Abstract
The genetic basis of prion disease incubation time is principally determined by polymorphisms in the prion protein gene, Prnp. However, it is now known that other genetic factors are important. Several quantitative trait loci (QTL) have been identified across the genome including a broad region of linkage on Mmu11. Monocyte chemoattractant protein 1 (MCP-1) maps to this region and has been associated with microglial activation and reduced survival in the ME7 mouse scrapie model of prion disease. We have identified 10 polymorphisms, 3 of which are nonsynonomous, in Mcp1 between “long” (CAST) and “short” (SJL or NZW) incubation-time mouse strains. Crosses between these strains and Mcp1−/− mice inoculated with the Chandler/RML mouse scrapie prion strain formed the basis of a quantitative complementation test. In these models loss of Mcp1 did not show an increase in incubation time suggesting that the effects of Mcp1 may be specific to the ME7 prion strain and that Mcp1 does not contribute to the QTL described on Mmu11.
PRION diseases or transmissible spongiform encephalopathies are fatal neurodegenerative disorders of humans and animals that include Creutzfeldt–Jakob disease (CJD), bovine spongiform encephalopathy (BSE), and scrapie (Collinge 2001). They are characterized by prolonged incubation periods, deposition of an abnormal form of the prion protein (PrPSc), and histologically by vacuolation (spongiosis) of the neuropil, gliosis, and neuronal loss.
The main genetic determinant of incubation time in mouse is variation in the prion protein gene, Prnp (Westaway et al. 1987; Carlson et al. 1988; Carlson et al. 1993; Moore et al. 1998); however, quantitative trait locus (QTL) mapping studies have successfully identified multiple loci across the genome that influence incubation time (Stephenson et al. 2000; Lloyd et al. 2001, 2002; Manolakou et al. 2001; Moreno et al. 2003).
Using the Chandler/RML mouse-adapted scrapie prion strain two independent studies identified QTL on chromosome 11 (Stephenson et al. 2000; Lloyd et al. 2001). Both studies mapped broad and overlapping regions of linkage that may contain multiple QTL. Stephenson et al. (2000) used a CAST/Ei and SJL/J F2 mouse intercross with the peak of linkage at D11Mit219 while a larger F2 intercross with CAST/Ei and NZW/OlaHsd found the peak of linkage near D11Mit36 (Lloyd et al. 2001). The extent of the overlap between these two regions suggests that they may share at least some QTL especially as CAST/Ei was used in both cases. Identifying individual candidate genes within these large regions is especially challenging. However it has been reported that monocyte chemoattractant protein-1 (MCP-1), which maps within this region of chromosome 11, plays a role in the onset of late-stage clinical signs of prion disease (Felton et al. 2005).
MCP-1 belongs to the CC family of chemokines and is thought to have a pro-inflammatory role within the CNS recruiting monocytes and activating resident microglia (Gu et al. 1997, 1999). Prion diseases display an atypical inflammatory response dominated by microglial activation suggesting a role for MCP-1 in this process (Perry et al. 2002). Although the inflammatory response is an early occurrence in disease pathogenesis (Giese et al. 1998), preceding neuronal death and the onset of severe clinical signs by several weeks, Felton et al. (2005) showed that late-stage clinical signs were delayed by 4 weeks and survival time increased by 2–3 weeks in Mcp1−/− mice inoculated intracerebrally with the ME7 prion strain. The onset of early behavioral changes was not delayed in the knockout mice as compared to wild type; neither were there any differences in microglial activation or neuronal death. It is suggested that MCP-1 is not necessarily required for microglial priming but may stimulate their further activation thus exacerbating neuronal damage (Felton et al. 2005).
To assess whether Mcp1 is a potential quantitative trait gene (QTG) for prion disease incubation time we identified polymorphisms between our “long” (CAST/Ei) and “short” (NZW/OlaHsd and SJL/J) incubation-time mouse strains. Mcp1−/− mice were also used in a quantitative complementation test to ascertain whether Mcp1 is a QTG or influences the phenotype independently of the QTL (Flint and Mott 2001).
