Abstract
Interlocus gene conversion is considered a crucial mechanism for generating novel combinations of polymorphisms in duplicated genes. The importance of gene conversion between duplicated genes has been recognized in the major histocompatibility complex and self-incompatibility genes, which are likely subject to diversifying selection. To theoretically understand the potential role of gene conversion in such situations, forward simulations are performed in various two-locus models. The results show that gene conversion could significantly increase the number of haplotypes when diversifying selection works on both loci. We find that the tract length of gene conversion is an important factor to determine the efficacy of gene conversion: shorter tract lengths can more effectively generate novel haplotypes given the gene conversion rate per site is the same. Similar results are also obtained when one of the duplicated genes is assumed to be a pseudogene. It is suggested that a duplicated gene, even after being silenced, will contribute to increasing the variability in the other locus through gene conversion. Consequently, the fixation probability and longevity of duplicated genes increase under the presence of gene conversion. On the basis of these findings, we propose a new scenario for the preservation of a duplicated gene: when the original donor gene is under diversifying selection, a duplicated copy can be preserved by gene conversion even after it is pseudogenized.
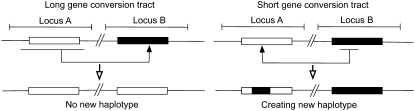
INTERLOCUS gene conversion plays significant roles in shaping the pattern of polymorphism and divergence in duplicated genes (Ohta 1980; Li 1997; Innan 2003; Teshima and Innan 2004). Gene conversion exchanges DNA segments between duplicated genes, which is usually described as a copy-and-paste event (Figure 1). With this mechanism, duplicated genes undergo nonindependent molecular evolution. There are at least two major outcomes of gene conversion, which seem somewhat conflicting. One is the phenomenon called “concerted evolution,” in which gene conversion reduces sequence variation between duplicates (Ohta 1980; Zimmer et al. 1980; Dover 1982; Li 1997). The other is that gene conversion creates and enhances genetic variation within a gene family (Miyata et al. 1980; Baltimore 1981; Ohta 1991, 1992, 1997). This discrepancy between the two conflicting outcomes can be explained when considering the role of selection and how “genetic variation” is defined.
Figure 1.—
Illustration of gene conversion events. See text for details.
Concerted evolution is the outcome that can be more intuitively understandable, because it is obvious that a gene conversion event makes the sequence identical for the converted region regardless of its tract length (Figure 1). When gene conversion occurs frequently, the sequence identity between duplicated genes is likely high, while there would be some divergence from their orthologous genes in a different species. This phenomenon was first observed in the rDNA genes (Brown et al. 1972; Coen et al. 1982), and the genomewide significance of the role of gene conversion has been recently emphasized in many organisms (Semple and Wolfe 1999; Gao and Innan 2004; Ezawa et al. 2006; Wang et al. 2007). Theoretically, the gene conversion rate is the major factor to determine the identity level of DNA sequences under neutrality (Nagylaki and Petes 1982; Ohta 1982; Nagylaki 1983; Innan 2002, 2003).
Thus, gene conversion indeed reduces the variation between duplicates, when it is defined as the average nucleotide divergence between paralogs. However, the situation would be different if genetic diversity is measured in terms of haplotype or a stretch of DNA sequence. Suppose that we define haplotypes on the basis of the sequences in a particular region (the boxed regions in Figure 1). In the right side of Figure 1, a gene conversion event creates a new chimeric haplotype because the gene conversion occurs within the boxed region. On the other hand, when the gene conversion tract completely covers the boxed region as illustrated in the left side of Figure 1, it does not introduce any new haplotypes into a population. It should be noted that in both cases, the average divergence is reduced by gene conversion. Nevertheless, the gene conversion event introduces a novel variant (haplotype) in the population in the former case (right side in Figure 1). This role of creating new haplotypes is emphasized at a locus where a high level of haplotype diversity is required, such as animal major histocompatibility complex (MHC) genes (Baltimore 1981; Ohta 1991, 1997; Parham and Ohta 1996; Martinsohn et al. 1999) and plant self-incompatibility (SI) genes (Sato et al. 2002; Charlesworth et al. 2003). In those loci, it is well known that strong selection prefers a large amount of genetic variation at the haplotype level. In such a situation, haplotype variation can be enhanced by gene conversion especially when the gene conversion tract length is small.
In this article, we focus on the second outcome of gene conversion, that is, creating novel haplotypes. Our purpose is to elucidate the advantageous effect of gene conversion for loci under diversifying selection. We are especially interested in the effect of gene conversion tract length, as we predict that smaller conversion tracts can create new haplotypes more efficiently. We first consider a two-locus model in which a pair of duplicated loci is stably maintained in a population (model I). We also use a model that allows copy-number variation (model II): the first locus is fixed in the population, while the second locus can appear and disappear by mutations (duplication and loss). The second locus can be either functional or a pseudogene. In either case, we predict gene conversion between duplicates has an advantageous effect in terms of creating haplotype variation. Therefore, it might be expected that having a duplicate is advantageous for the population even when it is a pseudogene as long as gene conversion is active between them. Using these two models, we investigate the effect of gene conversion tract length on the following quantities: quantity (Q)1, haplotype variation maintained in the population in model I; Q2, the fixation probability of a duplicated gene in model II; and Q3, the longevity of a fixed duplicate in model II.
The first quantity, the number of haplotypes and haplotype diversity, has been investigated by Ohta (1991, 1997). She used a nine-locus model to make the situation similar to the human MHC genes. It is assumed that each gene consists of 50 infinite-allele sites and that the average gene conversion tract length is fixed to be half of the gene length (i.e., 25 sites). In this article, on the basis of our prediction, we use various tract lengths to investigate their effect, although we specify the models to be two-locus ones.
More importantly, the second and third quantities are investigated for addressing the questions on the maintenance mechanism of duplicated genes. Various scenarios for the preservation of duplicated genes have been proposed; i.e., one of the duplicated genes loses its function (pseudogenization or nonfunctionalization), one obtains a novel function (neofunctionalization), and both copies are preserved to complement the ancestral function (subfunctionalization) (for a review, see Walsh 2003). It is believed that the major fate of a duplicated gene is the first one: the extra duplicated copy is pseudogenized shortly after duplication and disappears from the genome. Here, we show that in a special occasion where the original donor gene is under diversifying selection, a duplicated copy can be preserved even after it is pseudogenized. This is based on our prediction that when a gene is under diversifying selection, having an extra copy would be advantageous because the duplicated copy could enhance the variation in the original gene through gene conversion. For this purpose, it should not matter whether the duplicate is a functional gene or a pseudogene. The new scenario is supported by our large amount of simulations under various conditions.
GENETIC VARIATION AND HAPLOTYPE STRUCTURE IN THE MHC AND SI LOCI
This section shows that gene conversion plays significant roles in shaping the standing haplotype variations in the MHC and SI genes, classic example genes of diversifying selection (e.g., Wright 1939; Takahata 1990). In both cases, very strong balancing selection is operating to maintain a number of haplotypes in a species. In the next section, we design simple two-locus models according to the observations introduced here.
