Abstract
Transmission ratio distortion (TRD) is frequently observed in inter- and intraspecific hybrids of plants, leading to a violation of Mendelian inheritance. Sex-independent TRD (siTRD) was detected in a hybrid between Asian cultivated rice and its wild ancestor. Here we examined how siTRD caused by an allelic interaction at a specific locus arose in Asian rice species. The siTRD is controlled by the S6 locus via a mechanism in which the S6 allele acts as a gamete eliminator, and both the male and female gametes possessing the opposite allele (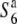 ) are aborted only in heterozygotes (
) are aborted only in heterozygotes (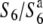 ). Fine mapping revealed that the S6 locus is located near the centromere of chromosome 6. Testcross experiments using near-isogenic lines (NILs) carrying either the S6 or
). Fine mapping revealed that the S6 locus is located near the centromere of chromosome 6. Testcross experiments using near-isogenic lines (NILs) carrying either the S6 or 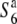 alleles revealed that Asian rice strains frequently harbor an additional allele (
alleles revealed that Asian rice strains frequently harbor an additional allele (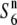 ) the presence of which, in heterozygotic states (
) the presence of which, in heterozygotic states (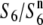 and
and 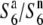 ), does not result in siTRD. A prominent reduction in the nucleotide diversity of S6 or
), does not result in siTRD. A prominent reduction in the nucleotide diversity of S6 or 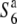 carriers relative to that of
carriers relative to that of 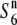 carriers was detected in the chromosomal region. These results suggest that the two incompatible alleles (S6 and
carriers was detected in the chromosomal region. These results suggest that the two incompatible alleles (S6 and 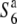 ) arose independently from
) arose independently from 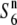 and established genetically discontinuous relationships between limited constituents of the Asian rice population.
and established genetically discontinuous relationships between limited constituents of the Asian rice population.
TRANSMISSION ratio distortion (TRD) refers to a naturally occurring phenomenon in which the two alleles at a heterozygous locus do not transmit equally to the progeny (Crow 1988; Lyttle 1991; Temin et al. 1991). TRD is induced by a variety of mechanisms, including the nonrandom segregation of chromosomes during meiosis (Pardo-Manuel de Villena and Sapienza 2001; Birchler et al. 2003; Fishman and Willis 2005), preferential dysfunction of gametes in hybrids (Lyttle 1991; Temin et al. 1991; Silver 1993; Moyle and Graham 2006; Úbeda and Haig 2005), and preferential success of gametes in fertilization (Price 1997; Diaz and MacNair 1999). TRD is detected not only within a given population but also between populations and/or species. Genomewide surveys have frequently revealed significant TRD in intra- and interspecific hybrids (Moyle and Graham 2006). Because TRD dramatically alters the frequencies of alleles in a population by disrupting Mendelian segregation, it has been hypothesized that TRD is the driving force that contributes to the rise of reproductive barriers (Frank 1991; Hurst and Pomiankowski 1991; Orr and Irving 2004).
In plants, preferential dysfunction of gametes can take place in either the male (Cameron and Moav 1957; Loegering and Sears 1963; Sano 1983) or the female (Maguire 1963; Scoles and Kibirge-Sebnya 1983), or in both (Rick 1966; Endo and Tsunewaki 1975; Sano et al. 1979; Finch et al. 1984). Among these types, sex-independent TRD (siTRD) exerts the strongest effect on segregation distortion, since the abortion of both male and female gametes carrying one of the two alleles is induced by the presence of the opposite allele in the heterozygote (Rick 1966; Endo and Tsunewaki 1975; Sano et al. 1979; Finch et al. 1984).
In rice, extensive studies have been carried out on the genetic basis of isolating barriers in intra- and interspecific hybrids, since barriers often present a serious problem in hybridization efforts in breeding programs. A variety of genetic mechanisms have been proposed to explain hybrid sterility (reviewed by Koide et al. 2008a), including those involving combinations of chromosomes bearing cryptic structural differences (Li et al. 1997), nuclear and cytoplasmic genomes of different origin (Shinjo 1984), recessive alleles of duplicate genes (Oka 1974, 1988), two complementary dominant genes (Chu and Oka 1972; Li et al. 1997), and different alleles in a single gene (Ikehashi and Araki 1986; Morishima et al. 1992). Among these, the interaction between alleles is most frequently observed as the cause of preferential dysfunction of gametes leading to TRD (Ikehashi and Araki 1986; Morishima et al. 1992; Koide et al. 2008a). Recent genomewide surveys have revealed the presence of a number of chromosomal regions that are involved in TRD in inter- and intraspecific crosses in rice (Xu et al. 1997; Harushima et al. 2001). However, there has been no report to date that addresses how each TRD system has arisen and become fixed in the rice populations.
We have previously reported that a gamete eliminator (S6) functions in a hybridization between Oryza sativa and O. rufipogon (Sano 1992). The hybrid plants between T65wx (O. sativa ssp. japonica) and near-isogenic lines (NILs) carrying a segment of chromosome 6 derived from W593 (O. rufipogon) in the genetic background of T65wx exhibited a reduced rate of seed setting. When the hybrids were reciprocally crossed with T65wx, all the resultant progeny exhibited a reduced seed-setting rate, while the progeny derived from self-pollination of the hybrid plants exhibited a normal seed-setting rate (Sano 1992). This phenomenon was due to an interaction between the gene designated S6 in the chromosomal segment derived from W593 and its opposite allele (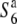 ) in T65wx. The S6 allele acts as a “gamete eliminator,” and both male and female gametes possessing the
) in T65wx. The S6 allele acts as a “gamete eliminator,” and both male and female gametes possessing the 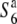 allele are aborted only in the heterozygote (
allele are aborted only in the heterozygote (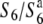 ) (Sano 1992). This locus thus affords an opportunity to examine the genetic basis and evolution of the siTRD system. In the present study, we focus on siTRD caused by the allelic interaction of the S6 locus and report the first detailed characterization of the siTRD system. We describe the presence of an additional allele (
) (Sano 1992). This locus thus affords an opportunity to examine the genetic basis and evolution of the siTRD system. In the present study, we focus on siTRD caused by the allelic interaction of the S6 locus and report the first detailed characterization of the siTRD system. We describe the presence of an additional allele (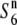 ) that induces no preferential abortion in heterozygotes with either the S6 or
) that induces no preferential abortion in heterozygotes with either the S6 or 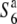 allele, which was revealed by testcrosses examining allelic distribution in wild and cultivated rice accessions. We also report the histological analysis of gametogenesis, genetic mapping of the S6 gene, and the analysis of nucleotide diversity around the S6 locus. On the basis of the findings, the involvement of the siTRD system in the formation of isolating barriers in the evolution of the Asian rice population is discussed.
allele, which was revealed by testcrosses examining allelic distribution in wild and cultivated rice accessions. We also report the histological analysis of gametogenesis, genetic mapping of the S6 gene, and the analysis of nucleotide diversity around the S6 locus. On the basis of the findings, the involvement of the siTRD system in the formation of isolating barriers in the evolution of the Asian rice population is discussed.
MATERIALS AND METHODS
Genetic stocks:
Three NILs, T65wx, T65S6 (W593), and T65WxSe1 (Pat), were made and used for genetic mapping (Table 1). T65S6 (W593) and T65WxSe1 (Pat) harbor the short arm of chromosome 6 from W593 (O. rufipogon from Malaysia) and Patpaku (ssp. indica of O. sativa from Taiwan), respectively, in the background of T65wx. The detailed genotypes of these three NILs are described in Matsubara et al. (2003). To examine the allelic distribution at the S6 locus, two other NILs, 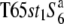 and T65st1S6, were also established and used for the analysis (Table 1). These two NILs carried stripe1 (st1) as a visible marker, which is linked with the S6 locus (P = 0.13, Sano 1992).
and T65st1S6, were also established and used for the analysis (Table 1). These two NILs carried stripe1 (st1) as a visible marker, which is linked with the S6 locus (P = 0.13, Sano 1992). 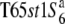 carried the
carried the 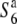 allele from T65wx. T65st1S6 was selected from an F2 population of
allele from T65wx. T65st1S6 was selected from an F2 population of 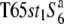 × T65S6 (W593). To fine-map the S6 gene, another NIL, A58S6 was established using A58 (ssp. japonica of O. sativa) as the recurrent parent because of the short life cycle of A58 (Table 1). A58S6 carries the S6 allele introduced from T65S6 (W593).
× T65S6 (W593). To fine-map the S6 gene, another NIL, A58S6 was established using A58 (ssp. japonica of O. sativa) as the recurrent parent because of the short life cycle of A58 (Table 1). A58S6 carries the S6 allele introduced from T65S6 (W593).
TABLE 1.
Genetic stocks of NILs and recurrent parents used in this study
| Genotype
|
|||||
|---|---|---|---|---|---|
| Lines | st1 | S6 | Donor (species) | Backcross generation | Reference |
| T65S6 (W593)a | + | S6 | W593 (O. rufipogon) | BC8 | Sano (1992) |
| T65 WxSe1 (Pat)b | + |
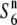 c c
|
Patpaku (O. sativa ssp. indica) | BC8 | Dung et al. (1998) |
| A58S6 | + | S6 | W593 (O. rufipogon) | BC4 and BC5 | This study |
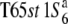 |
st1 | 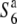 |
F1176 (O. sativa ssp. japonica) | BC7 | This study |
| T65st 1S6 | st1 | S6 | F1176 (O. sativa ssp. japonica) and W593 (O. rufipogon) | BC7 | This study |
| T65wx | + | 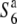 |
Kinoshita-mochi (O. sativa ssp. japonica) | BC12 | Oka (1974) |
| A58 | + | 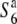 |
Kokusyokuto-2 (O. sativa ssp. japonica) | ||
Formerly named W593A (Sano 1992).
Formely named T65 (Wx-pat) (Dung et al. 1998).
The allelic state was determined in this study.
Seeds were germinated in petri dishes at 30° in late April, and each of the seedlings was transplanted into plastic pots and grown in a greenhouse. The plants were placed in short-day fields (10.5 hr) 8 weeks after sowing due to photoperiod sensitivity, as needed.