MATERIALS AND METHODS
Mice:
Mcp1 knockout mice (B6.129S4-Ccl2tm1Rol/J) were obtained as homozygotes from The Jackson Laboratory (Bar Harbor, ME). These mice were created by B. Rollins (Dana-Farber Cancer Institute, Harvard Medical School, Boston) and backcrossed to C57BL/6J for 10 generations (Lu et al. 1998). Control C57BL/6J mice were also obtained from The Jackson Laboratory to ensure uniformity of genetic background. CAST/Ei mice were obtained from the Medical Research Council (MRC) Mammalian Genetics Unit (Harwell, UK); NZW/OlaHsd and SJL/JOlaHsd were obtained from Harlan (Bicester, UK).
Hemizygous mice were generated by crossing Mcp1−/− mice to C57BL/6J using both males and females from each strain. These F1 progeny (males and females from both directions of the cross) were further crossed to CAST/Ei, NZW/OlaHsd, or SJL/JOlaHsd to produce mice that were hemizygous or wild type at the Mcp1 locus on a genetic background that was 50% C57BL/6J and 50% from one of the other strains.
Genotyping:
DNA was extracted from 0.5 cm tail biopsies using a Promega DNA extraction kit and resuspended in 50 μl TE (10 mm Tris–HCL, 1 mm EDTA, pH 7.5). A 1:10 dilution of this stock was used as template for subsequent PCRs. DNA for SJL/J was purchased from The Jackson Laboratory and used at a concentration of 10 ng/μl in subsequent PCRs. All PCRs were carried out using a PTC-225 (MJ Research) thermal cycler. Wild-type and knockout alleles were distinguished using two PCR reactions as designed by The Jackson Laboratory. Reaction A amplifies an 888-bp product from the wild-type allele while reaction B amplifies an ∼1300-bp product specific to the knockout allele. Ten-microliter PCR reactions were carried out in MegaMix Blue (Microzone) according to the manufacturer's instructions using 5 pmol of forward and reverse primers. Both reactions share the same forward primer (Forward: GGAGCATCCACGTGTTGGC) but are differentiated by the reverse primer. Reverse primer reaction A: ACAGCTTCTTTGGGACACC; reverse primer reaction B: TCCTCGTGCTTTACGGTATCG. Reaction A cycling conditions were: 94° for 3 min, 45° for 30 sec, 72° for 40 sec, for one cycle; 94° for 60 sec, 55° for 30 sec, 72° for 3 min, for 30 cycles; and 72° for 2 min. For reaction B, cycling conditions were: 94° for 3 min, 45° for 30 sec, 72° for 40 sec, for one cycle; 94° for 60 sec, 62° for 30 sec, 72° for 3 min, for 35 cycles; and 72° for 2 min. Fragments were resolved on a 1% agarose gel and visualized with ethidium bromide.
Sequencing:
PCR products were generated as above using primers and conditions as detailed in supplemental Table 1. Cycling conditions were determined empirically but in general were 94° for 15 min; 94° for 30 sec, 60° for 45 sec, 72° for 60 sec for 40 cycles; 72° for 5 min. PCR products were cleaned using Microclean (Microzone) according to the manufacturer's instructions and resuspended in H2O. One hundred to 200 ng PCR product was added to a 15-μl sequencing reaction including 5 pmol of either the forward or reverse primer, 1 μl BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems), and 5 μl Better Buffer (Microzone). Cycling conditions were 95° for 30 sec, 50° for 15 sec, 60° for 120 sec, for 30 cycles. Reactions were ethanol precipitated, washed in 70% ethanol and resuspended in 10 μl MegaBACE loading solution (Amersham Biosciences). Products were detected on a MegaBACE1000 capillary sequencer (Amersham Biosciences). Samples were injected at 3 kV for 40 sec and run at 9 kV for 100 min.
