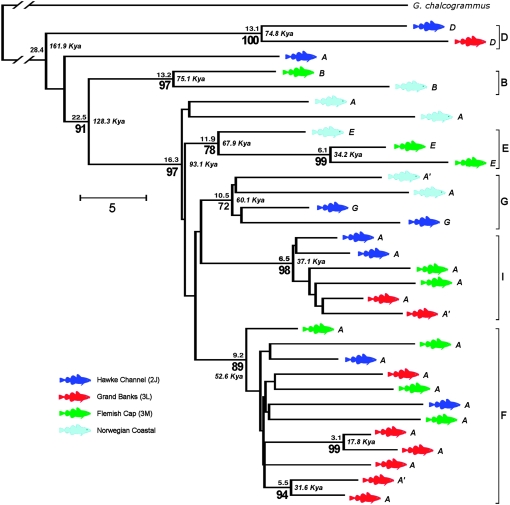
Mitochondrial DNA genome phylogeography of Atlantic cod. A neighbor-joining tree (see materials and methods) rooted with respect to the most closely related species, G. (= Theragra) chalcogrammus (Coulson et al. 2006), is shown. For each clade supported by >70% of 10,000 bootstrap replications (number below vertex), the mean number of nucleotide substitutions to its most recent common ancestor (MRCA) (number above vertex) was used to estimate the time of coalescence (number beside vertex, in italics, in thousands of years), on the basis of an estimated fixation rate of 5703 years per substitution. Distance to the MRCA was calculated using a linearized maximum likelihood model with the molecular clock constraint enforced. The correspondence between single-locus haplotypes (italics) (Árnason 2004) and haplogroup designations (boldface type) is discussed in the text. (A′ haplotypes are single-substitution variants of A. Haplotypes F and I, previously observed only in one fish each, are recycled as designations for the two most abundant genomic haplogroups, as is B).
RESULTS
There were 298 single-nucleotide polymorphisms (SNPs) among 32 fish over 15,655 bp each (total > 500 kbp), of which 98 are phylogenetically informative (sensu Nei 1987). Each of the fish examined has a unique mtDNA sequence: pairwise differences range from 6 to 60. From the mean genetic distance (excluding the CR) between G. morhua and its closest relative Alaska pollock [G. (= Theragra) chalcogrammus] (Coulson et al. 2006) of 0.040 substitutions/site, and on the assumption that the Atlantic cod diverged from its Pacific relative at the time of the last opening of the Bering Strait 3.5 MYA (Grant and Ståhl 1988; Vermeij 1991; Coulson et al. 2006), we calculate a divergence rate of 1.12 × 10−8 substitutions/site/year. This indicates a temporal interval of 5703 years/substitution, which is slightly slower than the interval of 5140 years calculated for the most abundant haplogroup (H) in Homo (Mishmar et al. 2003; Achilli et al. 2004).
Phylogenetic analysis identified 11 haplogroup clades that are supported in >70% of bootstrap replications. Of these, three are pairs of fish within more inclusive clades, four (E, F, G, and I) along with two ungrouped fish constitute a superclade EFGI that includes >84% of the fish examined, and which together with B constitutes a yet more inclusive superclade BEFGI. Haplogroup D is the outgroup to all other fish (Figure 3). The distribution of population samples does not differ significantly from random, either among the six primary clades B, D, E, F, G, and I (χ2 = 20.43, d.f. = 15, P = 0.14), or among these six and the two inclusive superclades EFGI and BEFGI (χ2 = 20.13, d.f. = 21, P = 0.54).