MHC genes in primates:
The MHC is a large genomic region containing multiple MHC genes, which play the major role in the immune system of vertebrates. The proteins encoded by the MHC genes are involved in the immune response to various pathogens. The MHC genes are classified into two classes, the class I and class II MHCs. Both classes of MHC genes have a peptide-binding region (PBR) of ≈50 amino acids in the second exon for class II and in the second and third exons for class I, which recognizes nonself peptides. It has been considered that overdominant selection is operating in the PBR because individuals having various kinds of recognition specificity to outsider peptides would be selectively advantageous (Doherty and Zinkernagel 1975a). Therefore, those genes typically exhibit exceptionally high levels of nucleotide variation (Doherty and Zinkernagel 1975b; Klein 1986; Hughes and Nei 1988; Gaudieri et al. 2000).
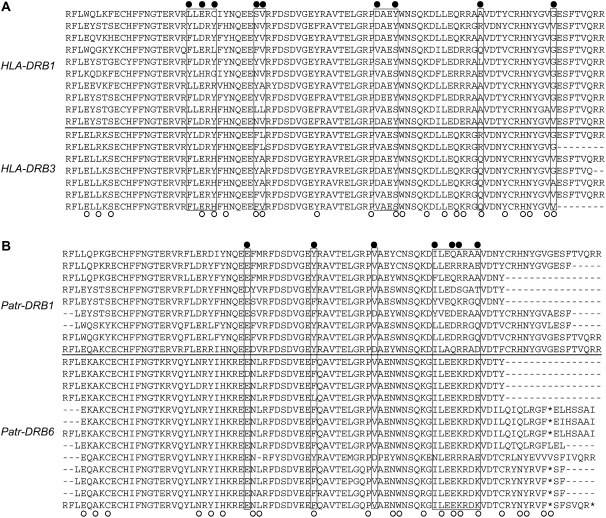
Human MHC genes are referred to as the HLA (human leukocyte antigen) genes. There are at least 9 protein-coding genes and 17 pseudogenes for the class I HLA, which are designated as HLA-A–HLA-Z. There are at least 15 coding genes and 9 pseudogenes for the class II HLA, denoted by, for example, HLA-DRA, HLA-DRB1, and HLA-DQA1 (Robinson et al. 2003; Horton et al. 2004). Several lines of evidence show that gene conversion occurs frequently between the class II HLA genes (Gorski and Mach 1986; Wu et al. 1986; Parham and Ohta 1996), although clear evidence is not available for the class I HLA loci (Hughes and Nei 1989; Nei et al. 1997). To demonstrate the point, HLA-DRB1 and -DRB3 are used as examples, which are those with particularly high levels of polymorphism in the HLA-DRB cluster. Figure 2A shows the multiple alignment of various haplotypes in the second exon of the two genes. It is shown that a number of polymorphic sites at the nucleotide level are shared by the two genes, indicating the role of interlocus gene conversion (Innan 2003). The observed number of such shared polymorphic sites is far beyond that explained by multiple mutations. There are a number of shared polymorphic sites among other HLA-DRB genes (not shown), indicating that gene conversion has been active in the HLA-DRB cluster, thereby contributing to the introduction of new types of PBR.
Figure 2.—
Amino acid sequence alignments of alleles in duplicated DRB genes in humans and chimpanzees. Data are from the IMGT/HLA and IMGT/MHC databases (Robinson et al. 2003). The open circles indicate amino acids involved in peptide-binding regions (PBRs) in humans (Brown et al. 1993). The solid circles represent shared polymorphic sites, which is a strong signature of gene conversion (Innan 2003). Putative gene conversion tracts are boxed. (A) DRB1 vs. DRB3 in humans. (B) DRB1 vs. DRB6 (pseudogene) in chimpanzees.
Very similar patterns are also observed in other primates (Go et al. 2003; Abbott et al. 2006; Doxiadis et al. 2006). Of particular interest is chimpanzee's ortholog of HLA-DRB6 (represented by Patr-DRB6), which has a very high level of variation, although it is a pseudogene. This gene has a number of shared polymorphic sites with other functional genes such as Patr-DRB1 (Figure 2B), indicating that gene conversion is heavily involved in shaping the observed pattern of polymorphism. A similar pattern also holds for macaque's DRB6 (data not shown), while the amount of variation is not very high in humans.
SI genes in plants:
SI is a mechanism for preventing self-fertilization in plants. SI is controlled by two tightly linked SI genes. The two SI genes encode the molecules for pistil- and pollen-side self recognitions. Since both genes have multiple alleles and recombination between the two genes is strongly suppressed, the specific term, “S haplotype,” is commonly used instead of the classical terminology, “S allele” (Nasrallah and Nasrallah 1993). The interaction between the pistil and pollen molecules encoded by the same S haplotypes prevents fertilization. Under this system, individuals with minor S haplotypes have high mate availabilities, and therefore a very large number of S haplotypes can be maintained in the population by frequency-dependent selection (Wright 1939).
In Brassica, the pistil- and pollen-side recognitions are controlled by two tightly linked genes, SRK (Stein et al. 1991; Takasaki et al. 2000) and SP11 (also named SCR) (Schopfer et al. 1999; Suzuki et al. 1999), respectively. The SRK gene possesses a hypervariable (HV) region of ≈100 amino acids, in which a number of nonsynonymous variations are likely observed. This HV region is considered to be involved in recognition specificity (Stein et al. 1991; Hinata et al. 1995).
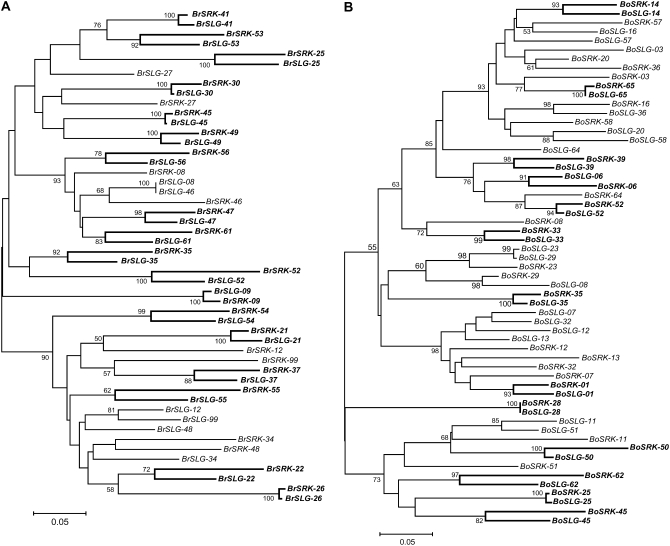
It is known that in Brassica SRK has a tightly linked paralog called SLG. SLG is a partial duplicate of SRK because it has only the 5′ half of the coding region (including the HV region) of SRK. While the expression of SLG has been confirmed, SLG may not be directly involved in the SI reaction (Suzuki et al. 2000). Therefore, we can consider that SLG is a nonfunctional gene in terms of the function of SI (it does not matter whether SLG has other functions), and we call it a pseudogene for simplicity. The situation could be similar to that of the DRB1–DRB6 (pseudogene) pair in chimpanzees (Figure 2B). The level of nucleotide variation in the HV region in the functional gene, SRK, is very high, and its putatively nonfunctional duplicated copy, SLG, also has a high level of variation in the corresponding part of the HV region perhaps through gene conversion (Sato et al. 2002). The action of interlocus gene conversion can be clearly demonstrated in the allelic trees in Figure 3. For two Brassica species (Brassica rapa and B. oleracea), we determined the sequences for several strains, and these data are pooled with publicly available data for the construction of the allelic trees (see supplemental material for details). S haplotypes, for which the sequences of both SRK and SLG are available, are used. It is found that for roughly half of S haplotypes, the two paralogs are most closely related to each other (emphasized by thick lines in Figure 3), which is a typical observation for gene pairs subject to gene conversion. The data also allow us to estimate the ratio of the gene conversion rate to the mutation rate, which turns out to be 30–40, indicating a high gene conversion rate in these regions. The estimation is based on two quantities: the average pairwise nucleotide differences within and between two loci, πw and πb (Innan 2002). πw = 0.1741 and πb = 0.1759 for B. rapa, and πw = 0.1665 and πb = 0.1677 for B. oleracea. The original estimation method (Innan 2002) is designed for a neutral case, but we believe we can apply the method to the SRK–SLG pair as a very special case. This is because allelic genealogy under multiallelic balancing selection can be roughly described by a neutral coalescent process when time is rescaled (Takahata 1990). This logic should hold for SRK and also for SLG when the two loci are completely linked.