Inoculation and phenotyping:
The inoculum was generated from Chandler/RML mouse-adapted scrapie (obtained from A. Aguzzi, Institute of Neuropathology, University of Zurich, Zurich) by a single passage in CD-1 Swiss mice. Brains from terminally sick mice were used to generate a 1% homogenate in PBS (Lloyd et al. 2001). This was used as the inoculum in all subsequent experiments. Mice were anesthetized with isofluorane/O2 and inoculated intracerebrally into the right parietal lobe with 30 μl inoculum. All groups of mice included both males and females derived from each direction of the crosses. Incubation time was calculated retrospectively after a definite diagnosis of scrapie had been made and defined as the number of days from inoculation to the onset of clinical signs. This was assessed by daily examination for neurological signs of disease. Early indicators of prion disease include erect ears, rigid tail, piloerection, and ungroomed appearance, slight hunched posture, and clasping of hind limbs when lifted; however, a definitive diagnosis was not made until a confirmatory sign was seen such as ataxia, generalized tremor, loss of righting reflex, or limb paralysis. Animals were culled as soon as clinical scrapie was confirmed or if showing signs of distress or loss of up to 20% of body weight. All procedures were conducted in accordance with UK regulations and international standards on animal welfare.
RNA extraction:
RNA was extracted from whole brains from either uninfected or RML terminally sick mice. In both cases, tissue was homogenized using a Ribolyser according to the manufacturer's instructions. RNA from uninfected brains was prepared using the RNeasy Maxi (QIAGEN) kit according to the manufacturer's instructions. RNA from prion infected tissue was extracted using TRIreagent (Ambion) according to the manufacturer's instructions. Samples were treated with DNaseI (QIAGEN) and purified further using RNeasy Mini (QIAGEN) columns according to the manufacturer's instructions.
Real-time RT–PCR:
Four micrograms of total RNA was reversed transcribed with AMV reverse transcriptase and random primers from the Reverse Transcription System (Promega) according to the manufacturer's instructions. Reactions with no reverse transcription were also carried out for each sample to ensure no genomic DNA contamination. Mcp1 real-time PCR was carried out on a 7500 Fast Real-time PCR System (Applied Biosystems) in a total volume of 15 μl using 1 μl cDNA (200–300 ng) and ROX MegaMix∼Gold (Microzone) according to the manufacturer's instructions. Primers (6 pmol) and probe (3 pmol) were as described by Felton et al. (2005) and supplied by Sigma Genosys. Rodent GAPDH (Applied Biosystems) was duplexed within the reaction as an endogenous control according to the manufacturer's instructions. All reactions were carried out in triplicate. Standard curves were derived for both probes and used to calculate the quantity of gene-specific cDNA in the reaction. Mcp1 values were normalized by dividing with the quantity of GAPDH.
Neuropathology and immunohistochemistry:
Tissue was fixed in 10% buffered formal saline (BFS) and prion infectivity was inactivated by incubation in 98% formic acid for 1 hr. After further washing for 24 hr in 10% BFS, tissue samples were processed and paraffin wax embedded. Sagittal sections were cut at a nominal thickness of 4 μm, treated with 98% formic acid for 5 min, and then boiled in EDTA-Tris-citrate buffer pH 7.8 for 20 min. Immunohistochemical staining was performed with anti-PrP monoclonal antibody ICSM35 (D-Gen, London) (Asante et al. 2002) for prion distribution, glial fibrillary acid protein (GFAP) (Dako) for gliosis on a Ventana automated immunohistochemical staining machine using a basic diaminobenzidine detection system according to the manufacturer's instructions (Ventana Medical Systems, Tucson, AZ). Spongiosis was determined using a serial section stained with hematoxylin and eosin (H&E). Activated microglia were stained with anti-Iba1 polyclonal antibody (Wako) on an automated immunohistochemical staining machine as above.