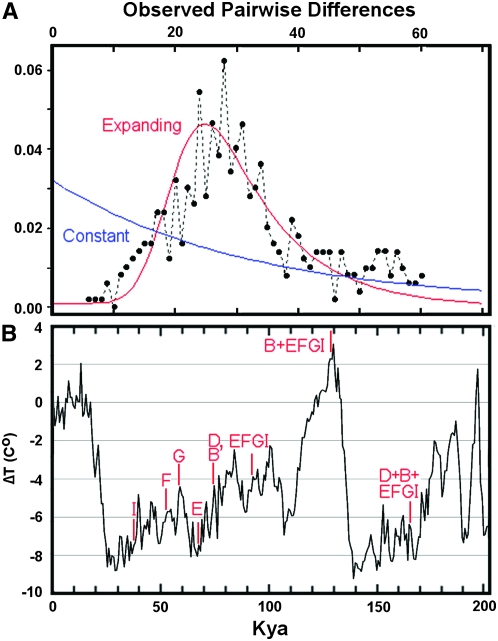
The null hypothesis of constant population size can be evaluated by the correlation of the mean pairwise sequence difference and the number of segregating sites and is rejected both by Tajima's (1989) and Fu and Li's (1993) D statistics (D = −2.23, P < 0.02, and D = −3.26, P < 0.01, respectively). The pairwise mismatch distribution (Rogers and Harpending 1992) (Figure 4A) is dominated by a peak (τ = 20.6) (Rozas and Rozas 1995) that corresponds to expansion of the EFGI superclade 59 KYA. Coalescence times within the EFGI superclade range from 34 to 68 KYA, and the superclade itself originates 93 KYA, (Figure 4B). The most recent common maternal ancestor of all cod examined dates to 162 KYA, corresponding to the divergence of haplogroup D from all others.
Figure 4.—
(A) Pairwise mismatch distributions of Atlantic cod. The mean number of substitutions is 31.1, and ranges from 27.9 to33.3 within the North Cape and Hawke Channel samples, respectively. The null hypothesis of constant population size (smooth curve) is rejected in favor of a population expansion 59 KYA (peaked curve) (Rozas and Rozas 1995), corresponding to the upward inflection of the peak. (B) Reconstruction of the temperature trend from deuterium concentrations in the Vostok ice core over the past 200 KY (Petit et al. 1999, 2001), which is closely tied to analogous data from Greenland (North Greenland Ice Core Project 2004; European Project for Ice Coring in Antarctica Community Members 2006). The geological and biological timescales are calibrated by the estimated evolutionary rate of one substitution/5.7 KY/lineage, so that a pairwise difference of 70 substitutions corresponds to a divergence 200 KYA. The estimated τ = 20.6 (Rozas and Rozas 1995) then implies a population expansion beginning 58.7 KYA. The coalescence times of the major clades are indicated: most occur during the middle of the Wisconsinan/Weichsel glaciation. The second oldest of these (B + EFGI) occurs 128 KYA at the peak of the Sangamon/Würm interglacial, and includes the MRCA of the Norwegian coastal and Flemish Cap populations. The codmother (D + B + EFGI) dates to 162 KYA at the middle of the Illinoian/Saale glaciation.
DISCUSSION
The historical and contemporary distributions of codfish are heavily influenced by the availability of appropriate depths of the continental shelves, which have varied greatly according to sea level shifts during periods of glaciation (Rose 2007; Bigg et al. 2008). The coalescent of all codfish dates to 162 KYA, and the outgroup to all other codfish, haplogroup D, has thus far been observed only in two fish from two populations in the western Atlantic. The coalescent that includes all Norwegian coastal fish (EFGI + B) is more recent, and its origin 128 KYA dates to the very peak of the Sangamon/Würm interglacial (Figure 3). Subsequent expansion at 59 KYA of the EFGI superclade that is responsible for the majority of the observed genomic biodiversity all occur within the middle of the Wisconsinan glaciation (Petit et al. 1999, 2001) (Figure 4B). The smallest observed divergence (six substitutions, between two Grand Banks fish) is threefold greater than the expected two substitutions for a post-Wisconsinan expansion 11.4 KYA. Such an event would predict one or more large cohorts of fish with near-identical mtDNA genome sequences, a phenomenon that has not been observed.