Figure 3.—
Gene trees inferred from the HV regions of SRK and SLG in B. rapa (A) and B. oleracea (B). The OTUs represent the registered names of S haplotypes. When the two duplicates in the same S haplotype are most closely related, the branch between them is emphasized in a thick line. The bootstrap values in percentiles (>50%) are also shown. The bar represents genetic distance estimated by amino acid sequences.
A similar pattern has been also observed in Arabidopsis lyrata. The SRK gene in this species has multiple SLG-like paralogs, and evidence for gene conversion among them is available. Some paralogs would have complete gene structures and their expression has been empirically confirmed, but there may be no evidence that those paralogs are involved in the SI reaction and are subject to diversifying selection, like Brassica SLG (Charlesworth et al. 2003; Prigoda et al. 2005).
MODELS AND SIMULATION
On the basis of the observations in the MHC and SI genes, we design two-locus models as follows (summarized in Table 1). Two linked duplicated loci, A and B (Figure 1), are considered in a random-mating population with N diploids. As briefly mentioned above, model I assumes that the two loci are fixed in the population; that is, all chromosomes have both A and B. In model II, it is assumed that A is fixed, while presence/absence polymorphism is allowed for locus B. Each gene is represented by L bp of DNA sequences, corresponding to the PBR of the MHC genes and the HV region of SRK. At each site, four allelic states, 0, 1, 2, 3, are allowed, representing four nucleotides, “A,” “T,” “G,” and “C.” Codons (triplets of nucleotides) are assigned in the nucleotide sequences. For simplicity, we assume any mutation at the first and second positions causes amino acid changes, while it does not at the third position.
TABLE 1.
Summary of the models and results
| Model I: | Model II: | ||||
|---|---|---|---|---|---|
| Case | Representative | Locus A | Locus B | Haplotype variation | Fixation probability and longevity |
| I | MHC | Selected | Selected | K: Figure 4 | Figure 7 |
| H: supplemental Figure 1 | |||||
| II | MHC | Selected | Pseudogene | K: Figure 5a | Figure 8 |
| H: supplemental Figure 2 | |||||
| II′ | SI | Selected | Pseudogene | K: Figure 6a | Figure 9 |
| H: supplemental Figure 3 |
K, number of haplotypes; H, haplotype diversity.
Results of the simulations with deleterious mutations are shown in supplemental Figures 4 and 5.
To simulate the molecular evolution of the two loci, we incorporate point mutation, recombination between A and B, recombination within each gene, and gene conversion between A and B as mutational mechanisms. Point mutations are introduced with a fixed rate, where μ is mutation rate per site. Recombination between the two loci occurs at rate rb per diploid per generation, and in a similar way, intragenic (allelic) recombination within each gene is allowed at rate rw per base pair per generation. Note that intergenic recombination can occur in any individual, regardless of the presence of locus B. On the other hand, intragenic recombination in locus B occurs only in diploids, in which locus B is present in both chromosomes. Gene conversion occurs between the two loci at the same rate. We use a gene conversion model that assumes that the gene conversion tract length is a random variable from the geometric distribution with parameter q (Wiuf and Hein 2000; Teshima and Innan 2004). A gene conversion event can be initiated at any site in the duplicated region at rate g per generation, so that the gene conversion rate per site (c, the rate that a site experiences gene conversion defined in Innan 2002, 2003) is given by c = g/Q, where Q = qL. L/Q represents the average tract length of gene conversion. It should be noted that gene conversion can be initiated out of the simulated region, because our simulated region should be a part of the duplicated region. We assume that the duplicated regions are much larger than L bp. Table 2 summarizes the parameters used in this study.
TABLE 2.
Summary of parameters
| L | Number of nucleotides in the simulated region representing PBD and HV |
| θ | 4Nμ: population mutation parameter, where μ is the nucleotide mutation rate per site |
| Ns | Population selection parameter, where s is the selection coefficient |
| C | 4Nc: population gene conversion parameter, where c is the gene conversion rate per site |
| 1/Q | Expected tract length of gene conversion in units of L bp |
| Rb | 4Nrb: population recombination parameter, where rb is the interlocus recombination rate between duplicated genes |
| Rw | 4Nrw: population recombination parameter, where rw is the intragenic recombination rate per base pair |
| V | 4Nv: population deletion parameter, where v is the rate of deletion per gene |
In our simulations, point mutation, recombination, and gene conversion are introduced at each generation. Then, the fitness of each diploid individual is determined (see below for details), according to which the next generation is randomly generated under the Wright–Fisher model. Throughout this study, we fix N = 100 and θ (4Nμ) = 0.01 because computational time with a large N is very long. Nevertheless, our results hold for a large population, which was confirmed with limited parameter sets (not shown).
Model I:
Model I investigates the haplotype variation in the simulated region when the two loci are stably maintained in the population. The haplotype variation is considered at the amino acid level: in our simplified sequence model, a pair of sequences is considered as distinct haplotypes when they have at least one heterogeneous sites at the first and second positions in the codons. As a measure of haplotype variation, the number of haplotypes (K) in a population and the haplotype diversity (H, the probability that randomly chosen haplotypes are different) are calculated. Throughout this article, because the results for K and H are consistent, only the results for K are shown (see supplemental Figures 1–3 for the results for H).
We consider two cases: in case I, the two genes are functional, while locus B is assumed to be a pseudogene in case II. A pseudogene is defined such that it has no fitness effect on the host genome whether or not it is transcribed. Both cases can be applied to the MHC types of genes, while only case II can be applied to the SI types of genes. We first describe the simulation procedure for the MHC types of genes. Following the human MHC genes, L is assumed to be 150 bp, which is similar to the typical length of the PBR of the MHC genes. Let i be the number of different haplotypes in a diploid individual, which determines the fitness of the host individual. Note that only haplotypes in the active genes are counted, so that i = 1, 2, 3, 4 for case I and i = 1, 2 for case II. We assume that the selection effect is additive; that is, the fitness is given by
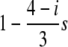 |
(1) |
in case I and
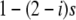 |
(2) |
in case II.