RESULTS
Mcp1 polymorphisms:
To determine whether Mcp1 is a plausible QTG for prion disease incubation time we established its physical position on Mmu11 as 81,851,769–81,853,421 (NCBI Build 36). This locates Mcp1 between D11Mit219 (72,135,165) and D11Mit36 (83,658,790), which represent the peaks of linkage as determined by Stephenson et al. (2000) and Lloyd et al. (2001), respectively. These data are consistent with Mcp1 being a candidate QTG.
Mcp1 (NM_011333) is also known as chemokine (C-C motif), ligand 2 (Ccl2), and small inducible cytokine A2 (Scya2), spans 1.65 kb of genomic DNA, and has a predicted mRNA transcript of 806 bp that encodes a protein of 148 amino acids. If Mcp1 has a role in influencing incubation time in the F2 intercrosses described then one would expect to see polymorphisms between the long incubation-time strain (CAST/Ei) and the short incubation-time strains (NZW/OlaHsd or SJL/J). Therefore we sequenced all three exons of the gene including some flanking sequence to ensure coverage of the whole transcript including untranslated regions, open reading frames, and splice sites. In addition we sequenced a 240 bp enhancer region, 2.3 kb upstream of the transcription start site, that contains two platelet-derived growth factor (PDGF)-responsive promoter motifs that bind NF-κB (Freter et al. 1995).
Ten polymorphisms were identified, 2 in the 5′-UTR and 8 within the open reading frame, 3 of which cause amino acid changes (Table 1). The insertion/deletion polymorphism in the 5′-UTR occurs within a putative α-CP2 binding site (Kim et al. 1990). Although this transcription factor has been implicated in the activation of the α-globin gene it is uncertain whether it would influence the transcription of Mcp1. The variation seen between the three strains is however consistent with the possibility that this polymorphism is a functional variant in both crosses.
TABLE 1.
Mcp-1 polymorphisms
| Polymorphism (amino acid)
|
|||||
|---|---|---|---|---|---|
| Region | Position | Codon | NZW/OlaHsd (108 ± 1) | CAST/Ei (188 ± 3) | SJL/J (122 ± 1) |
| 5′-UTR | 81,851,811 | — | C | T | C |
| 81,851,812–81,851,817 | — | NC | ACCAGC Del | ACCAGC Ins | |
| Exon 1 | 81,851,873 | 6 | T (Met) | C (Thr) | T (Met) |
| 81,851,887 | 11 | C | C | T | |
| Exon 2 | 81,852,688 | 29 | C | C | T |
| 81,852,742 | 47 | G | A | G | |
| 81,852,749 | 50 | A (Ser) | G (Gly) | G (Gly) | |
| Exon 3 | 81,853,194 | 90 | C | T | C |
| 81,853,199 | 91 | T | C | T | |
| 81,853,201 | 92 | G (Arg) | G (Arg) | A (Gln) | |
Numbering is based on genome sequence NCBI Build 36 (NM_011333). Prion disease incubation times are shown for each strain following intracerebral inoculation of Chandler/RML prions ± SEM (Lloyd and Collinge 2005). Amino acids are shown only for nonsynonomous polymorphisms. NC, no change when compared to database sequence for Mcp-1; Del, deletion; Ins, insertion.
The three nonsynonymous polymorphisms were found at codons 6, 50, and 92. The S50G change occurs between NZW/OlaHsd and CAST/Ei but is not seen in SJL/J suggesting that it could be a candidate QTN for the NZW × CAST F2 cross but not the SJL × CAST F2 cross. Conversely, the R92Q variant is seen only between CAST/Ei and SJL/J, which makes it a candidate only for the SJL × CAST QTL. In contrast, the M6T polymorphism could be a candidate for both crosses with the threonine seen in the long and the methionine in the short incubation-time strains, respectively. Alternatively, these codons may not act independently but may influence incubation time together thus providing different haplotypes (MSR, TGR, and MGQ) and incubation times for each of the three mouse strains.
One polymorphism in the 5′-UTR and five in the ORF were silent changes. Although no known function could be ascribed to these nucleotides they cannot be excluded as potential QTN as they may influence other unexplored functions such as gene expression.