Cytological observations:
Spikelets were sampled from panicles before heading. Samples were fixed in FAA (formalin:glacial acetic acid:70% ethanol = 1:1:18) and stored in 70% ethanol until use. Pollen fertility was estimated from the percentages of pollen grains stainable with potassium iodine solution (I2-KI). Ovaries were dehydrated in a graded ethanol-butanol series, embedded in Paraplast Plus (Oxford Labware, St. Louis), and then cut into 10-μm thick sections. Sections were stained with safranin and fast green (Sylvester and Ruzin 1993) and observed under light microscopy (BH-2, Olympus, Tokyo).
Mapping of the S6 gene:
To map the S6 locus, a total of 1886 segregating plants of A58 × A58S6 were genotyped. Since the S6 allele kills female gametes possessing the 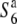 allele in heterozygotes (
allele in heterozygotes (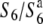 ), the seed-setting rate was analyzed to detect the heterozygotes of
), the seed-setting rate was analyzed to detect the heterozygotes of 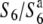 according to the method described below. In addition, three recombinant lines (P-1, P-2, and P-3) were obtained from the derivatives of T65wx × T65WxSe1 (Pat) to determine the allelic state at the S6 locus of Patpaku.
according to the method described below. In addition, three recombinant lines (P-1, P-2, and P-3) were obtained from the derivatives of T65wx × T65WxSe1 (Pat) to determine the allelic state at the S6 locus of Patpaku.
For genotyping the alien segments with molecular markers, genomic DNA was isolated from a small piece of frozen leaf according to the method of Monna et al. (2002) with slight modifications. Six markers on chromosome 6 (Wx, OsC1, Hd1, RG264, RM3498, and G2028) were used according to the method of Matsubara et al. (2003). Two microsatellite markers (RM3183 and RM3498) were selected from the public database (http://www.gramene.org/). In addition, five cleaved amplified polymorphic sequence (CAPS) markers (S14439, P139, R111C, G05, and C133A) were designed on the basis of sequences in the public database (accession nos. AP003763, AP003574, AP005656, AP005967, and AP005450). The primers for PCR amplification were as follows: 5′-ccg aaa aga gtc ctc cga ag-3′ and 5′-cca cct aag aag cca gca cc -3′ for S14439; 5′-gaa atg cca ctg gcc tac at-3′ and 5′-ttc agg cga gca att tag gt-3′ for P139; 5′-tca ggg cta atc aat ggc gaa g-3′ and 5′-tta gtg gat gcc tgg acg atg a-3′ for R111C; 5′-cca ttc ctc cgt cca aac aca t-3′ and 5′-ccc aaa tca cac aca tcg tgc t-3′ for G05; and 5′-cct aaa cgc aag cca ctg tc-3′ and 5′-gca ttg cat gtt cag ttt tc-3′ for C133A. To detect the polymorphisms in the CAPS markers (S14439, P139, R111C, G05, and C133A), the amplified products were digested with HpyCH4, AluI, HinfI, Sau3A, and MspI, respectively. The recombination values were estimated on the basis of the maximum likelihood method.
Survey for allelic distribution at the S6 locus:
To examine the distribution of the S6 allele in Asian rice strains, 9 strains of O. sativa and 14 strains of O. rufipogon were surveyed (Table 2). The strains of O. sativa included 3 strains of ssp. japonica and 6 ssp. indica. Since intervarietal crossings frequently produce semi-sterility as well as TRD in rice (Oka 1988), the siTRD specific to the S6 locus was evaluated on the basis of the level of seed setting of F1 plants and distorted segregation for a marker (st1) linked to S6. The rate of seed setting was determined by counting the number of fertile and sterile spikelets of two panicles for each plant. Strains carrying the S6 allele are expected to yield a low seed-setting rate in F1 hybrids and a segregation distortion for st1 in F2 when crossed with 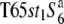 , while strains carrying the
, while strains carrying the 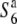 allele are expected to exhibit these abnormalities when crossed with T65st1S6. Furthermore, when no difference is found between the crosses with both
allele are expected to exhibit these abnormalities when crossed with T65st1S6. Furthermore, when no difference is found between the crosses with both 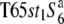 and T65st1S6, the strain is expected to carry an additional allele (
and T65st1S6, the strain is expected to carry an additional allele ( ), the presence of which in heterozygotic states (
), the presence of which in heterozygotic states (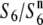 and
and 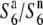 ) does not result in siTRD, as reported in the tomato by Rick (1966, 1971). To determine the allelic state of W1807 (O. rufipogon), segregation distortion was examined for a molecular marker, P139, closely linked to S6 (Figure 2).
) does not result in siTRD, as reported in the tomato by Rick (1966, 1971). To determine the allelic state of W1807 (O. rufipogon), segregation distortion was examined for a molecular marker, P139, closely linked to S6 (Figure 2).
TABLE 2.
Seed setting and segregation patterns of the marker st1 observed in testcrosses for allelic identification in the S6 locus
| Crossing with T65st1S6
|
Crossing with 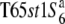
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Strain | Subspecies or type | Origin | F1 seed setting (A)a | F2st1 (%) | F1 seed setting (B)a | F2st1 (%) | A/B | Putative allele |
| O. sativa | T65 | ssp. japonica | Taiwan | 34.1 | 58.4b,** | 72.3 | 23.9 | 0.47 | 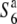 |
| Koshihikari | ssp. japonica | Japan | 41.5 | 60.2b,** | 88.0 | 22.6 | 0.47 | 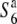 |
|
| A58 | ssp. japonica | Japan | 46.2 | 57.3b,** | 86.0 | 22.8 | 0.53 | 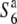 |
|
| PTB10 | ssp. indica | India | 16.7 | 20.3 | 14.0 | 17.8 | 1.19 | 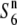 |
|
| IR36 | ssp. indica | Philippines | 80.2 | 17.1** | 76.7 | 17.2 | 1.05 | 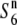 |
|
| Patpaku | ssp. indica | Taiwan | 72.7 | 23.3 | 67.0 | 21.3 | 1.09 | 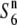 |
|
| Acc27590 | ssp. indica | Bangladesh | 82.1 | — | 79.0 | — | 1.04 | 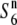 |
|
| Acc27591 | ssp. indica | Bangladesh | 31.7 | 9.2** | 44.1 | 24.1 | 0.72 | 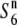 |
|
| 444 | ssp. indica | India | 65.2 | 17.8 | 54.9 | 26.5 | 1.19 | 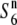 |
|
| O. rufipogon | W107 | Annual | India | 42.1 | 29.7 | 55.1 | 20.5 | 0.76 | 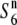 |
| W2002 | Annual | Myanmar | 80.3 | 21.6 | 74.4 | 22.2 | 1.08 | 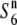 |
|
| W630 | Annual | Myanmar | 47.6 | 12.5** | 42.7 | — | 1.11 | 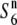 |
|
| W2048 | Perennial | China | 20.4 | 45.1** | 25.7 | 23.5 | 0.79 |  |
|
| W1718 | Perennial | China | 52.5 | — | 40.8 | — | 1.29 | 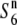 |
|
| W1943 | Perennial | China | 71.0 | — | 73.3 | — | 0.97 | 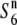 |
|
| W1944 | Perennial | China | 31.1 | 18.4 | 23.5 | 26.3 | 1.32 | 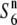 |
|
| W1945 | Perennial | China | 54.6 | 20.9 | 47.6 | 20.5 | 1.15 | 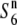 |
|
| W1952 | Perennial | China | 47.6 | 13.6* | 42.3 | — | 1.13 | 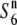 |
|
| W1681 | Perennial | India | 82.1 | — | 74.9 | — | 1.10 | 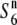 |
|
| W593 | Perennial | Malaysia | 81.4 | 15.0* | 39.1 | 2.7** | 2.08 | S6 | |
| W172 | Perennial | Thailand | 67.2 | 11.2 | 25.5 | 3.6** | 2.64 | S6 | |
| W1294 | Perennial | Philippines | 67.9 | 20.6 | 28.0 | 2.6** | 2.43 | S6 | |
| W1807c | Perennial | Sri Lanka | 77.1 | 26.7 | 26.6 | 0.1** | 2.90 | S6 | |
* and ** indicate significant deviation from Mendelian inheritance at 5% and 1% levels, respectively.
The rate of seed setting was determined by counting fertile and sterile spikelets of two panicles for each plant.
When more tightly linked marker, R111C, was used, the frequency of the W593-derived allele was close to 100% in the F2 populations derived from A58 × A58S6 and T65wx × T65S6 (W593) (see text).
The allelic state of W1807 was analyzed by crossings the line with T65WxS6 (W593) and T65wx. The frequency of the alleles in the progeny was determined by the marker P139 linked with the S6 locus.
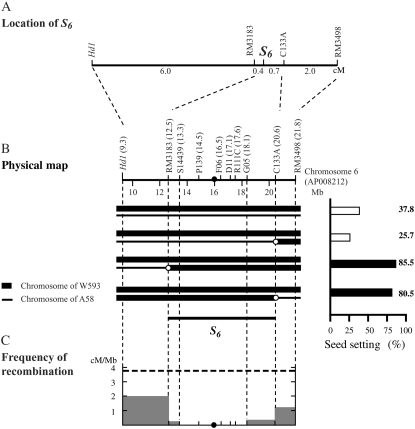
Figure 2.—
Localization of the S6 gene on the rice linkage map. (A) Location of the S6 gene on choromosome 6. Genetic distance was estimated on the basis of 1886 segregating plants of A58 × A58S6 (BC4 and BC5) except for the genetic distance between Hd1 and RM3183, which was estimated using 216 BC4 F2 plants from the same population. (B) Graphical genotypes of chromosome 6 of recombinant plants and the rates of seed setting of the respective plants through self-fertilization. The physical map of chromosome 6 is based on the Rice Genome Research Program (http://rgp.dna.affrc.go.jp). The map position of each marker along chromosome 6 (AP008212) is shown in parentheses (in megabases). A solid circle represents the centromere. Chromosomal regions derived from W593 (carrying the S6 allele) and A58 (carrying the 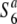 allele) are indicated by thick and thin lines, respectively. Open circles represent the A58-derived alleles. The estimated range of the position of the S6 gene is shown by a bar below the graphical genotypes. The rate of seed setting was determined by counting the number of fertile and sterile spikelets of two panicles for each plant. The data represent the average values obtained from at least two individual plants for each genotype. The rate of seed setting is shown as a percentage. Solid and open bars indicate seed fertility above and below 50%, respectively. (C) Regional recombination frequency on chromosome 6. The frequency of recombination (in centimorgans/megabases) was estimated using the mapping populations. The average recombination value of chromosome 6 (3.87 cM/Mb; Wu et al. 2003) is shown by a dashed line.
allele) are indicated by thick and thin lines, respectively. Open circles represent the A58-derived alleles. The estimated range of the position of the S6 gene is shown by a bar below the graphical genotypes. The rate of seed setting was determined by counting the number of fertile and sterile spikelets of two panicles for each plant. The data represent the average values obtained from at least two individual plants for each genotype. The rate of seed setting is shown as a percentage. Solid and open bars indicate seed fertility above and below 50%, respectively. (C) Regional recombination frequency on chromosome 6. The frequency of recombination (in centimorgans/megabases) was estimated using the mapping populations. The average recombination value of chromosome 6 (3.87 cM/Mb; Wu et al. 2003) is shown by a dashed line.