For each parameter set, a single run of simulation is performed. The initial state of the population is that the allelic state is 0 at all 2 × L sites for all chromosomes. A prerun of 5000N generations is carried out to accumulate variation. After the prerun, K and H are computed in every N generations. The simulation run is continued to accumulate 105 observations (i.e., 107 generations), from which the average K and H are obtained for each locus. The K and H of the two genes for case I are almost identical because of the symmetry of the model, while they are different in case II where selection works only on locus A. Therefore, we pool the data from the two loci in case I, while in case II we investigate the levels of variation in loci A and B separately.
Only case II may be applied to the SI types of genes and this special case is called case II′. There are some parameters specific to case II′. According to the Brassica SRK genes, L = 300 bp is assumed. Because homozygotes are usually lethal, s = 1 is assumed. Because the recombination between S haplotypes is strongly suppressed (reviewed in Uyenoyama 2005), Rb = Rw = 0 is assumed. Brassica species employ a sporophytic self-incompatibility (SSI) system, in which both the pistil- and pollen-side recognition specificities are controlled by parental diploid genotypes (Takayama and Isogai 2005). In other words, because all individuals have two S haplotypes as heterozygotes, both pistil and pollen exhibit two recognition specificities. Our simulation model follows the SSIcod model of Vekemans et al. (1998). In practice, two individuals are randomly chosen in the population of the previous generation, representing female (pistil) and male (pollen) parents. If the two parents share at least one haplotype in locus A, these two parents are discarded, and another pair of parents is randomly chosen again. If the male and female parents do not share any haplotype, a new progeny is generated. This procedure is repeated until N progenies are obtained. Note that our model assumes complete codominance of the S haplotypes. The effect of dominance among the S haplotypes is ignored for simplicity. Although its possibility has been pointed out, the effect on our conclusion may not be large because our primary interest is in gene conversion. Dominance would work to reduce the effect of selection (Schierup et al. 1997; Vekemans et al. 1998).
Model II:
In model II, presence/absence polymorphism of the locus B is allowed, while all 2N chromosomes have locus A. The model can be applied to both MHC and SI types of genes. The model is to investigate the fixation probability and longevity of the duplicated gene, locus B. To evaluate the fixation probability, a simulation starts with the initial condition where no chromosome in the population has locus B. After a prerun (200N generations) for accumulating variation in locus A, a single chromosome is randomly selected, where a duplicate of locus A arises as locus B. Accordingly, this chromosome has two copies of exactly the same sequences, and the other 2N – 1 have only locus A. After this duplication event, no duplication or deletion event is incorporated, so that we are able to trace the fate of this duplicate: fixation or extinction. From at least 5 × 105 replications of this simulation, we obtain the fixation probability of locus B.
Similarly we investigate how long a fixed locus B can be maintained in the population when it is subject to deletion. The system starts with the state where locus B is fixed in the population. The simulation includes a prerun (20,000N generations), the procedure of which is the same as that of model I (no deletion occurs in the prerun). After the prerun ends at time t = 0, locus B is subject to deletion at rate V. Then, the simulation continues until locus B becomes extinct from the population. From a number of replications of this simulation (105), the average time from t = 0 to extinction is computed.
RESULTS
Haplotype variation:
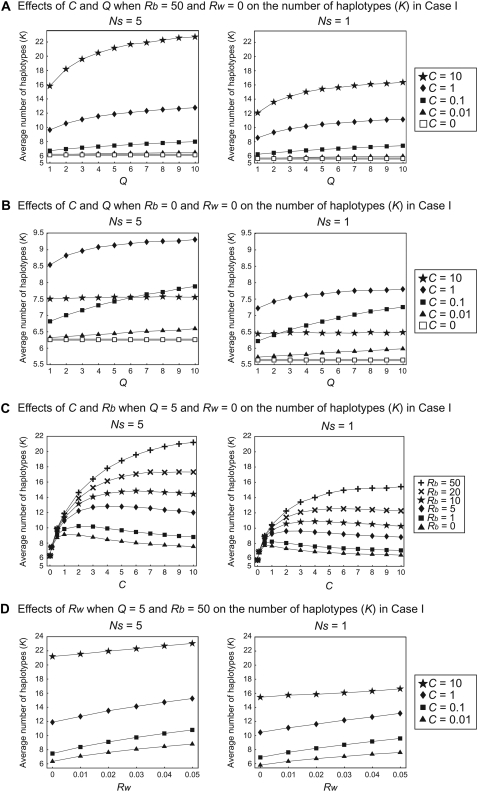
We first focus on the haplotype variation in case I in model I. In Figure 4A, the average number of haplotypes per gene, K, is plotted against Q, the parameter to determine the size of the gene conversion tract: the expected length is long for a small Q. Because the two loci are functional in this model, K in Figure 4A is the average over the two loci. Two selection intensities (Ns = 5 and 1) are used, and Rb = 50 and Rw = 0 are assumed. Four values of the gene conversion rate are considered (C = {0.01, 0.1, 1, 10}) and C = 0 is also included as a control. It is clearly demonstrated that Q and the average number of haplotypes are positively correlated for all gene conversion rates, supporting our hypothesis of high efficacy of generating novel haplotypes when the conversion tract is small. This positive correlation is also observed when Rb = 0 (Figure 4B).
Figure 4.—
Results of simulation of case I for the number of haplotypes, K. (A) The effects of C and Q on K when Rb = 50 and Rw = 0. (B) The effects of C and Q on K when Rb = 0 and Rw = 0. (C) The effects of C and Rb on K when Q = 5 and Rw = 0. (D) The effects of Rw on K when Q = 5 and Rb = 50.
Overall, the average number of haplotypes is larger for Ns = 5 than for Ns = 1 (Figure 4, A and B), indicating that diversifying selection at the two loci works to favor a high level of haplotype diversity, in agreement with classic theory under a single-locus model (Takahata 1990). Recombination is helpful in terms of shuffling the variation in the two loci. K is positively correlated with the recombination rates between the two loci (Figure 4C) and within each locus (Figure 4D), although the effect of the latter is weak.
The effect of the gene conversion rate on the number of haplotypes is not simple as demonstrated in Figure 4C. When C is relatively small, K and C are positively correlated, but they are in a negative correlation if C exceeds a certain threshold. It seems that there is an optimum C in terms of enhancing haplotype variation. The optimum C should depend on other parameters, among which the recombination rate between the two loci might be of special importance: the threshold is high for a large Rb. When C is large, the effect of gene conversion on homogenizing haplotypes between the two loci might be too large, so that the effect of creating new haplotypes may be canceled out. However, even with a very high C, K is larger than that expected with no gene conversion (open rectangles with shaded lines in Figure 4, A and B).