Quantitative complementation of Mcp1:
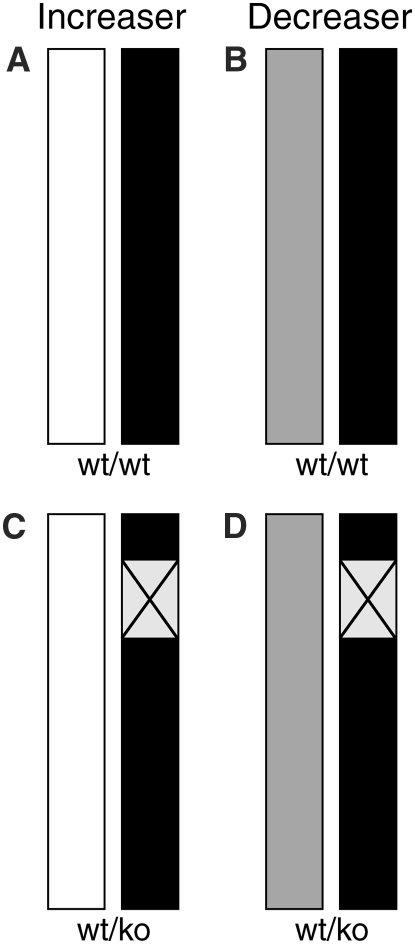
Quantitative complementation has been used successfully by others to determine whether a candidate gene can interact with known functional alleles to influence a quantitative phenotype (Yalcin et al. 2004). Therefore, to test whether Mcp1 is a QTG for prion disease incubation time a quantitative complementation test was designed (Figure 1). Mcp1−/− were crossed to C57BL6/J mice to produce hemizygous mice Mcp1−/+ on a C57BL/6J genetic background. These mice were further crossed to CAST/Ei, NZW/OlaHsd, and SJL/JOlaHsd. From previous work it is known that CAST carries the chromosome 11 allele driving a longer incubation time (“increaser” QTL) while NZW and SJL carry the shorter incubation time alleles (“decreaser” QTL). The resulting offspring from this cross will have either one or two copies of Mcp1 (Figure 1). Where two copies are present, one will be derived from the reference strain (C57BL/6J) while the other will carry either an increaser or decreaser allele (Figure 1, A and B). In the hemizygous mice the single Mcp1 copy will either be an increaser or decreaser allele (Figure 1, C and D). If Mcp1 knockout has no effect on incubation time then no difference will be observed between the wild-type and hemizygous mice for each genetic background (no difference between A and C or B and D). If Mcp1 affects the phenotype but not the QTL there should be no difference between the increaser and decreaser alleles. If Mcp1 affects the QTL there will not only be a difference between the wild-type and hemizygous mice but there should also be a difference between the effects of the increaser and decreaser QTL alleles (Flint and Mott 2001).
Figure 1.—
Schematic of chromosome 11 for the offspring of quantitative complementation crosses. All mice are the result of a cross between Mcp1−/+ mice (C57BL/6J background, solid bar) and an inbred mouse strain carrying either an increaser (CAST, open bars, A and C) or decreaser (NZW or SJL, shaded bar, B and D) QTL. Mice represented by A and B have two copies of Mcp1: one allele from C57BL6/J and the other representing an increaser (A) or decreaser (B) allele. C and D depict mice carrying only one Mcp1 allele derived from the increaser (C) or decreaser (D) strain, respectively. The box with the 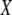 represents the null Mcp1 allele on a C57BL/6J background. If Mcp1 acts as a QTG an increase in incubation time would be expected in C relative to A and in D relative to B. Further, the incubation time would be expected to increase where the increaser allele is present (C) compared to where the decreaser allele is present (D). If Mcp1 does not act as a QTG no differences would be expected between A and C or between B and D.
represents the null Mcp1 allele on a C57BL/6J background. If Mcp1 acts as a QTG an increase in incubation time would be expected in C relative to A and in D relative to B. Further, the incubation time would be expected to increase where the increaser allele is present (C) compared to where the decreaser allele is present (D). If Mcp1 does not act as a QTG no differences would be expected between A and C or between B and D.