Diversity survey:
Nine strains of O. sativa and 14 strains of O. rufipogon were used for the analysis of the DNA polymorphisms (Table 2). The nucleotide sequences of five regions (P139, F06, D11, R111C, and C133A) around the S6 gene were determined by direct sequencing of the polymerase chain reaction (PCR) products. The following primers were used for the PCR: 5′-ccg aaa aga gtc ctc cga ag-3′ and 5′-cca cct aag aag cca gca cc-3′ for F06; and 5′-gaa atg cca ctg gcc tac at-3′ and 5′-ttc agg cga gca att tag gt-3′ for D11. These primers were designed on the basis of sequences in the public database (accession nos. AP003545 and AP003512). One loosely linked locus, Hd3a, which is located on the short arm of chromosome 6, and two unlinked loci, sd1 (semi-dwarf1), and qSH4, which are located on chromosome 1 and chromosome 4, respectively, were used as references. The nucleotide sequences of sd1 and qSH4 were determined according to the methods of Nagano et al. (2005) and Onishi et al. (2007), respectively. For qSH4, the combined sequences of the two regions (43A12-161k and 04A17-9k) were used. For Hd3a, the primers used were 5′-agc tag ata gct gcc tct atc aca gta t-3′ and 5′-cta gct tca tga gag acc tta gcc-3′, 5′-ccc tgc acc aca cac agt tc-3′ and 5′-tgt ctg aac ctg caa tgt at-3′ or 5′-agc tag ata gct gcc tct atc aca gta t-3′ and 5′-tat ata tgt tgt gtg tcg aga atc att tc-3′. These primers were designed on the basis of sequences in the public database (accession no. AP007223). Each accession was sequenced in both directions using a Big Dye Terminator Cycle sequencing kit (Applied Biosystems, Foster City, CA) on an ABI310 automatic sequencer (Applied Biosystems). The DNA sequences determined in this study are available from DDBJ/GenBank/EMBL (accession nos. AB433361–AB433506, AB433508–AB433511, and AB433513–AB433529). The sequence alignment was done using the CLUSTAL W computer program (Thompson et al. 1994) with minor modifications by visual inspection. Molecular population genetic analysis was conducted using DnaSP version 3.14 (Rozas and Rozas 1999). We compared the level of nucleotide diversity per silent site based on π (Nei 1987) and θw (Watterson 1975) and calculated Tajima's D statistic for testing neutrality (Tajima 1989). Among the five regions around the S6 gene, D11 and C133A had portions of protein coding regions predicted from the public database (Rice Genome Research Project, http://rgp.dna.affrc.go.jp). D11 contained a part of the first exon and all of the second and third exons of a putative 2′-hydroxyisoflavone reductase. C133A contained the fifth exon of the putative mlo2 protein. To rule out selection acting on the functional regions, the silent sites were used to compare nucleotide diversity.
RESULTS
Effects of allelic interaction at the S6 locus on gametogenesis:
A previous study indicated that the S6 gene, derived from the wild rice (O. rufipogon) strain W593, induced the preferential abortion of both male and female gametes possessing its allelic alternative (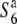 ) from cultivated rice (O. sativa) strain T65 only in the heterozygote, and as a result, no
) from cultivated rice (O. sativa) strain T65 only in the heterozygote, and as a result, no 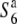 allele was transmitted to the progeny (Sano 1992). To examine the female gametogenesis in the heterozygote (
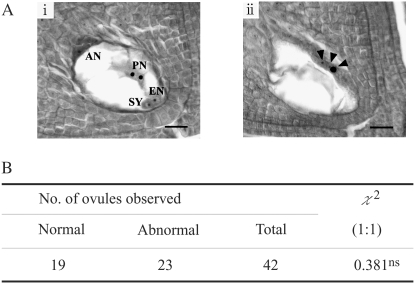
allele was transmitted to the progeny (Sano 1992). To examine the female gametogenesis in the heterozygote (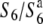 ) which was causing the siTRD, histological investigations were carried out (Figure 1). If dysfunction of the female gametes carrying the
) which was causing the siTRD, histological investigations were carried out (Figure 1). If dysfunction of the female gametes carrying the 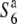 allele is induced in the presence of the S6 allele in the heterozygote, half of the embryo sacs should degenerate. Of 42 ovules, 23 in fact did exhibit an abnormality in the embryo sac structure, while the remaining 19 ovules had a mature seven-celled structure like that found in the parental strain (T65wx) (Figure 1). The frequency of arrested embryo sacs was 0.55 (23/42), indicating that the embryo sacs carrying the
allele is induced in the presence of the S6 allele in the heterozygote, half of the embryo sacs should degenerate. Of 42 ovules, 23 in fact did exhibit an abnormality in the embryo sac structure, while the remaining 19 ovules had a mature seven-celled structure like that found in the parental strain (T65wx) (Figure 1). The frequency of arrested embryo sacs was 0.55 (23/42), indicating that the embryo sacs carrying the 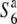 allele were aborted, which is consistent with the notion that transmission of the
allele were aborted, which is consistent with the notion that transmission of the 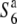 allele is extremely reduced in the heterozygote (
allele is extremely reduced in the heterozygote ( ). On the other hand, no abnormalities were detected in mature pollen grains in the heterozygote (
). On the other hand, no abnormalities were detected in mature pollen grains in the heterozygote (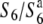 ), as also in T65wx. To confirm the TRD through male gametes, the segregation of F2 plants derived from T65wx × T65S6 (W593) was analyzed by using a molecular marker, R111C, tightly linked with the S6 locus (see below). Almost all F2 plants (84/98) were homozygotes for the W593-derived allele and no homozygotes for the T65wx-derived allele were detected, indicating that transmission of the
), as also in T65wx. To confirm the TRD through male gametes, the segregation of F2 plants derived from T65wx × T65S6 (W593) was analyzed by using a molecular marker, R111C, tightly linked with the S6 locus (see below). Almost all F2 plants (84/98) were homozygotes for the W593-derived allele and no homozygotes for the T65wx-derived allele were detected, indicating that transmission of the 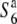 allele is reduced not only through female but also male gametes, as has been inferred from backcrosses (Sano 1992).
allele is reduced not only through female but also male gametes, as has been inferred from backcrosses (Sano 1992).
Figure 1.—
Embryo sac abnormality in the heterozygotes of 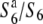 . (A) Normal (i) and abnormal (ii) embryo sacs in T65wx/T65S6 (W593). Arrow heads indicate nuclei. EN, egg nucleus; SY, synergid cell; PN, polar nuclei; AN, antipodal cell nuclei. Bar, 20 μm. (B) Frequencies of normal and abnormal embryo sacs in T65wx/T65S6 (W593). ns indicates nonsignificant deviation from 1:1 ratio.
. (A) Normal (i) and abnormal (ii) embryo sacs in T65wx/T65S6 (W593). Arrow heads indicate nuclei. EN, egg nucleus; SY, synergid cell; PN, polar nuclei; AN, antipodal cell nuclei. Bar, 20 μm. (B) Frequencies of normal and abnormal embryo sacs in T65wx/T65S6 (W593). ns indicates nonsignificant deviation from 1:1 ratio.
Fine mapping:
To roughly map the S6 gene, 216 segregating plants from A58 × A58S6 were genotyped using four molecular markers (Hd1, RM3183, C133A, and RM3498) (Figure 2A). A58 and A58S6 carry the 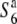 and S6 alleles, respectively (Table 1). The rate of seed setting of each F2 plant was scored to examine the phenotypic segregation of the S6 locus, since the dysfunction of female gametes in the heterozygote (
and S6 alleles, respectively (Table 1). The rate of seed setting of each F2 plant was scored to examine the phenotypic segregation of the S6 locus, since the dysfunction of female gametes in the heterozygote (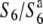 ) is reflected in the productivity of the F2 plants. The F2 plants were clearly segregated into two distinct groups: one exhibiting <50% (32.8 ± 4.4%) seed-setting rate and the other exhibiting >70% (80.2 ± 0.4%), and no F2 plant exhibited a seed-setting rate between these values. Plants of the former and the latter groups were classified as heterozygotes and homozogotes, respectively. The allelic states of the above markers indicated that all plants classified as homozygotes were those of the S6 allele. As a consequence of this analysis, the estimated recombination values of RM3183-S6, S6-C133A, and C133A-RM3498 were calculated to be 0.0040 ± 0.0010, 0.0070 ± 0.0014, and 0.0201 ± 0.0023, respectively (Figure 2A). The frequencies of the W593-derived alleles were significantly higher than those expected from Mendelian inheritance, especially at RM3183 (208/216, 96.3%) and C133A (206/216, 95.4%), which confirmed the notion that the S6 allele is preferentially transmitted through both male and female gametes.