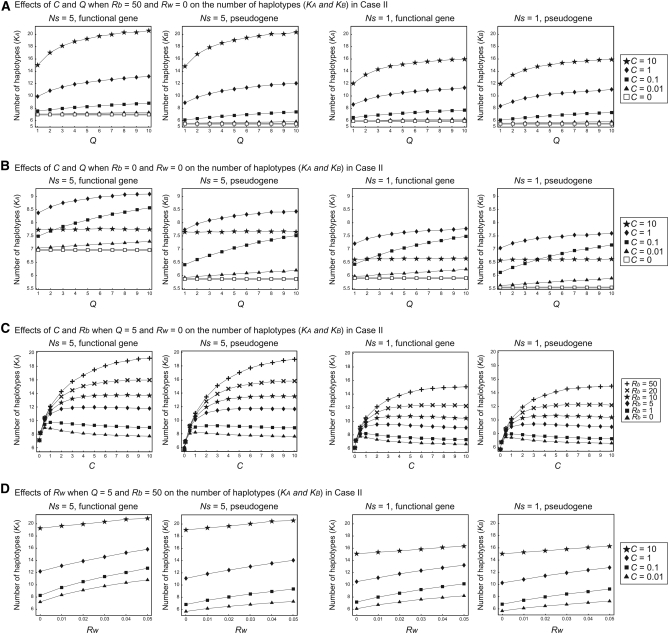
The results in case II (Figure 5) are essentially identical to those in case I (Figure 4), except that the average numbers of haplotypes for the two loci (represented by KA and KB) are different: because of no selection at locus B, the expectation of KB is smaller than that of KA. Nevertheless, KB is much larger than that expected without gene conversion (C = 0) because high variability in locus A is transferred to locus B. KA and KB become similar as C increases. The overall patterns observed in case I hold in Figure 5: KA and KB are positively correlated with Ns, Q, Rb, and Rw. Again we observe that there is an optimum C to maximize the effect of creating new haplotypes. The optimum C is large for a large Rb.
Figure 5.—
Results of simulation of case II for the number of haplotypes, K. (A) The effects of C and Q on K when Rb = 50 and Rw = 0. (B) The effects of C and Q on K when Rb = 0 and Rw = 0. (C) The effects of C and Rb on K when Q = 5 and Rw = 0. (D) The effects of Rw on K when Q = 5 and Rb = 50.
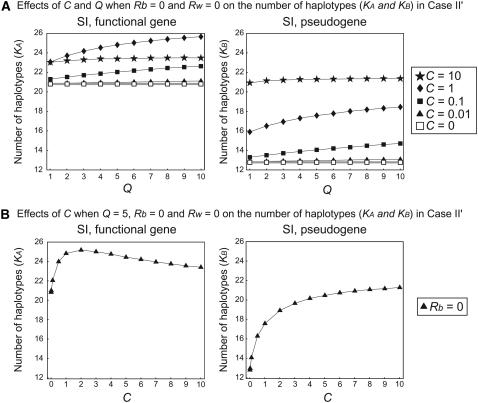
As a special case of case II, we consider case II′ (Figure 6). The pattern is very similar to those observed in Figures 4 and 5, although the effects of Rb, Rw, and Ns are not investigated in case II′ because we use fixed values for those (Rb = Rw = 0 and s = 1).
Figure 6.—
Results of simulation of case II′ for the number of haplotypes, K. (A) The effects of C and Q on K when Rb = 0 and Rw = 0. (B) The effects of C on K when Q = 5, Rb = 0, and Rw = 0.
Thus, it can be summarized that there is an overall advantage of interlocus gene conversion in creating haplotype variation in loci under diversifying selection. Its quantitative effects are as follows:
The effect of gene conversion is large when the gene conversion tract is small (large Q).
There is an optimum gene conversion rate (C).
The effect of gene conversion is enhanced by intra- and interlocus recombination.
These quantitative effects also hold for the haplotype diversity, H, as shown in supplemental Figures 1–3.
Deleterious mutations including nonsense mutations are important factors in case II and case II′, but not incorporated in the above simulations. Such deleterious mutations should be quickly eliminated in case I so that their evolutionary contribution is negligible. However, the effect cannot be ignored in case II and case II′ because it is likely that premature stop codons and deleterious amino acids can be accumulated in locus B (pseudogene), where there is no purifying selection. When deleterious mutations in locus B are transferred by gene conversion, it has a significant impact on fitness. It is known that a number of gene conversion-mediated human diseases involve nonsense mutations in pseudogenes (Chen et al. 2007). In the Brassica SI system, it has been reported that gene conversion from SLG to SRK could cause nonfunctionalization of SRK (Fujimoto et al. 2006). Additional simulations with deleterious mutations are also performed, and we obtain almost identical results (supplemental Figures 4 and 5). In all parameter sets investigated, we confirm the above three quantitative effects of gene conversion. We observe a slight reduction in K and H as the effect of deleterious mutations. This can be understood if we consider that deleterious mutations in locus B work to reduce the effective gene conversion rate from B to A because of its deleterious effect (i.e., such deleterious gene conversion would be quickly eliminated from the population). Note that it is obvious that gene conversion in the other direction is neutral.
Fixation probability and longevity:
It can be easily imagined that when the duplicated gene (locus B) is functional, there is a direct advantage of it, which offers a potential source for having more haplotypes for the host individual (Figure 4). More interestingly, we show that even when locus B is nonfunctional, its existence can help to introduce new haplotypes in locus A (Figures 5 and 6). In this section, to investigate the evolutionary significance of the existence of locus B, we investigate the second and third quantities listed in the Introduction (i.e., fixation probability and longevity) by using model II. To demonstrate the point, intragenic recombination and deleterious mutations are ignored because they have minor effects (data not shown).
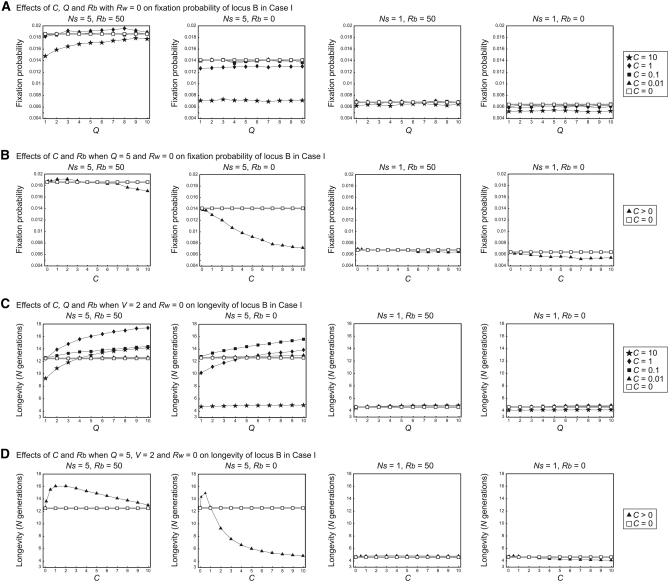
We first focus on case I, where both loci are functional. As mentioned above, there is a direct advantage of having the extra copy (i.e., locus B), which is seen as elevated fixation probabilities in comparison with the case of no selection (i.e., 1/2N= 0.005 in Figure 7, A and B). It seems that there is almost no positive effect of gene conversion to enhance the fixation of locus B (Figure 7, A and B). The effect is rather negative: the fixation probability in the cases of C > 0 is overall lower than that of no gene conversion (C = 0, open rectangles with shaded lines in Figure 7). This could be because the effect of gene conversion is positive only when locus B accumulates a high level of variation; otherwise the other negative outcome of homogenizing variation may dominate the positive one. Usually, the fixation process of a new duplicate occurs in a relatively short time (much shorter than the time required to accumulate a high level of polymorphism). In this model, because the fixation of locus B is already advantageous, the negative effect of gene conversion may be emphasized.
Figure 7.—
Results of simulation of case I for fixation probability and longevity of locus B. (A) The effects of C, Q, and Rb on fixation probability when Rw = 0. (B) The effects of C and Rb on fixation probability when Q = 5 and Rw = 0. (C) The effects of C, Q, and Rb on longevity when Rw = 0. (D) The effects of C and Rb on longevity when Q = 5 and Rw = 0.