The predominance in the western Atlantic of the single-locus haplotype A is thus not a consequence of population expansion after the most recent glaciation, as previously hypothesized (Carr and Marshall 1991; Árnason 2004). Instead, A is a paraphyletic assemblage of diverse genomic lineages, including some that separated close to the base of the mtDNA tree (Figure 3). The high frequency of haplotype A in the west conceals cryptic lineages not seen in the single-locus data (e.g., F and I) that are absent in the east. Although Norwegian fish with the A haplotype are on average slightly more differentiated from each other (28.3 ± 4.6 substitutions) than are non-Norwegian A fish (23.5 ± 8.8 substitutions), the difference is not significant (t.05[∞] = 1.32, P > 0.1], and the maximum difference is greater among the latter (44 vs. 34 substitutions). Overall, in contrast with the single-locus data, Norwegian coastal cod mtDNA genomes are no more diverse (nucleotide diversity π = 2.05 × 10 −3) than western samples (π = 1.78 or 2.13 × 10−3 at North Cape and Hawke Channel, respectively).
Except for A, the principal remaining single-locus mtDNA haplotypes can each be identified with one or the other of the genomic haplogroups. Haplotype D occurs in the pair of fish that forms the outgroup to all others (which we therefore designate as haplogroup D), haplotype E is equivalent to haplogroup E that occurs on both sides of the Atlantic, and haplotype G occurs in a pair of fish within haplogroup G (Figure 3). Thus, the fish examined here appear to include most if not all of the known major cod mtDNA lineages identified by single-locus studies.
The provenance and fate of the coalescent ancestor of all codfish examined (the codmother) and her descendants can be compared and contrasted with those of the human mitochondrial “Eve” and her children, the only other species to date with an extensive whole-mtDNA-genome phylogeography. The placement of the oldest and most diverse human clades in sub-Saharan Africa is consistent with the hypothesis of an African origin for the human species (Ingman et al. 2000). The coalescence of all non-African mtDNA genomes into two clades 52 KYA that underwent a large-scale expansion 38.5 KYA supports the out-of-Africa model of the recent radiation of modern human continental groups and coincides with a cultural shift in Europe.
Given this model, does the observation that the most basal clade comprises two fish from the western Atlantic support a western codmother and an “out-of-Newfoundland” hypothesis of population expansion? The origin of the major coalescent that includes all Norwegian coastal cod coincides with the peak of the previous interglacial 128 KYA, a period of maximum ocean depth and availability of isobaths on the continental shelves that would facilitate transcontinental movement. On the basis of a paleoclimate model, Bigg et al. (2008) also conclude that observed genetic diversity in cod indicates that populations on both sides of the Atlantic had persisted through the last glacial maximum, and that standing diversity probably dates to the previous glacial minimum. Critical genomic evaluation of this hypothesis is at present limited by the restricted numerical and geographic sampling. Árnason (2004) showed that haplotype D is distributed throughout the range of cod across the Atlantic and into the North and Baltic Seas, with its highest frequency in the mid-Atlantic. If all D haplotypes are indeed always part of the same genomic haplogroup lineage as the haplogroup D identified here, genomic dissection of this haplotype should determine its exact phylogeographic structure. Two general scenarios can be proposed. One or more geographically localized subsets of D-clade fish may occur as monophyletic lineages nested inside a more diverse phylogeography. This is the case with Greenland cod (G. macrocephalus ogac), which are a monophyletic lineage within the more widely distributed Pacific cod (Coulson et al. 2006), or with another gadine, Norwegian pollock [G. (= Theragra) finnmarchica], which mitogenomic analysis has shown to be a subpopulation of the more widespread G. chalcogrammus (Ursvik et al. 2007), though much older than the previous example (Carr and Marshall 2008). If this is the case for Atlantic cod, the basal distribution should indicate the species' provenance, as the human data favor the out-of-Africa hypothesis (Ingman et al. 2000). For example, nesting of the D haplogroup identified here within a more widespread mid- or eastern-Atlantic cod would favor an “out-of-Norway” hypothesis. Alternatively, Árnason's (2004) nested clade analysis suggests D has undergone long-distance dispersal. If this were a contemporary, postglacial phenomenon, we might expect to see individual transatlantic D-clade fish associated with multiple tips of a tree that is rooted elsewhere in the species' range.