) is reflected in the productivity of the F2 plants. The F2 plants were clearly segregated into two distinct groups: one exhibiting <50% (32.8 ± 4.4%) seed-setting rate and the other exhibiting >70% (80.2 ± 0.4%), and no F2 plant exhibited a seed-setting rate between these values. Plants of the former and the latter groups were classified as heterozygotes and homozogotes, respectively. The allelic states of the above markers indicated that all plants classified as homozygotes were those of the S6 allele. As a consequence of this analysis, the estimated recombination values of RM3183-S6, S6-C133A, and C133A-RM3498 were calculated to be 0.0040 ± 0.0010, 0.0070 ± 0.0014, and 0.0201 ± 0.0023, respectively (Figure 2A). The frequencies of the W593-derived alleles were significantly higher than those expected from Mendelian inheritance, especially at RM3183 (208/216, 96.3%) and C133A (206/216, 95.4%), which confirmed the notion that the S6 allele is preferentially transmitted through both male and female gametes.
For fine mapping, 1670 additional segregating plants were surveyed with four additional markers (S14439, P139, R111C, and G05). However, no recombination was detected between S14439 and G05, indicating that the frequency of recombination is extremely low near the centromere. The ratio of genetic distance to physical distance (cM/Mb) greatly decreased with decreasing distance from the centromere, confirming the restriction of recombination near the centromere (Figure 2C). Due to a deficit of recombinants in this region, the S6 locus was delimited to a region of >8000 kb between RM3183 and C133A, even with a large mapping population (Figure 2B).
Distribution of the S6 and S6a alleles in Asian rice accessions:
To examine the allelic distribution at the S6 locus, two NILs carrying the S6 or 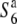 allele, namely, the T65st1S6 or
allele, namely, the T65st1S6 or 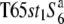 lines, respectively, were established (Table 1). The allelic distribution at the S6 locus in 23 strains of Asian cultivated rice and wild rice were investigated by testcrosses with these NILs (Table 2). Because st1 is linked with the S6 locus, and its phenotype in homozygotes is manifested at the young seedling stage, it was used as a visible marker of the segregation distortions caused by the S6 locus.
lines, respectively, were established (Table 1). The allelic distribution at the S6 locus in 23 strains of Asian cultivated rice and wild rice were investigated by testcrosses with these NILs (Table 2). Because st1 is linked with the S6 locus, and its phenotype in homozygotes is manifested at the young seedling stage, it was used as a visible marker of the segregation distortions caused by the S6 locus.
The F1 hybrids between three strains of O. sativa ssp. japonica and T65st1S6 all exhibited a low seed-setting rate, but the F1 hybrids between these strains and 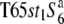 did not, but rather, exhibited a normal rate (Table 2). In addition, a marked excess of st1 homozygotes in the F2 populations was observed when the three strains were crossed with T65st1S6. These results indicate that the three strains carry the
did not, but rather, exhibited a normal rate (Table 2). In addition, a marked excess of st1 homozygotes in the F2 populations was observed when the three strains were crossed with T65st1S6. These results indicate that the three strains carry the 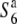 allele at the S6 locus. In contrast, the F1 hybrids between the four wild strains (W593, W172, W1294, and W1807) and either
allele at the S6 locus. In contrast, the F1 hybrids between the four wild strains (W593, W172, W1294, and W1807) and either 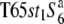 or T65wx exhibited a low seed-setting rate but the F1 hybrids between these strains and T65st1S6 or T65S6 (W593) did not, indicating that the four wild strains carry the S6 allele at the S6 locus. A marked distortion of the linked st1 marker in the F2 populations supported the conclusion that the four wild strains harbor the S6 allele.
or T65wx exhibited a low seed-setting rate but the F1 hybrids between these strains and T65st1S6 or T65S6 (W593) did not, indicating that the four wild strains carry the S6 allele at the S6 locus. A marked distortion of the linked st1 marker in the F2 populations supported the conclusion that the four wild strains harbor the S6 allele.
The other six O. sativa ssp. indica and nine O. rufipogon strains (W107, W2002, W630, W1718, W1943, W1944, W1945, W1952, and W1681) produced a pattern different from that described above (Table 2). There was no substantial difference in the seed-setting rate in the F1 hybrids derived from the two testcrosses, although the seed-setting rate varied depending on the crossing parents. A marked excess of st1 homozygotes was detected in neither of the F2 populations derived from the two testcrosses. Therefore, it was suspected that these strains carry an additional allele (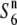 ; “n” refers to “neutral”; Rick 1966, 1971), which induces no preferential abortion in the heterozygotes (
; “n” refers to “neutral”; Rick 1966, 1971), which induces no preferential abortion in the heterozygotes (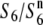 and
and 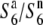 ). Subsequent analysis indicated that the estimated chromosomal location of the
). Subsequent analysis indicated that the estimated chromosomal location of the 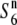 gene overlapped with that of the S6 gene, suggesting that the
gene overlapped with that of the S6 gene, suggesting that the 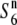 gene is allelic to the S6 gene (see Figure 3).
gene is allelic to the S6 gene (see Figure 3).
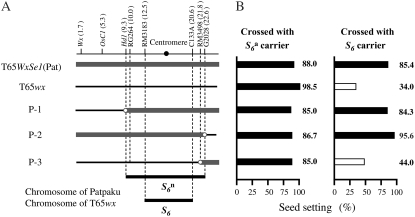
Figure 3.—
Localization of the 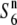 gene on the rice linkage map. (A) Location of the
gene on the rice linkage map. (A) Location of the 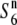 gene on chromosome 6. The physical map of chromosome 6 is based on the Rice Genome Research Program (http://rgp.dna.affrc.go.jp). The map position of each marker along chromosome 6 (AP008212) is shown in parentheses (in megabases). Graphical genotypes of NILs used for mapping the
gene on chromosome 6. The physical map of chromosome 6 is based on the Rice Genome Research Program (http://rgp.dna.affrc.go.jp). The map position of each marker along chromosome 6 (AP008212) is shown in parentheses (in megabases). Graphical genotypes of NILs used for mapping the  gene are shown below the linkage map. Chromosomal regions derived from Patpaku (carrying the
gene are shown below the linkage map. Chromosomal regions derived from Patpaku (carrying the 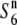 allele) and T65wx (carrying the
allele) and T65wx (carrying the 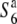 allele) are indicated by thick shaded lines and thin solid lines, respectively. A solid circle represents the centromere. Open circles represent T65wx-derived alleles. The estimated ranges of the positions of the
allele) are indicated by thick shaded lines and thin solid lines, respectively. A solid circle represents the centromere. Open circles represent T65wx-derived alleles. The estimated ranges of the positions of the 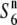 and S6 genes are shown by solid bars below the graphical genotypes. (B) The rate of seed setting in F1 plants derived from crosses between NILs for the
and S6 genes are shown by solid bars below the graphical genotypes. (B) The rate of seed setting in F1 plants derived from crosses between NILs for the 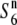 gene and S6 or
gene and S6 or 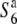 carriers. For the analysis of seed setting, see legend to Figure 2. The rate of seed setting is shown as a percentage (average of two individual plants). Solid and open bars indicate rates of seed setting above and below 50%, respectively.
carriers. For the analysis of seed setting, see legend to Figure 2. The rate of seed setting is shown as a percentage (average of two individual plants). Solid and open bars indicate rates of seed setting above and below 50%, respectively.
The F1 hybrids between the remaining strain W2048 and the two tester lines exhibited a low seed-setting rate. When the seed-setting rate of a hybrid is low, TRD would be expected to be a more reliable indicator for determining the allelic state at the S6 locus. Unlike the other wild rice strains, W2048 displayed the segregation distortion: a marked excess of st1 homozygotes was detected in the F2 population derived from a cross between W2048 and T65st1S6, but not that from a cross between W2048 and 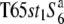 . These results indicate that W2048 carries the
. These results indicate that W2048 carries the 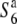 allele. Taken together, these results reveal that plants carrying the
allele. Taken together, these results reveal that plants carrying the 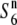 allele are present at a high frequency and are widely distributed in Asia. The S6 allele was detected in four strains from South to Southeast Asia, and the
allele are present at a high frequency and are widely distributed in Asia. The S6 allele was detected in four strains from South to Southeast Asia, and the 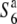 allele was detected in three cultivated (O. sativa ssp. japonica) and one wild (O. rufipogon) rice strain from East Asia.
allele was detected in three cultivated (O. sativa ssp. japonica) and one wild (O. rufipogon) rice strain from East Asia.
The additional allele (S6n) at the S6 locus:
As mentioned above, of the 23 Asian wild rice and cultivated rice strains examined, 15 strains exhibited no TRD, as evaluated by the st1 phenotype in the crosses with either T65st1 S6 or 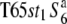 (Table 2), suggesting the presence of an additional allele (
(Table 2), suggesting the presence of an additional allele (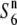 ) at the S6 locus, which induces no preferential abortion in the heterozygotes (
) at the S6 locus, which induces no preferential abortion in the heterozygotes (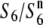 and
and 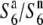 ), as proposed in the tomato (Rick 1966, 1971). To test this possibility, a NIL, T65WxSe1 (Pat), was characterized. Because the testcross indicated that Patpaku carried the
), as proposed in the tomato (Rick 1966, 1971). To test this possibility, a NIL, T65WxSe1 (Pat), was characterized. Because the testcross indicated that Patpaku carried the 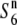 gene (Table 2), the chromosomal fragment transgressed from Patpaku should contain the
gene (Table 2), the chromosomal fragment transgressed from Patpaku should contain the 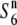 gene, if
gene, if 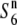 is allelic to the S6 gene (Figure 3). Hybrid plants between T65WxSe1 (Pat) and T65wx (
is allelic to the S6 gene (Figure 3). Hybrid plants between T65WxSe1 (Pat) and T65wx (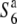 carrier), and those between T65WxSe1 (Pat) and T65S6 (W593), both exhibited a high seed-setting rate (88.0 and 85.4%, respectively). This shows that neither the S6 nor the
carrier), and those between T65WxSe1 (Pat) and T65S6 (W593), both exhibited a high seed-setting rate (88.0 and 85.4%, respectively). This shows that neither the S6 nor the 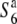 alleles induces abortion of gametes carrying the Patpaku-derived allele in the heterozygotes. The fragment introduced from Patpaku was further segmented by selfing the heterozygote plants. Three F3 lines with distinct homozygous introgressions (P-1, P-2, and P-3; Figure 3) in the S6 region were selected on the basis of the six markers (Wx, OsC1, Hd1, RG264, RM3498, and G2028) and were used to map the
alleles induces abortion of gametes carrying the Patpaku-derived allele in the heterozygotes. The fragment introduced from Patpaku was further segmented by selfing the heterozygote plants. Three F3 lines with distinct homozygous introgressions (P-1, P-2, and P-3; Figure 3) in the S6 region were selected on the basis of the six markers (Wx, OsC1, Hd1, RG264, RM3498, and G2028) and were used to map the 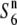 gene. When the three lines were crossed with both the
gene. When the three lines were crossed with both the 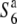 and S6 carriers, only the hybrid plants between P-3 and T65S6 (W593) exhibited a low seed-setting rate (44.0%) (Figure 3). This result indicates that if a plant harbors the chromosomal region between Hd1 and G2028 from Patpaku, the plant induces no preferential abortion in response to the S6 and
and S6 carriers, only the hybrid plants between P-3 and T65S6 (W593) exhibited a low seed-setting rate (44.0%) (Figure 3). This result indicates that if a plant harbors the chromosomal region between Hd1 and G2028 from Patpaku, the plant induces no preferential abortion in response to the S6 and 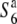 alleles. Since the region between Hd1 and G2028 contained the region in which the S6 gene was located, we concluded that Patpaku possesses the
alleles. Since the region between Hd1 and G2028 contained the region in which the S6 gene was located, we concluded that Patpaku possesses the 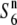 allele at the S6 locus.
allele at the S6 locus.