Hemizygous and wild-type mice for each mouse cross were inoculated intracerebrally with 30 μl 1% Chandler/RML inoculum. Additional groups of Mcp1−/−, C57BL/6J, and their F1 progeny, Mcp−/+ were inoculated as controls. C57BL/6J were also inoculated with PBS as a negative control. All groups challenged with prions developed classical signs of mouse scrapie. No clinical differences were observed between any of the groups. These signs included early nonspecific symptoms such as erect ears, rigid tail, piloerection, and ungroomed appearance progressing to more definitive signs such as ataxia and limb paralysis. Animals were culled at this stage and not allowed to develop more severe signs of disease. No clinical signs were observed in the PBS control group.
Group incubation times (Table 2) were compared by Kaplan–Meier log-rank survival analysis. For the control C57BL/6J background, no significant difference was observed between the hemizgygous and wild-type mice. Unexpectedly, the knockout mice showed a reduced incubation time (161 ± 0.5 days) compared to the wild-type controls (166 ± 3.9 days) which was statistically significant (P = 0.0175). This is in direct contrast to the results reported by Felton et al. (2005) for the ME7 model of murine prion disease where they described a significant increase in survival for Mcp1−/− mice. A significant difference was also seen between the hemizygous (167 ± 3.0) and knockout mice (P = 0.0027). For each of the increaser (CAST) and decreaser (NZW and SJL) crosses no significant differences were observed between the hemizygous and Mcp1 wild-type mice. Taken together these data suggest that in the Chandler/RML model total knockout of Mcp1, but not loss of a single copy, results in a small (mean difference of 5 days) decrease in incubation time and is unlikely to contribute to the observed Mmu11 QTL.
TABLE 2.
Incubation times for Chandler/RML prions
| Mouse strain | Genotype | Mean (days) ± SEM (n) |
|---|---|---|
| Mcp1−/− | ko/ko | 161 ± 0.5 (10) |
| Mcp1−/+ | wt/ko | 167 ± 3.0 (9) |
| C57BL/6 | wt/wt | 166 ± 3.9 (8) |
| CAST × C57BL/6 | wt/wt | 206 ± 5.8 (9) |
| wt/ko | 205 ± 2.0 (9) | |
| NZW × C57BL/6 | wt/wt | 181 ± 3.8 (9) |
| wt/ko | 178 ± 9.5 (9) | |
| SJL × C57BL/6 | wt/wt | 148 ± 2.6 (10) |
| wt/ko | 152 ± 4.3 (10) |
wt, wild type; ko, knockout.
Neuropathology and immunocytochemistry:
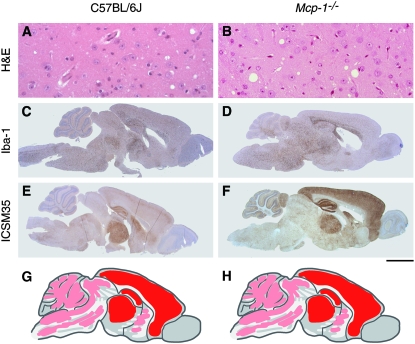
The neuropathological phenotype was also compared between Mcp1−/−, Mcp1−/+, and C57BL/6J groups. Mice were culled and the brains removed at the terminal stage of disease once a definitive diagnosis of mouse scrapie had been made. PrPSc deposition was determined by immunohistochemistry with the monoclonal antibody ICSM35 (Asante et al. 2002). No differences were seen between the groups (Figure 2). PrPSc is deposited extensively throughout the brain with the most intense staining seen in the cortex, hippocampus, and thalamus with less deposition in the cerebellum, brain stem, tectum, and basal ganglia (Figure 2, E–H). This PrPSc distribution pattern is characteristic of the Chandler/RML prion strain in inbred lines of mice such as C57BL/6J. Gliosis, (GFAP staining, data not shown) and spongiosis (Figure 2, A and B, H&E staining) mirrored the distribution pattern of PrPSc and did not differ between groups. Activated microglia were specifically detected with an anti-Iba1 (polyclonal) antibody (Figure 2, C and D). The distribution mirrored that of the general gliosis and PrPSc and showed no differences between the groups. No neuronal loss was detected in the hippocampus by H&E staining (data not shown).