Pairwise divergences in new population samples continue to reflect diversity that arises with the midglacial expansion of BFGI at 59 KYA. This is the case with trans-Laurentian Channel populations on the Georges Bank and Scotia Bank, east and northeast, respectively, of Cape Cod (S. M. Carr and H. D. Marshall, unpublished data). The identification of the two unrecognized monophyletic haplogroups F and I within the A haplotype, both of which are at present confined to the northwest Atlantic, also suggests that finer distinctions are still to be found, as has been the case for the widespread H haplogroup in Homo (Achilli et al. 2004). Further investigation may also reveal a pattern not yet discovered, for example, the existence of one or several cohorts of fish with identical or near-identical genome sequences as evidence of contemporary expansions, either as part of recoveries from true postglacial refugia (Hardie et al. 2006) or due to localized success in postcollapse recruitment (Smedbol and Wroblewski 2002). High-resolution mtDNA genome sampling of Amerindian lineages has been shown to discriminate postglacial vicariance and ecological events (Achilli et al. 2008).
One test of the directionality of cod movement is to count the cost of vicariance phylogeographic models in which the species originated in one area and spread to the others, either in an “island model” where movement between any pair of locations is equally likely, or in a “stepping-stone model” where movement between populations is more structured (Figure 5). For example, if the populations are arranged linearly in order of their occurrence across the North Atlantic, the out-of-Newfoundland and out-of-Norway hypotheses require, respectively, 20 and 22 steps in a weighted linear stepping-stone model, as compared with 12 and 13 steps in an unweighted island model. In the stepping-stone model, a Flemish Cap origin requires 24 events, and a North Cape origin, 26. None of the alternative models can be rejected (all P > 0.5 by a two-tailed binomial test; Zar 1999).
Figure 5.—
Alternative phylogeographic hypotheses for the origins of Atlantic cod. Suppose that fish arose in one area and spread historically to the others in either an island or linear stepping-stone model of population structure (Wright 1969). In the former, movement between any pair of the four populations requires a single step; in the latter, movement between nonadjacent populations requires a number of steps equal to their separation in the series. The out-of-Newfoundland and out-of-Norway hypotheses then require, respectively, 20 and 22 steps in the linear stepping-stone model (as shown), or 12 and 13 steps in an island model (one step at each node). If the eastern population is connected with those in the west only by way of Flemish Cap (dotted line), the stepping-stone model requires 22 events with either origin.
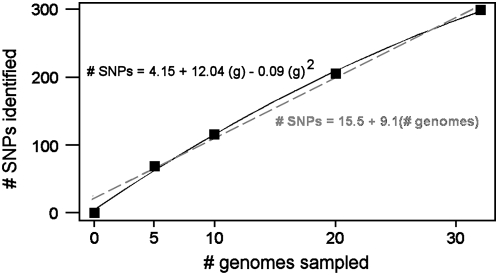
Complete mtDNA genome sequences clearly provide superior phylogeographic and phylogenetic resolution than do single-locus studies (Ingman et al. 2000; Coulson et al. 2006; Achilli et al. 2008). In Homo, the extensive homoplasy in the hypervariable control region sequences means that haplogroups are recognized by polythetic signatures, including plesiomorphic absences of restriction sites, rather than phylogenetic synapomorphies; this blurs resolution. In Gadus, there is a good correspondence between the major cytochrome b haplotypes and some of the genomic haplogroups (D, E, and G), but other groups are undetected (B, F, I, and superclade EFGI), and the major type A is paraphyletic. Given the limitations of a single locus, is it possible to survey the segregating sites in the complete genome from a population subsample, and thereafter to screen the larger population for just those SNPs (Dong et al. 2001)? The linear regression in Figure 6 suggests that the number of SNPs identifiable in codfish increases at least initially almost linearly, with each new genome adding ∼9 SNPs. The quadratic regression suggests a plateau at ∼400 SNPs from ∼60 genomes. These predictions can be tested empirically. [Additional cod genomes from the trans-Laurentian populations continue to be unique (S. M. Carr and H. D. Marshall, unpublished data).] Where there is strong phylogeographic structure, population-specific SNP surveys may seriously underestimate diversity in the other groups. For example, of a total of 655 SNP sites in the complete mtDNA genomes of 53 cosmopolitan humans (Ingman et al. 2000), 365 (56%) are variable in 21 Africans, 357 (55%) are variable in 32 non-Africans, and only 67 (10% of the total) are variable in both groupings. That is, a priori limitation of a SNP survey to just those sites variable in either grouping would fail to detect >80% of the variants in the other grouping.