Survey of nucleotide diversity in chromosomal regions around the S6 locus:
Owing to the presence of the additional allele, association between genetic diversity and allelic state at the S6 locus was analyzed on the basis of the relative nucleotide diversity between plants carrying the incompatible alleles (S6 or 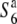 ) and those carrying the additional allele (
) and those carrying the additional allele ( ). We analyzed the nucleotide sequences at the five loci (P139, F06, D11, R111C, and C133A) encompassing the centromeric region (Figure 4), and compared Watterson's θ (θw) and π between these loci (Table 3). Silent sites were used for comparing nucleotide diversity to rule out selection acting on the functional regions. Tajima's D statistic revealed no significant deviations from neutrality at these loci (Table 3). The
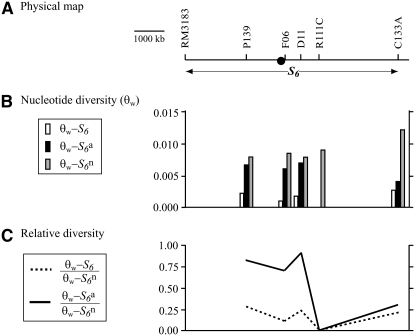
). We analyzed the nucleotide sequences at the five loci (P139, F06, D11, R111C, and C133A) encompassing the centromeric region (Figure 4), and compared Watterson's θ (θw) and π between these loci (Table 3). Silent sites were used for comparing nucleotide diversity to rule out selection acting on the functional regions. Tajima's D statistic revealed no significant deviations from neutrality at these loci (Table 3). The 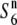 carriers showed values ranging from 7.78 × 10−3 (D11) to 10.50 × 10−3 (C133A) for θw, and from 6.75 × 10−3 (P139) to 8.66 × 10−3 (F06) for π, which is comparable to the values for the nucleotide diversity previously reported in nuclear genes of rice (Yoshida et al. 2004; Yoshida and Miyashita 2005; Rakshit et al. 2007; Zhu et al. 2007). At these five loci, the estimates of θw and π were lower in the S6 and
carriers showed values ranging from 7.78 × 10−3 (D11) to 10.50 × 10−3 (C133A) for θw, and from 6.75 × 10−3 (P139) to 8.66 × 10−3 (F06) for π, which is comparable to the values for the nucleotide diversity previously reported in nuclear genes of rice (Yoshida et al. 2004; Yoshida and Miyashita 2005; Rakshit et al. 2007; Zhu et al. 2007). At these five loci, the estimates of θw and π were lower in the S6 and 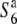 carriers than those in the
carriers than those in the 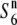 carriers (Table 3 and Figure 4B). In particular, the estimates were zero at the R111C locus in the S6 and
carriers (Table 3 and Figure 4B). In particular, the estimates were zero at the R111C locus in the S6 and 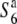 carriers. Since the F06 and D11 loci were physically closer to the centromere than the R111C locus, but a higher level of diversity was detected at these loci compared to the diversity at the R111C locus, the low nucleotide diversities of the S6 and
carriers. Since the F06 and D11 loci were physically closer to the centromere than the R111C locus, but a higher level of diversity was detected at these loci compared to the diversity at the R111C locus, the low nucleotide diversities of the S6 and 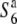 carriers in the R111C locus is evidently not due solely to the effect of low recombination near the centromere. The relative diversities (
carriers in the R111C locus is evidently not due solely to the effect of low recombination near the centromere. The relative diversities (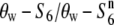 and
and 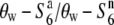 ) increased depending on the physical distance from the R111C locus (Figure 4C). The levels of nucleotide diversity at the P139 and C133A loci were similar to those at the unlinked loci sd1 and qSH4 and the loosely linked locus Hd3a (Table 3), suggesting that the reduction in nucleotide diversity is specific to the chromosomal region around the R111C locus, and this is not accounted for by either demographic effects or selection acting on the sites. These results indicate an association between nucleotide diversity in the chromosomal region and the allelic state at the S6 locus, and are consistent with the notion that the S6 and
) increased depending on the physical distance from the R111C locus (Figure 4C). The levels of nucleotide diversity at the P139 and C133A loci were similar to those at the unlinked loci sd1 and qSH4 and the loosely linked locus Hd3a (Table 3), suggesting that the reduction in nucleotide diversity is specific to the chromosomal region around the R111C locus, and this is not accounted for by either demographic effects or selection acting on the sites. These results indicate an association between nucleotide diversity in the chromosomal region and the allelic state at the S6 locus, and are consistent with the notion that the S6 and 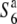 alleles arose independently from the
alleles arose independently from the 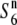 allele (discussed below).
allele (discussed below).
Figure 4.—
Survey of the nucleotide diversity around the centromere on chromosome 6. (A) A physical map of the candidate region for the S6 gene. The position of the centoromere is indicated by a solid circle. (B) Levels of nucleotide diversity (θw) for the S6, 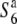 , and
, and 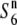 carriers at five sites in the chromosomal region. (C) The relative diversities of the S6 and
carriers at five sites in the chromosomal region. (C) The relative diversities of the S6 and 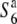 carriers to the
carriers to the 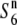 carriers at five sites in the chromosomal region.
carriers at five sites in the chromosomal region.
TABLE 3.
Levels of nucleotide diversity
|
S6 (n = 4)
|
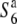 (n = 4) (n = 4)
|
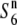 (n = 15) (n = 15)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of polymorphic positions | Nucleotide diversityb
|
Tajima's D | No. of polymorphic positions | Nucleotide diversityb
|
Tajima's D | No. of polymorphic positions | Nucleotide diversityb
|
Tajima's D | |||||
| Loci | Lengtha | (θw × 103) | (π × 103) | (θw × 103) | (π × 103) | (θw × 103) | (π × 103) | ||||||
| Chromosome 6 | |||||||||||||
| P139 | 748 | 3 | 2.18 ± 1.58 | 2.23 ± 0.59 | 0.16766 | 9 | 6.55 ± 3.97 | 6.01 ± 3.19 | −0.82943 | 19 | 7.81 ± 3.27 | 6.75 ± 0.95 | −0.55548 |
| F06 | 530 | 1 | 0.98 ± 0.98 | 0.90 ± 0.48 | −0.61237 | 6 | 5.95 ± 3.80 | 5.45 ± 2.89 | −0.80861 | 15 | 8.37 ± 3.42 | 8.66 ± 0.99 | 0.13696 |
| D11 | 688 | 1 | 1.72 ± 1.72 | 1.58 ± 0.84 | −0.75455 | 4 | 6.89 ± 4.70 | 6.31 ± 3.35 | −0.79684 | 8 | 7.78 ± 3.79 | 7.33 ± 1.52 | −0.25041 |
| R111C | 590 | 0 | 0.00 ± 0.00 | 0.00 ± 0.00 | — | 0 | 0.00 ± 0.00 | 0.00 ± 0.00 | — | 17 | 8.86 ± 3.77 | 6.68 ± 0.64 | −0.99328 |
| C133A | 892 | 7 | 4.59 ± 2.87 | 4.21 ± 1.88 | −0.81734 | 6 | 2.65 ± 1.81 | 2.43 ± 1.29 | −0.80861 | 32 | 10.50 ± 4.21 | 8.14 ± 1.87 | −1.03951 |
| Hd3a | 2448 | 18 | 4.59 ± 2.63 | 4.45 ± 1.94 | −0.31310 | 15 | 3.82 ± 2.22 | 3.50 ± 1.72 | −0.84729 | 43 | 6.30 ± 2.44 | 5.88 ± 0.51 | −0.28539 |
| Chromosome 1 | |||||||||||||
| sd1 | 1883 | 9 | 3.92 ± 2.37 | 3.59 ± 1.90 | −0.82943 | 7 | 3.04 ± 1.90 | 2.79 ± 1.48 | −0.82407 | 36 | 11.05 ± 4.45 | 8.55 ± 1.48 | −1.16932 |
| Chromosome 4 | |||||||||||||
| qSH4 | 1734 | 18 | 5.75 ± 3.29 | 5.46 ± 1.76 | −0.50485 | 18 | 5.72 ± 3.28 | 5.24 ± 2.78 | −0.85194 | 45 | 8.10 ± 3.13 | 8.23 ± 0.99 | −0.11812 |
Indels were ignored for computation.
No. of nucleotide positions analyzed.
Silent substitutions including synonymous substitutions and changes in the noncoding positions were used for the analysis.