In Figure 7, C and D, the longevity of locus B is investigated. Overall, the effect of gene conversion is not very negative unless C is very large. For some parameter sets, we see a slight positive effect of gene conversion to preserve locus B, probably because the process requires a much longer time than that for the fixation process.
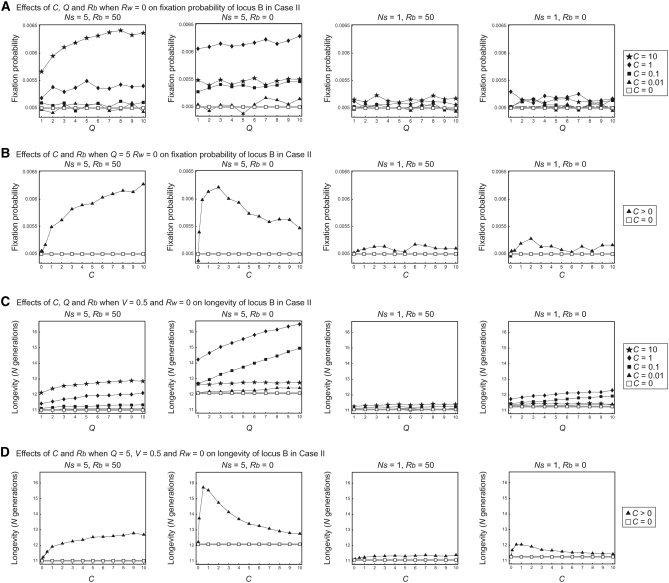
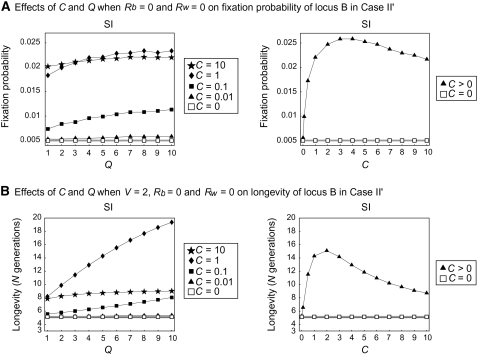
In contrast, the situation is quite different in case II, in which locus B is nonfunctional (Figure 8). If there is no gene conversion (open rectangles with shaded lines in Figure 8), there is no evolutionary advantage for having locus B; therefore, the fixation probability is 1/2N and the longevity depends on the deletion rate, V. In Figure 8, we observe the effect of gene conversion in the positive direction for preserving locus B. The fixation probability and longevity for C > 0 are larger than those for no gene conversion (Figure 8). The quantitative effects of gene conversion on these two quantities are in agreement with the first and second points in the previous section, Haplotype variation (although the relationship with the recombination rate may be complicated). The fixation probability and longevity are positively correlated with Q and Ns, and there seems to be an optimum C to maximize the fixation probability and longevity. This pattern also holds for case II′ (Figure 9).
Figure 8.—
Results of simulation of case II for fixation probability and longevity of locus B. (A) The effects of C, Q, and Rb on fixation probability when Rw = 0. (B) The effects of C and Rb on fixation probability when Q = 5 and Rw = 0. (C) The effects of C, Q, and Rb on longevity when Rw = 0. (D) The effects of C and Rb on longevity when Q = 5 and Rw = 0.
Figure 9.—
Results of simulation of case II′ for fixation probability and longevity of locus B. (A) The effects of C and Q on the fixation probability. (B) The effects of C and Q on longevity.
DISCUSSION
Simple two-locus models with interlocus gene conversion are used to investigate the effect of gene conversion on the haplotype variation in the two loci (model I) and the fixation probability and longevity of the second locus (model II). The models are designed such that we can investigate those three quantities in situations similar to the PBR in the MHC genes and the HV region in SI genes. For the former (MHC), we consider two situations: (1) both of the two loci are functional and (2) the first locus is functional and the second is nonfunctional, while for the latter (SI), it is automatically assumed that the second locus is a pseudogene because only one copy is usually involved in the SI system.
Ohta (1991, 1997) used similar models to our model I and demonstrated that gene conversion works to increase the number of haplotypes. Our results confirmed her conclusion (Figures 4 and 5). Her models assume that the average gene conversion tract length is fixed to be a half of the gene length. Here, we extended the models, in which the tract length follows a geometric distribution with parameter Q. This is because we predict that Q is an important factor to determine the efficacy of gene conversion on creating new haplotypes. We find that Q and the number of haplotypes, K, are in a positive correlation for the wide range of parameters investigated in cases I, II, and II′ (Figures 4–6), supporting our prediction.
Other parameters also affect K, including mutation, recombination, gene conversion rates, and selection intensity (Figures 4–6). Although the population mutation rate θ is fixed through this study, it is easy to imagine that θ and K should be in a positive correlation. For the selection coefficient, only two representative values are used (Ns = 1 and 5). It is found that K is always larger for Ns = 5, indicating a positive effect of Ns on K, which is expected from classic theoretical results in a single-locus model (Takahata 1990). The inter- and intralocus recombination rates (Rb and Rw) are also positively correlated with K. The former works to provide new pairs for gene conversion as theoretically demonstrated (Innan 2002, 2003), while the latter directly contributes to the introduction of new haplotypes (Takahata and Satta 1998). The relationship between C and K is not simple. There seems to be an optimum gene conversion rate to maximize its effect.
The fixation probability and longevity are also positively correlated with Q, indicating that short gene conversion tracts work efficiently to create new haplotypes when the gene conversion rate (C) is the same. Similar to the effect on the haplotype diversity, there seems to be an optimum C to maximize the fixation probability and longevity (Figures 7–9).
It is interesting that this positive effect of gene conversion in creating new haplotypes is observed when one of the two loci is a pseudogene (Figures 5 and 6), suggesting a possibility that the original functional gene under diversifying selection can be benefited by having an extra copy even when it is a pseudogene. As a consequence, a pseudogene created by duplication can be preserved by gene conversion and diversifying selection. We use model II to quantitatively evaluate this possibility. It is found that selection operating at locus A enhances the fixation probability and longevity of its nonfunctional duplicate, locus B (Figures 8 and 9). Q and Ns are positively correlated with the fixation probability and longevity, while there is an optimum C to maximize an advantageous effect. It is important to note that the fixation probability and longevity for C > 0 are always larger than those for C = 0.
This result can be contrasted with previous theoretical studies on the fates of duplicated genes. It is predicted that pseudogenization is the most likely fate of duplicated genes (Li 1980; Ohta 1987; Lynch and Conery 2000; Walsh 2003). It is usually considered that there is no reason for the genome to keep pseudogenes; subsequently pseudogenes should be degenerated and disappear relatively quickly. Our simulations demonstrate that when diversifying selection is working in the donor gene, there is a positive effect of having a duplicate on the host genome, even after it is pseudogenized.
It is usually considered that having a pseudogenized duplicate is deleterious when gene conversion occurs between them. This is because gene conversion from the pseudogene to the functional gene could transfer deleterious mutations accumulated in the pseudogene (Chen et al. 2007). A number of gene conversion-mediated human genetic diseases involve such events. However, when the donor gene is under diversifying selection, our simulations suggest that the effect of gene conversion might dominate the negative effect (supplemental Figures 4 and 5).