Figure 2.—
Neuropathology and immunohistochemistry for C57BL/6J (A, C, and E) and Mcp1−/− (B, D, and F) mice. The scale bar represents 50 μm for A and B and 2 mm for C–F. A and B are stained with hematoxylin and eosin and show evidence of spongiosis (vacuolation). Distribution of activated microglia is shown in C and D stained with polyclonal antibody Iba1. E and F are stained with monoclonal antibody ICSM-35 to show abnormal PrP deposition. PrP staining is diffuse in both strains. Schematic representation of PrP deposition in the brain is shown for C57BL/6J (G) and Mcp1−/− (H). Red represents areas of intense staining, dark pink areas of moderate staining, and light pink areas of light staining. For both strains, the cortex, hippocampus, and thalamus show intense staining (red) with light staining (light pink) in the cerebellum, brain stem, tectum, and basal ganglia. No significant differences are seen between the groups. Data for Mcp1−/+ mice not shown as results are indistinguishable from C7BL/6J and Mcp1−/−.
Mcp1 expression:
Mcp1 is known to play a role in the activation of microglia which are postulated to contribute to end-stage prion disease by exacerbating neuronal injury (Felton et al. 2005). According to this hypothesis, Mcp1−/− mice would be expected to exhibit an increase in incubation time. This was observed by Felton et al. (2005) using the ME7 model but the opposite was seen in this study with RML. Microglial activation was detected in both ME7 and RML models with no differences between wild-type (wt) and knockout mice suggesting that Mcp1 alone is not necessary for microglial activation. Nevertheless, it has been suggested that the presence of high levels of Mcp1 at later stages of disease may be required to drive the microglia to an increased level of activation sufficient to aggravate already damaged neurons. Therefore we determined the level of Mcp1 mRNA expression in our inbred mouse strains before inoculation and at end-stage disease to see whether the Mcp1 expression profile is comparable in the ME7 and RML models.
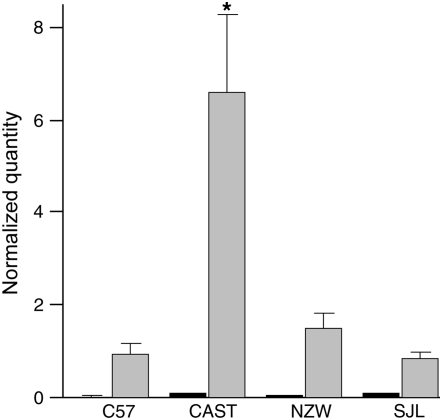
RNA was extracted from whole brains for normal and RML infected C57BL/6, CAST, NZW, and SJL mice. Following reverse transcription, cDNA was used for quantitative real-time PCR using primers and probes as previously described. Expression levels were normalized against GAPDH and expressed as a relative quantity (arbitrary units) (Figure 3). Mcp1 levels were very low in uninfected tissues but as described for ME7 (Felton et al. 2005), significantly raised in terminal disease (P ≤ 0.01 for all strains, Mann–Whitney test). Comparison of mouse strains showed no significant differences between long and short incubation-time strains for uninfected samples but did show significance (P = 0.01, Mann–Whitney test) for both CAST/NZW and CAST/SJL comparisons in RML infected samples.