DISCUSSION
The siTRD caused by the S6 locus and its independence from the effects of other loci associated with sexual affinity on the same chromosome:
TRD violates Mendel's rules by the preferential transmission of a particular chromosome or allele at the expense of its partner. Preferential gamete dysfunction has been frequently reported as a mechanism of TRD in a range of organisms (Lyttle 1991; Temin et al. 1991; Silver 1993; Úbeda and Haig 2005; Moyle and Graham 2006). Cytological observations revealed that about half of the female gametes degenerated in the 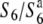 heterozygote (Figure 1). However, no defect was detected in the pollen grains by this method, although genetic analysis indicated that the transmission of both male and female gametes carrying the
heterozygote (Figure 1). However, no defect was detected in the pollen grains by this method, although genetic analysis indicated that the transmission of both male and female gametes carrying the 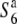 allele to the next generation was very low (Sano 1992; this study). This probably indicates that functional difference(s) between the male gametes with different alleles, which accounts for the TRD, occurs after the production of pollen grains. No reduction in the seed-setting rate was observed when the
allele to the next generation was very low (Sano 1992; this study). This probably indicates that functional difference(s) between the male gametes with different alleles, which accounts for the TRD, occurs after the production of pollen grains. No reduction in the seed-setting rate was observed when the 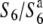 heterozygote was used as a male parent for crosses, which indicates that no aberration occurred after fertilization, although almost no
heterozygote was used as a male parent for crosses, which indicates that no aberration occurred after fertilization, although almost no 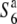 alleles were transmitted to the progeny (Sano 1992). Taken together, these observations suggest that the primary event(s) resulting in the TRD phenomenon through the male gametes in the
alleles were transmitted to the progeny (Sano 1992). Taken together, these observations suggest that the primary event(s) resulting in the TRD phenomenon through the male gametes in the 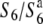 heterozygote occurs during a prezygotic process after the production of pollen grains. A plausible explanation for these observations is that TRD through the male gametes is caused by pollen tube competition (certation). Alternatively, it is possible that the dysfunction of the male gametes occurred at an early stage of gametogenesis and the observed pollen grains consist only of carriers of the S6 allele. In any event, the genetic effects of the S6 locus on the siTRD were altered in neither the female nor male gametes, even after repeated backcrosses, which indicates the stability of the phenomenon.
heterozygote occurs during a prezygotic process after the production of pollen grains. A plausible explanation for these observations is that TRD through the male gametes is caused by pollen tube competition (certation). Alternatively, it is possible that the dysfunction of the male gametes occurred at an early stage of gametogenesis and the observed pollen grains consist only of carriers of the S6 allele. In any event, the genetic effects of the S6 locus on the siTRD were altered in neither the female nor male gametes, even after repeated backcrosses, which indicates the stability of the phenomenon.
Genetic mapping revealed that the S6 locus is located near the centromere of chromosome 6. On the same chromosome, genes that affect the fertility of F1 hybrids have been identified. Among these are the cim (the cross-incompatibility reaction in the male) gene and the two Cif (the cross-incompatibility reaction in the female) genes responsible for the abortion of hybrid seeds, especially in endosperm, observed in a cross between W593 and T65wx (Matsubara et al. 2003; Koide et al. 2008c), which are the same parental strains of the materials used in the present study. The cim and one of the Cif genes, Cif2, have been mapped in a chromosomal region that overlaps with the S6 locus (Matsubara et al. 2003; Koide et al. 2008c). Another Cif gene, Cif1 is located on the short arm of chromosome 6. The plants carrying cim derived from T65wx exhibit cross-incompatibility when pollinated to the female plants carrying both Cif1 and Cif2 derived from W593. The genotypes of T65wx and W593 are cif1 cif2 cim/cif1 cif2 cim and Cif1 Cif2 Cim/Cif1 Cif2 Cim, respectively. These three genes (cim, Cif1, and Cif2) act sporophytically, so that the heterozygotes (Cif1 Cif2 Cim/cif1 cif2 cim) produce only gametes that are cross-compatible. As a consequence, the cross-incompatibility system controlled by the cim and Cif genes acts on the formation of hybrid seeds after fertilization (Matsubara et al. 2003; Koide et al. 2008c), whereas that by the S6 locus acts on gametes produced in the F1 plants. Therefore, the developmental phase of the S6 locus-mediated abortion and consequent segregation pattern are both unequivocally distinguished from those mediated by the Cif/cim genes, although the possibility cannot be ruled out that abortions at different developmental phases are caused by pleiotropic effects of a single gene, which would resemble a phenomenon caused by the Tcb1 locus in maize (Kermicle 2006).
The hybrid sterility loci, S1 (Sano 1990), S5 (Yanagihara et al. 1995), S8 (Singh et al. 2006), S10 (Sano et al. 1994), and S26 (Kubo and Yoshimura 2001), have also been mapped to chromosome 6, a mapping procedure carried out using different pairs of crossing parents. These genes were mapped in chromosomal regions apart from the S6 locus, and segregation analyses of marker genes tightly linked with these loci also indicated that the segregation distortion caused by these loci was independent of that by the S6 locus (our unpublished data). These results indicate that the siTRD phenomenon caused by the S6 locus is not affected by the allelic state at the other hybrid sterility loci on the same chromosome. The independence of the S6 locus-mediated siTRD ensures that the pattern of nucleotide diversity around the S6 locus (Table 3) depends exclusively on the allelic state at the S6 locus.
The S6n allele is widely distributed in Asian rice and functions in a locus-specific manner:
We examined the allelic distribution at the S6 locus in 23 strains of Asian rice (Table 2). In addition to the S6 locus, we have also analyzed the allelic distribution at the S1 locus that causes siTRD in the hybrids between the Asian and African rice species (Koide et al. 2008b). Although many loci causing TRD have been identified in the rice genome (see Koide et al. 2008a), no other attempts that analyze allelic distribution of a single locus in various rice accessions have been reported. A notable difference between the siTRD caused by the S6 and S1 loci is the presence or absence of an additional allele the presence of which, in heterozygotic states, does not result in siTRD; both the testcross experiments and subsequent genetic mapping indicated the presence of the 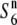 allele at the S6 locus (Table2; Figure 3), whereas no such additional allele has been detected regarding the S1 locus (Koide et al. 2008b). The testcross experiments also indicated that the
allele at the S6 locus (Table2; Figure 3), whereas no such additional allele has been detected regarding the S1 locus (Koide et al. 2008b). The testcross experiments also indicated that the 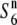 allele was distributed widely, and was predominant in Asian rice (Table 2). A phylogenetic analysis using the nucleotide sequence data from the five loci (P139, F06, D11, R111C, and C133A) around the S6 locus showed that both the S6 and
allele was distributed widely, and was predominant in Asian rice (Table 2). A phylogenetic analysis using the nucleotide sequence data from the five loci (P139, F06, D11, R111C, and C133A) around the S6 locus showed that both the S6 and 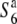 carriers clustered, and these clusters were separated from each other on the phylogenetic tree (data not shown). These results suggest that the
carriers clustered, and these clusters were separated from each other on the phylogenetic tree (data not shown). These results suggest that the 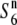 allele could be the ancestral state of the locus and the S6 and
allele could be the ancestral state of the locus and the S6 and 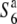 alleles evolved from the ancestral
alleles evolved from the ancestral 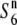 allele independently.
allele independently.
It has been frequently reported in varietal crosses that hybrid sterility in rice is caused by a single locus (Kitamura 1962; Oka 1964; Ikehashi and Araki 1986; Yanagihara et al. 1995; Liu et al. 1997). In these loci, a wide array of compatible alleles conferring compatibility to a wide range of strains has been proposed, and some have actually been mapped (Ji et al. 2005; Qiu et al. 2005; Wang et al. 2006). However, regarding the wide compatibility proposed in rice, whether these compatible alleles act specifically on the alleles at their own locus or on sterility genes at different loci, as reported in wheat (Tsujimoto and Tsunewaki 1985; Friebe et al. 2003) and Drosophila (Lyttle 1991; Temin et al. 1991), is not known. In the testcross experiments, the hybrids between the 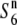 carriers and the tester lines did not necessarily exhibit a higher seed-setting rate than the hybrids between S6 or
carriers and the tester lines did not necessarily exhibit a higher seed-setting rate than the hybrids between S6 or 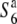 carriers and the tester lines (Table 2). These results suggest that, unlike alleles conferring wide compatibility, the
carriers and the tester lines (Table 2). These results suggest that, unlike alleles conferring wide compatibility, the 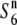 allele does not reduce the level of hybrid sterility that might be caused by other sterility loci, but rather interacts specifically with the S6 and
allele does not reduce the level of hybrid sterility that might be caused by other sterility loci, but rather interacts specifically with the S6 and 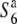 alleles.
alleles.
Chromosomal region-specific reduction in genetic diversity associated with allelic state:
The presence of the 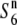 allele prompted us to survey the chromosomal region around the S6 locus in terms of nucleotide diversity and examine the association between the nucleotide diversity and allelic state at the S6 locus. Association-based mapping could identify regions that are associated with a particular phenotype (Thornsberry et al. 2001; Palaisa et al. 2003). If a new sequence variant underlying a given phenotype has emerged, then there would have been less time for recombination, and thus a significant association would be expected between phenotype and genotype in the vicinity of the causative mutation. As a result, a reduction in nucleotide diversity should occur around the causative mutation within a population of individuals exhibiting the same phenotype. If the S6 and
allele prompted us to survey the chromosomal region around the S6 locus in terms of nucleotide diversity and examine the association between the nucleotide diversity and allelic state at the S6 locus. Association-based mapping could identify regions that are associated with a particular phenotype (Thornsberry et al. 2001; Palaisa et al. 2003). If a new sequence variant underlying a given phenotype has emerged, then there would have been less time for recombination, and thus a significant association would be expected between phenotype and genotype in the vicinity of the causative mutation. As a result, a reduction in nucleotide diversity should occur around the causative mutation within a population of individuals exhibiting the same phenotype. If the S6 and 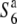 alleles have arisen independently from the
alleles have arisen independently from the 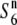 allele, a reduction in nucleotide diversities of the S6 and
allele, a reduction in nucleotide diversities of the S6 and 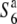 carriers relative to the diversity of the
carriers relative to the diversity of the 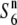 carriers should be observed near the locus, as the relative nucleotide diversities would tend to increase with physical distance from it due to the shuffling into different genetic backgrounds through recombination. In fact, the nucleotide diversities of the S6 and
carriers should be observed near the locus, as the relative nucleotide diversities would tend to increase with physical distance from it due to the shuffling into different genetic backgrounds through recombination. In fact, the nucleotide diversities of the S6 and 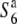 carriers were zero at the R111C locus and the relative diversities (θw-S6/θw-
carriers were zero at the R111C locus and the relative diversities (θw-S6/θw-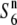 and
and 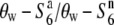 ) increased depending on the physical distance from the R111C locus (Figure 4B). Thus, a clear association between nucleotide diversity and allelic state at the S6 locus was detected by comparing nucleotide diversity of the S6 or
) increased depending on the physical distance from the R111C locus (Figure 4B). Thus, a clear association between nucleotide diversity and allelic state at the S6 locus was detected by comparing nucleotide diversity of the S6 or 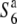 carriers with that of the
carriers with that of the 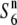 carriers, which most probably reflects independent lineages of the S6 and
carriers, which most probably reflects independent lineages of the S6 and 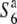 carriers. These results also suggest that the causative alleles are most likely located in a region close to the R111C locus among the loci examined.
carriers. These results also suggest that the causative alleles are most likely located in a region close to the R111C locus among the loci examined.