Figure 6.—
Accumulation of novel SNPs with increasing sample size. The number of SNPs was counted in resampled subsets of 5, 10, and 20 genomes, without replacement. Linear regression (dashed line) predicts that each new genome will add ∼9 novel SNPs. Quadratic regression (solid line) predicts a plateau of ∼400 SNP sites upon sampling 60 genomes. Both regression equations have r2 > 0.99.
Sequencing of complete mtDNA genomes by conventional dideoxy methods remains tedious for population geneticists, who cannot afford the luxury of automatic contig assembly from multiply redundant cloned molecules, as is common in larger genome studies (e.g., Ng et al. 2005). Redundancy is necessarily traded for increased sample size, and any given base is typically assayed only twice, from the forward and reverse strands. A new biotechnology, iterative DNA “resequencing” on microarrays offers an accurate and highly time- and cost-effective alternative that is especially suited to genomes with high SNP density (Maitra et al. 2004; Flynn and Carr 2007). Four of the Flemish Cap cod were sequenced by this method. Especially where parallel studies from multiple species in different taxonomic orders and classes can be multiplexed on a single “ArkChip” (Carr et al. 2008), large-scale mitochondrial genomics is practical as a standard approach to analysis of well-resolved intraspecific phylogeography.
Acknowledgments
The support, encouragement, and assistance of scientists and staff at the Northwest Atlantic Fisheries Science Centre in St. John's is gratefully acknowledged. We have particularly benefitted from discussions with J. Brattey, J. Goodyear, B. McCallum, and P. Pepin. S.M.C. benefitted from a discussion of glacial history of the island of Newfoundland at the Atlantic Canada Ice Dynamics workshop (J. Gosse, Chair). We thank G. Rose, G. Lilly, J. Ciszewska-Carr, Einar Árnason, and an anonymous reviewer for helpful comments on a previous draft of this article. The technical assistance of K. A. Johnstone, A. M. Pope, S. M. C. Flynn, and A. T. Duggan is gratefully acknowledged. H.D.M. was supported during the collection of these data by a postdoctoral fellowship from the Department of Fisheries and Oceans, which also supported the research described here as part of a University Research Partnerships Program with Memorial University of Newfoundland.
References
- Achilli, A., A. Oliveri, M. Pala, E. Metspalu, S. Fornarino et al., 2004. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Amer. J. Hum. Genet. 75 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilli, A., U. A. Perego, C. M. Bravi, M. D. Coble, Q.-P. Kong et al., 2008. The phylogeny of the four pan-American MtDNA haplogroups: implications for evolutionary and disease studies. PLoS ONE 3 e1764.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous, 2006. Order giving notice of decisions not to add certain species to the list of endangered species. Canada Gazette 8 61. [Google Scholar]
- Árnason, E., 2004. Mitochondrial cytochrome b DNA variation in the high-fecundity Atlantic cod: trans-Atlantic clines and shallow gene genealogy. Genetics 166 1871–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise, J. C., 2000. Phylogeography. Harvard University Press, Cambridge, MA.
- Bentzen, P., C. T. Taggart, D. E. Ruzzante and D. Cook, 1996. Microsatellite polymorphism and the population structure of Atlantic cod (Gadus morhua) in the northwest Atlantic. Can. J. Fish. Aquat. Sci. 53 2706–2721. [Google Scholar]
- Bigg, G. R., C. W. Cunningham, G. Ottersen, G. H. Pogson, M. R. Wadley et al., 2008. Ice-age survival of Atlantic cod: agreement between palaeoecology models and genetics. Proc. Biol. Sci. 275 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, W. M., M. George, Jr. and A. C. Wilson, 1979. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 76 1967–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr, S. M., and D. C. Crutcher, 1998. Population genetic structure in Atlantic Cod (Gadus morhua) from the North Atlantic and Barents Sea: Contrasting or concordant patterns in mtDNA sequence and microsatellite data?, pp. 91–103 in The Implications of Localized Fishery Stocks, edited by I. Hunt Von Herbing, I. Kornfield, M. Tupper and J. Wilson. Natural Resource, Agriculture and Engineering Service, Ithaca, NY.