These findings should be consistent with the real sequence data in duplicated genes under the effect of diversifying selection and gene conversion. As shown in Figures 2 and 3, the polymorphism data in the MHC and SI genes show a number of clear footprints of gene conversion between pseudogenes and their functional donors. Our results would explain why there are a number of pseudogenized duplicated genes in the MHC cluster.
Although we focused on the two very typical cases (the MHC and SI genes) throughout the article, our conclusion can be applied to any gene under diversifying selection (i.e., multiallelic overdominant selection and frequency-dependent selection). Examples include plants' disease-resistance genes (R genes). Similar to the MHC genes, the R multigenes usually form a cluster including many pseudogenes and gene conversion among them has been implicated (Parniske et al. 1997). Various surface antigen genes in the trypanosome could be another example, in which antigenic variation is considered to be generated via gene conversion from pseudogenes (Roth et al. 1989) and a number of pseudogenes (presumably >500) are retained in its genome (Berriman et al. 2005).
Furthermore, although our models cannot be directly applied, the results could explain the advantage of gene conversion in somatic cell divisions in the immunoglobulin genes (Miyata et al. 1980; Baltimore 1981; Ohta 1992). Immunoglobulin proteins are antibodies specific to foreign molecules. In general, the diversities of immunoglobulin heavy and light chains are generated by somatic recombination among a large number of segments (i.e., the V, D, and J segments) during the B cell maturation. This editing process creates high levels of variation in the antigen-binding domain of the immunoglobulin protein. In some species including chickens and rabbits, it is reported that pseudogenized duplicates are involved in this process probably through gene conversion (e.g., Reynaud et al. 1987; Thompson and Neiman 1987; Becker and Knight 1990; Roux et al. 1991). It is suggested that pseudogenes play an important role to generate somatic diversity and that selection might be operating to maintain those pseudogenes.
Acknowledgments
We appreciate Tomoko Ohta, Kosuke M. Teshima, Tomoyuki Kado, members of the Laboratory of Plant Breeding and Genetics in Tohoku University, and two anonymous reviewers for their various comments. S.T. is a Japan Society for the Promotion of Science (JSPS) postdoctral fellow. H.I. is supported by the JSPS, the National Science Foundation (USA), and an internal grant from the Graduate University for Advanced Studies.
References
- Abbott, K. M., E. J. Wickings and L. A. Knapp, 2006. High levels of diversity characterize mandrill (Mandrillus sphinx) MHC-DRB sequences. Immunogenetics 58 628–640. [DOI] [PubMed] [Google Scholar]
- Baltimore, D., 1981. Gene conversion: some implications for immunoglobulin genes. Cell 24 592–594. [DOI] [PubMed] [Google Scholar]
- Becker, R. S., and K. L. Knight, 1990. Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell 63 987–997. [DOI] [PubMed] [Google Scholar]
- Berriman, M., E. Ghedin, C. Hertz-Fowler, G. Blandin, H. Renauld et al., 2005. The genome of the african trypanosome Trypanosoma brucei. Science 309 416–422. [DOI] [PubMed] [Google Scholar]
- Brown, D. D., P. C. Wensink and E. Jordan, 1972. A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J. Mol. Biol. 63 57–73. [DOI] [PubMed] [Google Scholar]
- Brown, J. H., T. S. Jardetzky, J. C. Gorga, L. J. Stern, R. G. Urban et al., 1993. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature 364 33–39. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., B. K. Mable, M. H. Schierup, C. Bartolomé and P. Awadalla, 2003. Diversity and linkage of genes in the self-incompatibility gene family in Arabidopsis lyrata. Genetics 164 1519–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. M., D. N. Cooper, N. Chuzhanova, C. Férec and G. P. Patrinos, 2007. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8 762–775. [DOI] [PubMed] [Google Scholar]
- Coen, E. S., J. M. Thoday and G. Dover, 1982. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature 295 564–568. [DOI] [PubMed] [Google Scholar]
- Doherty, P. C., and R. M. Zinkernagel, 1975. a A biological role for the major histocompatibility antigens. Lancet 1 1406–1409. [DOI] [PubMed] [Google Scholar]
- Doherty, P. C., and R. M. Zinkernagel, 1975. b Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature 256 50–52. [DOI] [PubMed] [Google Scholar]
- Dover, G., 1982. Molecular drive: a cohesive mode of species evolution. Nature 299 111–117. [DOI] [PubMed] [Google Scholar]
- Doxiadis, G. G. M., M. K. H. van der Wiel, H. P. M. Brok, N. G. de Groot, N. Otting et al., 2006. Reactivation by exon shuffling of a conserved HLA-DR3-like pseudogene segment in a New World primate species. Proc. Natl. Acad. Sci. USA 103 5864–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezawa, K., S. O. Ota and N. Saitou, 2006. Genome-wide search of gene conversions in duplicated genes of mouse and rat. Mol. Biol. Evol. 23 927–940. [DOI] [PubMed] [Google Scholar]
- Fujimoto, R., T. Sugimura and T. Nishio, 2006. Gene conversion from SLG to SRK resulting in self-compatibility in Brassica rapa. FEBS Lett. 580 425–430. [DOI] [PubMed] [Google Scholar]
- Gao, L. Z., and H. Innan, 2004. Very low gene duplication rate in the yeast genome. Science 306 1367–1370. [DOI] [PubMed] [Google Scholar]
- Gaudieri, S., R. L. Dawkins, K. Habara, J. K. Kulski and T. Gojobori, 2000. SNP profile within the human major histocompatibility complex reveals an extreme and interrupted level of nucleotide diversity. Genome Res. 10 1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go, Y., Y. Satta, Y. Kawamoto, G. Rakotoarisoa, A. Randrianjafy et al., 2003. Frequent segmental sequence exchanges and rapid gene duplication characterize the MHC class I genes in lemurs. Immunogenetics 55 450–461. [DOI] [PubMed] [Google Scholar]
- Gorski, J., and B. Mach, 1986. Polymorphism of human Ia antigens: gene conversion between two DR β loci results in a new HLA-D/DR specificity. Nature 322 67–70. [DOI] [PubMed] [Google Scholar]
- Hinata, K., M. Watanabe, S. Yamakawa, Y. Satta and A. Isogai, 1995. Evolutionary aspects of the S-related genes of the Brassica self-incompatibility system: synonymous and nonsynonymous base substitutions. Genetics 140 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, R., L. Wilming, V. Rand, R. C. Lovering, E. A. Bruford et al., 2004. Gene map of the extended human MHC. Nat. Rev. Genet. 5 889–899. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., and M. Nei, 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335 167–170. [DOI] [PubMed] [Google Scholar]
- Hughes, A. L., and M. Nei, 1989. Ancient interlocus exon exchange in the history of the HLA-A locus. Genetics 122 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., 2002. A method for estimating the mutation, gene conversion and recombination parameters in small multigene families. Genetics 161 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., 2003. The coalescent and infinite-site model of a small multigene family. Genetics 163 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, J., 1986. Natural History of the Major Histocompatibility Complex. John Wiley & Sons, New York.
- Li, W. H., 1980. Rate of gene silencing at duplicate loci: a theoretical study and interpretation of data from tetraploid fishes. Genetics 95 237–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. H., 1997. Molecular Evolution. Sinauer Associates, Sunderland, MA.