The resolution of association analysis depends on the structure of the linkage disequilibrium (LD) across the entire genome. Garris et al. (2003) reported that significant LD was detected at sites which were at most 100 kb apart around the xa5 locus in cultivated rice strains. However, it has not established how far the LD extends in the region near the centromere where recombination rates are low. The results of the present study consequently suggest that association mapping is potentially useful even for chromosomal regions where recombination is restricted. Further analyses using more accessions and loci would reveal whether the local nucleotide diversity is directly associated with the S6 locus-mediated phenotype.
Allelic differentiation at the S6 locus and evolution of the siTRD system that serves as an isolating barrier:
The Dobzhansky–Muller model proposes that hybrid incompatibilities are generally caused by an interaction between genes that have functionally diverged in each population (Coyne and Orr 2004). This model assumes two or more interacting loci. TRD systems often involve alleles at a minimum of two closely linked loci, a distorter and its cis-acting target (Lyttle 1991). Examples of the TRD system of this type include the mouse t-haplotype (Silver 1993), segregation distorter (SD) of Drosophila (Temin et al. 1991), and Gametocidal 2 (Gc2) in wheat (Friebe et al. 2003).
Alternatively, the development of hybrid incompatibility might be explained by a simple one-locus stepwise mutation model (Nei et al. 1983). This model assumes changes in allelic state in two different populations: A0 mutates to A1 in one population and to A−1 in another population. If the heterozygotes for alleles that are two or more steps apart from each other are infertile, hybrid incompatibility appears in A1A−1 despite the fact that A1A0 and A−1A0 are completely fertile. This model was adopted for explaining allelic differentiation at the Gamete eliminater (Ge) locus in tomato (Rick 1971). Rick (1966, 1971) reported that there are three alleles (Gep, Gec, and Gen) at the locus. Gep induces abortion of the male and female gametes carrying Gec in the heterozygote (Gep/Gec). On the basis of their geographical distribution, it was suggested that Gep and Gec have arisen from the neutral allele Gen. Likewise, because of the presence of the 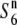 allele at the S6 locus in rice, the evolutionary process at the S6 locus can be explained by the one-locus stepwise mutation model. The results obtained so far are also consistent with the notion that the
allele at the S6 locus in rice, the evolutionary process at the S6 locus can be explained by the one-locus stepwise mutation model. The results obtained so far are also consistent with the notion that the 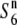 allele is the ancestral state of the locus, as described above.
allele is the ancestral state of the locus, as described above.
Recent studies have suggested that TRD, which is normally masked within a species, plays a role in establishing the sterility barriers between species (Dermitzakis et al. 2000; Tao et al. 2001; Orr and Irving 2004). There are at least two possible models for the formation of isolating barriers induced by normally masked TRD. Frank (1991) along with Hurst and Pomiankowski (1991) proposed that genes causing TRD are rapidly fixed in a population due to their “selfish nature,” but may often become suppressed within each population to alleviate their deleterious effects on fertility. These genes can become reexpressed in hybrids between individuals from allopatric populations and cause the sterility of hybrids.
Another possible explanation is that a transmission ratio distorter never did appear in the history of the two allopatric populations and TRD instead appeared as a consequence of an inappropriate interaction between genes that had diverged in these populations. These genes have no evident selfish behavior in so far as they are present in each population. In this model, genes of neither lineage pass through the adaptive valley represented by the driving genotype that results in TRD (Orr and Presgraves 2000). Assuming that the S6 and 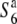 alleles have arisen independently from the
alleles have arisen independently from the 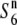 allele, the incompatible alleles (S6 and
allele, the incompatible alleles (S6 and 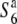 ) could have become fixed in different populations without carrying any selective disadvantage, which would favor the latter explanation. The predominance of the
) could have become fixed in different populations without carrying any selective disadvantage, which would favor the latter explanation. The predominance of the 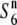 allele is consistent with the fact that the Asian wild rice population consists primarily of plants with interfertile relationships (Morishima et al. 1963; Chu et al. 1969). The siTRD system mediated by the allelic interaction between the S6 and
allele is consistent with the fact that the Asian wild rice population consists primarily of plants with interfertile relationships (Morishima et al. 1963; Chu et al. 1969). The siTRD system mediated by the allelic interaction between the S6 and 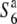 alleles established genetically discontinuous relationships between limited constituents (e.g., between O. sativa ssp. japonica and O. rufipogon in Southeast Asia) of the existing rice population.
alleles established genetically discontinuous relationships between limited constituents (e.g., between O. sativa ssp. japonica and O. rufipogon in Southeast Asia) of the existing rice population.
Acknowledgments
We thank M. Nakagahra for a gift of a seed stock (Fl 176). We thank Y. Kishima, I. Takamure, and H. Nagano for suggestions and comments. This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science, and Technology and the Ministry of Agriculture, Forestry, and Fisheries of Japan (integrated research project for plant, insect, and animal using genome technology GD-2001).
References
- Birchler, J. A., R. K. Dawe and J. F. Doebley, 2003. Marcus Rhoades, preferential segregation and meiotic drive. Genetics 164 835–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, D. R., and R. Moav, 1957. Inheritance in Nicotiana tabacum XXVII. Pollen killer, an alien genetic locus inducing abortion of microspores not carrying it. Genetics 42 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Y. E., and H. I. Oka, 1972. The distribution and effects of genes causing F1 weakness in Oryza breviligulata and O. glaberrima. Genetics 70 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Y. E., H. Morishima and H. I. Oka, 1969. Reproductive barriers distributed in cultivatedrice species and their wild relatives. Jpn. J. Genet. 44 207–223. [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Crow, J. F., 1988. The ultraselfish gene. Genetics 118 389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermitzakis, E. T., J. P. Masly, H. M. Waldrip and A. G. Clark, 2000. Non-mendelian segregation of sex chromosomes in heterospecific Drosophila males. Genetics 154 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, A., and M. R. MacNair, 1999. Pollen tube competitions as a mechanism of prezygotic reproductive isolation between Mimulus nasutus and its presumed progenitor M. guttatus. New Phytol. 144 471–478. [DOI] [PubMed] [Google Scholar]
- Dung, L. V., T. Inukai and Y. Sano, 1998. Dissection of a major QTL for photoperiod sensitivity in rice: its association with a gene expressed in an age-dependent manner. Theor. Appl. Genet. 97 714–720. [Google Scholar]
- Endo, T. R., and K. Tsunewaki, 1975. Sterility of common wheat with Aegilops triuncialis cytoplasm. J. Hered. 66 13–18. [Google Scholar]
- Finch, R. A., T. E. Miller and M. D. Bennett, 1984. “Cuckoo” Aegilops addition chromosome in wheat ensures its transmission by causing chromosome breaks in meiospores lacking it. Chromosoma 90 84–88. [Google Scholar]
- Fishman, L., and J. H. Willis, 2005. A novel meiotic drive locus almost completely distorts segregation in Mimulus (monkeyflower) hybrids. Genetics 169 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, S., 1991. Divergence of meiotic drive-suppression system as an explanation for sex-biased hybrid sterility and inviability. Evolution 45 262–267. [DOI] [PubMed] [Google Scholar]
- Friebe, B., P. Zhang, S. Nasuda and B. S. Gill, 2003. Characterization of a knock-out mutation at the Gc2 locus in wheat. Chromosoma 111 509–517. [DOI] [PubMed] [Google Scholar]
- Garris, A. J., S. R. Mccouch and S. Kresovich, 2003. Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., M. Nakagahra, M. Yano, T. Sasaki and N. Kurata, 2001. A genomewide survey of reproductive barriers in an intraspecific hybrid. Genetics 159 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, L. D., and A. Pomiankowski, 1991. Causes of sex ratio bias may account for unisexual sterility in hybrid: a new explanation of Haldane's rule and related phenomena. Genetics 128 841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehashi, H., and H. Araki, 1986. Genetics of F1 sterility in remote crosses of rice, pp. 119–130 in Rice Genetics, edited by International Rice Research Institute. International Rice Research Institute, Los Baños, Philippines.
- Ji, Q., J. Lu, Q. Chao, M. Gu and M. Xu, 2005. Delimiting a rice wide-compatibility gene S5n to a 50 kb region. Theor. Appl. Genet. 111 1495–1503. [DOI] [PubMed] [Google Scholar]
- Kermicle, J. L., 2006. A selfish gene governing pollen-pistil compatibility confers reproductive isolation between maize relatives. Genetics 172 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura, E., 1962. Genetic studies on sterility observed in hybrids between distantly related varieties of rice, Oryza sativa L. Bull. Chugoku Agr. Exp. Sta. Series A, 141–205.
- Koide, Y., K. Onishi, A. Kanazawa and Y. Sano, 2008. a Genetics of speciation in rice, pp. 247–259 in Rice Biology in the Genomics Era, edited by H. Y. Hirano, A. Hirai, Y. Sano and T. Sasaki. Springer-Verlag, Berlin.