- Carr, S. M., and H. D. Marshall, 1991. Detection of intraspecific DNA sequence variation in the mitochondrial cytochrome b gene of Atlantic cod (Gadus morhua) by the polymerase chain reaction. Can. J. Fisheries Aquat. Sc. 48 48–52. [Google Scholar]
- Carr, S. M., D. S. Kivlichan, P. Pepin and D. C. Crutcher, 1999. Molecular systematics of gadid fishes: implications for the biogeographic origins of Pacific species. Can. J. Zool. 77 19–26. [Google Scholar]
- Carr, S. M., and H. D. Marshall, 2008. Phylogeographic analysis of complete mtDNA genomes from Walleye Pollock shows an ancient origin of genetic biodiversity. Mito. DNA (in press). [DOI] [PubMed]
- Carr, S. M., H. D. Marshall, A. T. Duggan, S. M. C. Flynn, K. A. Johnstone et al., 2008. Phylogeographic genomics of mitochondrial DNA: highly-resolved patterns of intraspecific evolution and a multi-species, microarray-based DNA sequencing strategy for biodiversity studies. Comp. Biochem. Physiol. D Genomics Proteomics 3 1–11. [DOI] [PubMed] [Google Scholar]
- Carr, S. M., A. J. Snellen, K. A. Howse and J. S. Wroblewski, 1995. Mitochondrial DNA sequence variation and genetic stock structure of Atlantic cod (Gadus morhua) from bay and offshore locations on the Newfoundland continental shelf. Mol. Ecol. 4 79–88. [DOI] [PubMed] [Google Scholar]
- Coulson, M. W., H. D. Marshall, P. Pepin and S. M. Carr, 2006. Mitochondrial genomics of gadine fish: implications for taxonomy and biogeographic origins from whole-genome data sets. Genome 49 1315–1320. [DOI] [PubMed] [Google Scholar]
- Cross, T. F., and R. H. Payne, 1978. Geographic variation in Atlantic cod (Gadus morhua) off eastern North America: a biochemical systematics approach. J. Fish. Res. Bd. Can. 35 117–123. [Google Scholar]
- Department of Fisheries and Oceans, 2005. Stock assessment report on northern (2J + 3KL) cod. DFO Can. Sci. Advis. Sec. Sci. Advis. Rep. 2005 024. [Google Scholar]
- Dong, S., E. Wang, L. Hsie, Y. Cao, X. Chen et al., 2001. Flexible use of high-density oligonucleotide arrays for single-nucleotide polymorphism discovery and validation. Genome Res. 11 1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Project for Ice Coring in Antarctica Community Members, 2006. One-to-one coupling of glacial climate variability in Greenland and Antarctica. Science 444 195–198. [DOI] [PubMed] [Google Scholar]
- Flynn, S. M. C., and S. M. Carr, 2007. Species-specificity of SNP detection on DNA microarrays: efficiency and accuracy of re-sequencing of chimpanzee, gorilla, and codfish mtDNA genomes on a human-specific MitoChip. BMC Genomics 8 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y.-X., and W.-H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, W. S., and G. Ståhl, 1988. Evolution of Atlantic and Pacific cod: loss of genetic variation and gene expression in Pacific cod. Evolution 42 138–146. [DOI] [PubMed] [Google Scholar]
- Hardie, D. C., R. M. Gillett and J. S. Hutchings, 2006. The effects of isolation and colonization on the genetic structure of marine-relict populations of Atlantic Cod (Gadus morhua) in the Canadian Arctic. Can. J. Fish. Aquat. Sci 63 1830–1839. [Google Scholar]
- Horai, S., K. Hayasaka, R. Kondo, K. Tsugane and N. Takahata, 1995. Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs. Proc. Natl. Acad. Sci. USA 92 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings, J. A., 1996. Spatial and temporal variation in the density of northern cod and a review of hypotheses for the stock's collapse. Can. J. Fish. Aquat. Sci. 53 943–962. [Google Scholar]
- International Council for the Exploration of the Sea, 2006. Report of the workshop on the decline and recovery of cod stocks throughout the North Atlantic, including tropho-dynamic effects (WKDRCS), May, 2006, St. John's, NL, Canada.