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290 1151–1155. [DOI] [PubMed] [Google Scholar]
- Martinsohn, J. T., A. B. Sousa, L. A. Guethlein and J. C. Howard, 1999. The gene conversion hypothesis of MHC evolution: a review. Immunogenetics 50 168–200. [DOI] [PubMed] [Google Scholar]
- Miyata, T., T. Yasunaga, Y. Yamawaki-Kataoka, M. Obata and T. Honjo, 1980. Nucleotide sequence divergence of mouse immunoglobulin γ1 and γ2b chain genes and the hypothesis of intervening sequence-mediated domain transfer. Proc. Natl. Acad. Sci. USA 77 2143–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki, T., 1983. Evolution of a finite population under gene conversion. Proc. Natl. Acad. Sci. USA 80 6278–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagylaki, T., and T. D. Petes, 1982. Intrachromosomal gene conversion and the maintenance of sequence homogeneity among repeated genes. Genetics 100 315–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah, J. B., and M. E. Nasrallah, 1993. Pollen-stigma signaling in the sporophytic self-incompatibility response. Plant Cell 5 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., X. Gu and T. Sitnikova, 1997. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 94 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T., 1980. Evolution and Variation of Multigene Families. Springer-Verlag, New York.
- Ohta, T., 1982. Allelic and nonallelic homology of a supergene family. Proc. Natl. Acad. Sci. USA 79 3251–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T., 1987. Simulating evolution by gene duplication. Genetics 115 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T., 1991. Role of diversifying selection and gene conversion in evolution of major histocompatibility complex loci. Proc. Natl. Acad. Sci. USA 88 6716–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, T., 1992. A statistical examination of hypervariability in complementarity-determining regions of immunoglobulins. Mol. Phylogenet. Evol. 1 305–311. [DOI] [PubMed] [Google Scholar]
- Ohta, T., 1997. Role of gene conversion in generating polymorphisms at major histocompatibility complex loci. Hereditas 127 97–103. [DOI] [PubMed] [Google Scholar]
- Parham, P., and T. Ohta, 1996. Population biology of antigen presentation by MHC class I molecules. Science 272 67–74. [DOI] [PubMed] [Google Scholar]
- Parniske, M., K. E. Hammond-Kosack, C. Golstein, C. M. Thomas, D. A. Jones et al., 1997. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91 821–832. [DOI] [PubMed] [Google Scholar]
- Prigoda, N. L., A. Nassuth and B. K. Mable, 2005. Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol. Biol. Evol. 22 1609–1620. [DOI] [PubMed] [Google Scholar]
- Reynaud, C. A., V. Anquez, H. Grimal and J. C. Weill, 1987. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell 48 379–388. [DOI] [PubMed] [Google Scholar]
- Robinson, J., M. J. Waller, P. Parham, N. de Groot, R. Bontrop et al., 2003. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 31 311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, C., F. Bringaud, R. E. Layden, T. Baltz and H. Eisen, 1989. Active late-appearing variable surface antigen genes in Trypanosoma equiperdum are constructed entirely from pseudogenes. Proc. Natl. Acad. Sci. USA 86 9375–9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, K. H., P. Dhanarajan, V. Gottschalk, W. T. McCormick and R. W. Renshaw, 1991. Latent a1 VH germline genes in an a2a2 rabbit. Evidence for gene conversion at both the germline and somatic levels. J. Immunol. 146 2027–2036. [PubMed] [Google Scholar]
- Sato, K., T. Nishio, R. Kimura, M. Kusaba, T. Suzuki et al., 2002. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics 162 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., X. Vekemans and F. B. Christiansen, 1997. Evolutionary dynamics of sporophytic self-incompatibility alleles in plants. Genetics 147 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer, C. R., M. E. Nasrallah and J. B. Nasrallah, 1999. The male determinant of self-incompatibility in Brassica. Science 286 1697–1700. [DOI] [PubMed] [Google Scholar]
- Semple, C., and K. H. Wolfe, 1999. Gene duplication and gene conversion in the Caenorhabditis elegans genome. J. Mol. Evol. 48 555–564. [DOI] [PubMed] [Google Scholar]
- Stein, J. C., B. Howlett, D. C. Boyes, M. E. Nasrallah and J. B. Nasrallah, 1991. Molecular cloning of a putative receptor protein kinase gene encoded at the self-incompatibility locus of Brassica oleracea. Proc. Natl. Acad. Sci. USA 88 8816–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, G., N. Kai, T. Hirose, K. Fukui, T. Nishio et al., 1999. Genomic organization of the s locus: identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics 153 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., M. Kusaba, M. Matsushita, K. Okazaki and T. Nishio, 2000. Characterization of Brassica S-haplotypes lacking S-locus glycoprotein. FEBS Lett. 482 102–108. [DOI] [PubMed] [Google Scholar]
- Takahata, N., 1990. A simple genealogical structure of strongly balanced allelic lines and trans-species evolution of polymorphism. Proc. Natl. Acad. Sci. USA 87 2419–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., and Y. Satta, 1998. Footprints of intragenic recombination at HLA loci. Immunogenetics 47 430–441. [DOI] [PubMed] [Google Scholar]
- Takasaki, T., K. Hatakeyama, G. Suzuki, M. Watanabe, A. Isogai et al., 2000. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403 913–916. [DOI] [PubMed] [Google Scholar]
- Takayama, S., and A. Isogai, 2005. Self-incompatibility in plants. Annu. Rev. Plant Biol. 56 467–489. [DOI] [PubMed] [Google Scholar]
- Teshima, K. M., and H. Innan, 2004. The effect of gene conversion on the divergence between duplicated genes. Genetics 166 1553–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, C. B., and P. E. Neiman, 1987. Somatic diversification of the chicken immunoglobulin light chain gene is limited to the rearranged variable gene segment. Cell 48 369–378. [DOI] [PubMed] [Google Scholar]
- Uyenoyama, M. K., 2005. Evolution under tight linkage to mating type. New Phytol. 165 63–70. [DOI] [PubMed] [Google Scholar]
- Vekemans, X., M. H. Schierup and F. B. Christiansen, 1998. Mate availability and fecundity selection in multi-allelic self-incompatibility systems in plants. Evolution 52 19–29. [DOI] [PubMed] [Google Scholar]
- Walsh, B., 2003. Population-genetic models of the fates of duplicate genes. Genetica 118 279–294. [PubMed] [Google Scholar]
- Wang, X., H. Tang, J. E. Bowers, F. A. Feltus and A. H. Paterson, 2007. Extensive concerted evolution of rice paralogs and the road to regaining independence. Genetics 177 1753–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf, C., and J. Hein, 2000. The coalescent with gene conversion. Genetics 155 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1939. The distribution of self-sterility alleles in populations. Genetics 24 538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., T. L. Saunders and F. H. Bach, 1986. Polymorphism of human Ia antigens generated by reciprocal intergenic exchange between two DR β loci. Nature 324 676–679. [DOI] [PubMed] [Google Scholar]
- Zimmer, E. A., S. L. Martin, S. M. Beverley, Y. W. Kan and A. C. Wilson, 1980. Rapid duplication and loss of genes coding for the α chains of hemoglobin. Proc. Natl. Acad. Sci. USA 77 2158–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]