- Koide, Y., K. Onishi, D. Nishimoto, A. R. Baruah, A. Kanazawa et al., 2008. b Sex-independent transmission ratio distortion system responsible for reproductive barriers between Asian and African rice species. New Phytol. 179 888–900. [DOI] [PubMed] [Google Scholar]
- Koide, Y., M. Ikenaga, Y. Shinya, K. Matsubara and Y. Sano, 2008. c Two loosely linked genes controlling the female specificity for cross-incompatibility in rice. Euphytica (in press).
- Kubo, T., and A. Yoshimura, 2001. Linkage analysis of an F1 sterility gene in Japonica/Indica cross of rice. Rice Genet. News Lett. 18 52–54. [Google Scholar]
- Li, Z., S. R. M. Pinson, A. H. Paterson, W. D. Park and J. W. Stansel, 1997. Genetics of hybrid sterility and hybrid breakdown in an intersubspecific rice (Oryza sativa L.) population. Genetics 145 1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, K.D., J. Wang, H.B. Li, C.G. Xu, A.M. Liu et al., 1997. A genome-wide analysis of wide compatibility in rice and the precise location of the S5 locus in the molecular map. Theor. Appl. Genet. 95 809–814. [Google Scholar]
- Loegering, W. Q., and E. R. Sears, 1963. Distorted inheritance of stem-rust resistance of timstein wheat caused by a pollen-killing gene. Can. J. Genet. 5 65–72. [Google Scholar]
- Lyttle, T. W., 1991. Segregation distorters. Annu. Rev. Genet. 25 511–557. [DOI] [PubMed] [Google Scholar]
- Maguire, M. P., 1963. High transmission frequency of a Tripsacum chromosome in corn. Genetics 48 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, K., Khin-Thidar and Y. Sano, 2003. A gene block causing cross-incompatibility hidden in wild and cultivated rice. Genetics 165 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monna, L., H. X. Lin, S. Kojima, T. Sasaki and M. Yano, 2002. Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice. Theor. Appl. Genet. 104 772–778. [DOI] [PubMed] [Google Scholar]
- Morishima, H., K. Hinata and H. I. Oka, 1963. Comparison of modes of evolution of cultivated forms from two wild rice species, Oryza breviligulata and O. perennis. Evolution 17 170–181. [Google Scholar]
- Morishima, H., Y. Sano and H. I. Oka, 1992. Evolutionary studies in cultivated rice and its wild relatives. Oxford Surveys Evol. Biol. 8 135–184. [Google Scholar]
- Moyle, L. C., and E. B. Graham, 2006. Genome-wide association between hybrid sterility QTL and marker transmission ratio distortion. Mol. Biol. Evol. 23 973–980. [DOI] [PubMed] [Google Scholar]
- Nagano, H., K. Onishi, M. Ogasawara, Y. Horiuchi and Y. Sano, 2005. Genealogy of the “Green Revolution” gene in rice. Genes Genet. Syst. 80 351–356. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Nei, M., T. Maruyama and C.-I Wu, 1983. Models of evolution of reproductive isolation. Genetics 103 557–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, H. I., 1964. Considerations on the genetic basis of intervarietal sterility in Oryza sativa, pp.158–174 in Rice Genetics and Cytogenetics, edited by International Rice Research Institute. International Rice Research Institute, Los Baños, Philippines.
- Oka, H. I., 1974. Analysis of genes controlling F1 sterility in rice by the use of isogenic lines. Genetics 77 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, H. I., 1988. Origin of Cultivated Rice. Elsevier, Amsterdam.
- Onishi, K., K. Takagi, M. Kontani, T. Tanaka and Y. Sano, 2007. Different patterns of genealogical relationships found in the two major QTLs causing reduction of seed shattering during rice domestication. Genome 50 757–766. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., and S. Irving, 2004. Segregation distortion in hybrids between the Bogota and USA subspecies of Drosophila pseudoobscura. Genetics 169 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and D. C. Presgraves, 2000. Speciation by postzygotic isolation: forces, genes and molecules. BioEssays 22 1085–1094. [DOI] [PubMed] [Google Scholar]
- Palaisa, K. A., M. Morgante, M. Williams and A. Rafalski, 2003. Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 15 1795–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena, F., and C. Sapienza, 2001. Nonrandom segregation during meiosis: the unfairness of females. Mamm. Genome 12 331–339. [DOI] [PubMed] [Google Scholar]
- Price, C. S. C., 1997. Conspecific sperm precedence in Drosophila. Nature 388 663–666. [DOI] [PubMed] [Google Scholar]
- Qiu, S. Q., K. Liu, J. X. Jiang, X. Song, C. G. Xu et al., 2005. Delimitation of the rice wide compatibility gene S5n to a 40-kb DNA fragment. Theor. Appl. Genet. 111 1080–1086. [DOI] [PubMed] [Google Scholar]
- Rakshit, S., A. Rakshit, H. Matsumura, Y. Takahashi, Y. Hasegawa et al., 2007. Large-scale DNA polymorphism study of Oryza sativa and O. rufipogon reveals the origin and divergence of Asian rice. Theor. Appl. Genet. 114 731–743. [DOI] [PubMed] [Google Scholar]
- Rick, C. M., 1966. Abortion of male and female gametes in the tomato determined by allelic interaction. Genetics 53 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick, C. M., 1971. The tomato Ge locus: linkage relations and geographic distribution of alleles. Genetics 67 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15 174–175. [DOI] [PubMed] [Google Scholar]
- Sano, Y., 1983. A new gene controlling sterility in F1 hybrids of two cultivated rice species. J. Hered. 74 435–439. [Google Scholar]
- Sano, Y., 1990. The genic nature of gamete eliminator in rice. Genetics 125 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, Y., 1992. Genetic comparisons of chromosome 6 between wild and cultivated rice. Jpn. J. Breed. 42 561–572. [Google Scholar]
- Sano, Y., Y. E. Chu and H. I. Oka, 1979. Genetic studies of speciation in cultivated rice, 1. Genic analysis for the F1 sterility between O. sativa L. and O. glaberrima Steud. Jpn. J. Genet. 54 121–132. [Google Scholar]
- Sano, Y., R. Sano, M. Eiguchi and H.-Y. Hirano, 1994. Gamete eliminator adjacent to the wx locus as revealed by pollen analysis in rice. J. Hered. 85 310–312. [Google Scholar]
- Scoles, G. J., and I. N. Kibirge-Sebunya, 1983. Preferential abortion of gametes in wheat induced by an Agropyron chromosome. Can. J. Cytol. 25 1–6. [Google Scholar]
- Shinjo, C., 1984. Cytoplasmic male sterility and fertility restoration in rice having genome A, pp. 321–338 in Biology of Rice, edited by S. Tsunoda, and N. Takahashi. Elsevier, Amsterdam.
- Silver, L. M., 1993. The peculiar journey of a selfish chromosome: mouse t haplotypes and meiotic drive. Trends Genet. 9 250–254. [DOI] [PubMed] [Google Scholar]
- Singh, S. P., R. M. Sundaram, S. K. Biradar, M. I. Ahmed, B. C. Viraktamath et al., 2006. Identification of simple sequence repeat markers for utilizing wide-compatibility genes in inter-subspecific hybrids in rice (Oryza sativa L.). Theor. Appl. Genet. 113 509–517. [DOI] [PubMed] [Google Scholar]
- Sylvester, A. W., and S. E. Ruzin, 1993. Light microscopy I: dissection and microtechnique, pp. 83–95 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Tao, Y., D. L. Hartl and C. C. Laurie, 2001. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc. Natl. Acad. Sci. USA 98 13183–13188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin, R. G., B. Ganetzky, P. A. Powers, T. W. Lyttle, S. Pimpinelli et al., 1991. Segregation distortion in Drosophila melanogaster: genetic and molecular analyses. Am. Nat. 137 287–331. [Google Scholar]
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry, J. M., M. M. Goodman, J. Doebley, S. Kresovich, D. Nielsen et al., 2001. Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28 286–289. [DOI] [PubMed] [Google Scholar]
- Tsujimoto, H., and K. Tsunewaki, 1985. Gametocidal genes in wheat and its relatives. II. Suppressor of chromosome 3C gametocidal gene of Aegilops triuncialis. Can. J. Genet. Cytol. 27 178–185. [Google Scholar]
- Úbeda, F., and D. Haig, 2005. On the evolutionary stability of Mendelian segregation. Genetics 170 1345–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G. W., Y. Q. He, C. G. Xu and Q. Zhang, 2006. Fine mapping of f5-Du, a gene conferring wide-compatibility for pollen fertility in inter-subspecific hybrids of rice (Oryza sativa L.). Theor. Appl. Genet. 112 382–387. [DOI] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Pop. Biol. 7 256–276. [DOI] [PubMed] [Google Scholar]
- Wu, J., H. Mizuno, M. Hayashi-Tsugane, Y. Ito, Y. Chiden et al., 2003. Physical maps and recombination frequency of six rice chromosomes. Plant J. 36 720–730. [DOI] [PubMed] [Google Scholar]
- Xu, Y., L. Zhu, J. Xiao, N. Huang and S. R. McCouch, 1997. Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol. Gen. Genet. 253 535–545. [DOI] [PubMed] [Google Scholar]
- Yanagihara, S., S. R. McCouch, K. Ishikawa, Y. Ogi, K. Maruyama et al., 1995. Molecular analysis of the inheritance of the S-5 locus, conferring wide compatibility in indica/japonica hybrids of rice (O. sativa L.). Theor. Appl. Genet. 90 182–188. [DOI] [PubMed] [Google Scholar]
- Yoshida, K., and N. T. Miyashita, 2005. Nucleotide polymorphism in the Adh2 region of the wild rice Oryza rufipogon. Theor. Appl. Genet. 111 1215–1228. [DOI] [PubMed] [Google Scholar]
- Yoshida, K., N. T. Miyashita and T. Ishii, 2004. Nucleotide polymorphism in the Adh1 locus region of the wild rice Oryza rufipogon. Theor. Appl. Genet. 109 1406–1416. [DOI] [PubMed] [Google Scholar]
- Zhu, Q., X. Zheng, J. Luo, B. S. Gaut and S. Ge, 2007. Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Mol. Biol. Evol. 24 875–888. [DOI] [PubMed] [Google Scholar]