- Ingman, M., H. Kaessman, S. Pääbo and U. Gyllensten, 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408 709–713. [DOI] [PubMed] [Google Scholar]
- Inoue, J. G., M. Miya, K. Tsukamoto and M. Nishida, 2001. A mitogenomic perspective on the basal teleostean phylogeny: resolving higher-level relationships with longer DNA sequences. Mol. Phylogenet. Evol. 20 275–285. [DOI] [PubMed] [Google Scholar]
- Johnstone, K. A., H. D. Marshall and S. M. Carr, 2007. Biodiversity genomics for species at risk: patterns of DNA sequence variation within and among complete mitochondrial DNA genomes of three species of wolffish (Anarhichas spp.). Can. J. Zool. 85 151–158. [Google Scholar]
- Kocher, T. D., W. K. Thomas, A. Meyer, S. V. Edwards, S. Pääbo et al., 1989. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 86 6196–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison, W. P., and D. R. Maddison, 2000. MacClade version 4: Analysis of Phylogeny and Character Evolution. Sinauer Associates, Sunderland, MA.
- Maitra, A., Y. Cohen, S. E. Gillespie, M E. Mambo, N. Fukushima et al., 2004. The human MitoChip: a high throughput sequencing microarray for mitochondrial mutation detection. Genome Res. 14 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar, D., E. Ruiz-Pesini, P. Golik, V. Macaulay, A. G. Clark et al., 2003. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. USA 100 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya, M., H. Takeshima, H. Endo, N. B. Ishiguro, J. G. Inoue et al., 2003. Major patterns of higher teleostean phylogenies: a new perspective based on 100 complete mitochondrial DNA sequences. Mol. Phylogenet. Evol. 20 121–138. [DOI] [PubMed] [Google Scholar]
- Myers, R. A., N. J. Barrowman, J. A. Hutchings and A. A. Rosenberg, 1995. Population dynamics of exploited fish stocks at low population levels. Science 269 1106–1108. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Ng, S. H., C. G. Ariteri, I. E. Bosdet, R. Chiu, R. G. Danzmann et al., 2005. A physical map of the genome of Atlantic salmon, Salmo salar. Genomics 86 396–404. [DOI] [PubMed] [Google Scholar]
- North Greenland Ice Core Project, 2004. High-resolution record of Northern Hemisphere climate extending into the last interglacial period. Nature 431 137–142. [DOI] [PubMed] [Google Scholar]
- Nielsen, E., M. M. Hansen and D. Meldlrup, 2006. Evidence of microsatellite hitch-hiking selection in Atlantic cod (Gadus morhua L.): implications for inferring population structure in nonmodel organisms. Mol. Ecol. 15 3219–3229. [DOI] [PubMed] [Google Scholar]
- Pepin, P., and S. M. Carr, 1993. Morphological, meristic, and genetic analysis of stock structure in juvenile Atlantic cod (Gadus morhua) from the Newfoundland Shelf. Can. J. Fish. Aquat. Sci. 50 1924–1933. [Google Scholar]
- Petit, J. R., J. Jouzel, D. Raynaud, N. I. Barkov, J.-M. Barnola et al., 1999. Climate and atmospheric history of the past 420,000 years from the Vostok Ice Core, Antarctica. Nature 399 429–436. [Google Scholar]
- Petit, J. R., J. Jouzel, D. Raynaud, N. I. Barkov, J.-M. Barnola et al., 2001. Vostok ice core data for 420,000 years. IGBP PAGES/World Data Center for Paleoclimatology Data Contribution Series 2001–076. NOAA/NGDC Paleoclimatology Program, Boulder, CO.
- Pogson, G. H., and K. A. Mesa, 2004. Positive Darwinian selection at the pantophysin (Pan I) locus in marine gadid fishes. Mol. Biol. Evol. 21 65–75. [DOI] [PubMed] [Google Scholar]
- Rogers, A. R., and H. Harpending, 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9 552–569. [DOI] [PubMed] [Google Scholar]
- Rose, G. A., B. Deyoyng, D. W. Kulka, S. V. Goddard and G. L. Fletcher, 2001. Distribution shifts and overfishing the northern cod (Gadus morhua): a view from the ocean. Can J. Fish. Aquat. Sci. 57 644–663. [Google Scholar]
- Rose, G. A., 2004. Reconciling overfishing and climate change with stock dynamics of Atlantic cod (Gadus morhua) over 500 years. Can. J. Fish. Aquat. Sci. 61 1553–1557. [Google Scholar]
- Rose, G. A., 2007. Cod: The Ecological History of the North Atlantic Fisheries. Breakwater Books, St. John's, NL, Canada.
- Rozas, J., and R. Rozas, 1995. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput. Appl. Biosci. 11 621–625. [DOI] [PubMed] [Google Scholar]
- Ruzzante, D. E., C. T. Taggart and D. Cook, 1999. A review of the evidence for genetic structure of cod (Gadus morhua) populations in the NW Atlantic and population affinities of larval cod off Newfoundland and the Gulf of St. Lawrence. Fish. Res. 43 79–97. [Google Scholar]
- Ruzzante, D. E., C. T. Taggart, R. W. Doyle and D. Cook, 2001. Stability in the historical pattern of genetic structure of Newfoundland cod (Gadus morhua) despite the catastrophic decline in population size from 1964 to 1994. Conserv. Genet. 2 257–269. [Google Scholar]
- Shaw, J., 2006. Palaeogeography of Atlantic Canadian Continental Shelves from the Last Glacial Maximum to the present, with an emphasis on Flemish Cap. J. Northw. Atl. Fish. Sci. 37 119–126. [Google Scholar]
- Shelton, P. A., and B. P. Healey, 1999. Should depensation be dismissed as a possible explanation for the lack of recovery of the northern cod (Gadus morhua) stock? Can. J. Fish. Aquat. Sci. 56 1521–1524. [Google Scholar]
- Slatkin, M., and R. R. Hudson, 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin, M., and W. P. Maddison, 1989. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics 123 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedbol, R. K., and J. S. Wroblewski, 2002. Metapopulation theory and northern cod population structure: interdependency of subpopulations in recovery of a groundfish population. Fisheries Res. 55 161–174. [Google Scholar]
- Swofford, D. L., 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods). Sinauer Associates, Sunderland, MA.
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni, A., A. Achilli, V. Macaulay, M. Richards and H. J. Bandelt, 2006. Harvesting the fruit of the human mtDNA tree. Trends Genet. 22 339–345. [DOI] [PubMed] [Google Scholar]
- Ursvik, A., R. Breines, J. S. Christiansen, S.-E. Fevolden, D. H. Coucheron et al., 2007. A mitogenomic approach to the taxonomy of pollocks: Theragra chalcogramma and T. finnmarchica represent one single species. BMC Evol. Biol. 7 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeij, G. J., 1991. Anatomy of an invasion: the trans-Arctic interchange. Paleobiology 17 281–307. [Google Scholar]
- Wahlund, S., 1928. Zusammensetzung von population und korrelationserscheinungen von standpunkt der vererbungslehre ans betrachtet. Hereditas 11 65–106. [Google Scholar]
- Wilson, A. C., R. L. Cann, S. M. Carr, M. George Jr., U. B. Gyllensten et al., 1985. Mitochondrial DNA and two perspectives on evolutionary genetics. Biol. J. Linn. Soc. 26 375–400. [Google Scholar]
- Wright, S., 1969. Evolution and the Genetics of Natural Populations. Volume 2, The Theory of Gene Frequencies. University of Chicago Press, Chicago.
- Zar, J. H., 1999. Biostatistical Analysis, Ed. 4. Prentice Hall, Upper Saddle River, NJ.