Figure 3.—
Quantification of Mcp1 mRNA expression in whole brain by real-time RT–PCR for C57BL/6, CAST, NZW, and SJL mouse strains. Dark and light shaded bars represent uninfected mice and mice at the terminal stage of disease following inoculation with Chandler/RML scrapie prions, respectively. For each group n = 6 except for CAST–RML (n = 4) and NZW–RML (n = 5). All samples were tested in triplicate. Units are arbitary and represent quantity of Mcp1 transcript normalized by quantity of GAPDH. Error bars represent the standard error of the mean. The asterisk signifies P = 0.01 when compared to other strains for infected samples only (Mann–Whitney test). No highly significant differences were detected between strains for uninfected mice. Mcp1 mRNA expression is significantly increased in end-stage disease in all strains (P ≤ 0.01 Mann–Whitney test).
DISCUSSION
Mcp1 has been implicated in various models of neuronal damage such as prion disease, stroke, and experimental allergic encephalomyelitis (Teuscher et al. 1999; Hughes et al. 2002; Felton et al. 2005). The finding that Mcp1−/− mice show a delay in the onset of late-stage clinical signs and an increased survival time after ME7 prion infection (Felton et al. 2005) and the localization of Mcp1 within QTL regions of linkage (Stephenson et al. 2000; Lloyd et al. 2001) strongly suggested that Mcp1 was a plausible candidate QTG for prion disease incubation time. Sequence analysis of Mcp1 in inbred mice further supported this suggestion with polymorphisms in both 5′-UTR and ORF segregating between mouse strains in patterns consistent with the observed incubation times.
Quantitative complementation is a strategy based on techniques developed in Drosophila and adapted and used successfully for identifying QTL in mice (Flint and Mott 2001; Mackay 2001; Yalcin et al. 2004). Our data show that in the Chandler/RML model of murine prion disease, Mcp1 does not delay onset of clinical signs of disease nor increase survival time but rather shows a small decrease in incubation time. Although statistically significant, the mean difference in incubation time between the two groups was only 5 days. Although highly speculative, it is possible that this difference occurs because chemokines affect different neuronal populations which are differentially targeted in the ME7 and Chandler/RML model. No differences were seen between wild-type and knockout crosses suggesting that Mcp1 has no effect on the phenotype and does not contribute to the QTL. Based on the control data, any effect of Mcp1 in this model would only be seen in full knockout and not hemizygous mice. Our original studies (Lloyd et al. 2001) identified a broad region of linkage on Mmu11 and suggested the potential for multiple modifiers in this region. If the influence of a single gene such as Mcp1 on incubation time in this model is small it is possible that the quantitative complementation assay presented here may not be sensitive enough to detect such an effect especially as the effect of C57BL6/J alleles on the Mmu11 modifiers is unknown. Thus, it is not possible to completely exclude Mcp1 as a QTG; however, it remains unlikely that Mcp1 is an important QTG because the genotype mean incubation times calculated for linked markers on Mmu11 are consistent with an additive model of inheritance while Mcp1 appears to be dominant (Lloyd et al. 2001). Although all crosses in this study were set up using both males and females in each direction and both sexes were used in the inoculated groups, the numbers were of necessity small and therefore we are unable to exclude any imprinting effects.
Although differences in Mcp1 expression were observed at end-stage disease between different mouse strains this does not correlate with incubation time as the highest level of expression was observed in CAST/Ei and the model of MCP1 action would suggest that higher levels of Mcp1 mRNA would reduce rather than increase the incubation time. However, it should be noted that mRNA levels may not necessarily correlate with protein levels. Significant differences (P = 0.01, Mann–Whitney test) in Mcp1 expression at end stage were seen between CAST and both NZW and SJL which correlates with some of the polymorphisms described; therefore, although Mcp1 does not seem to be a major gene affecting incubation time these polymorphisms may influence a differential response to scrapie infection.
The increased survival seen by Felton et al. (2005) in Mcp1−/− mice was not replicated in our study. Not only was the onset of clinical signs not delayed, but our knockout mice displayed the full range of typical scrapie symptoms whereas in the previous study not all the knockout mice displayed signs of scrapie but rather were sacrificed following loss of >15% body weight. One possible explanation is the mouse model itself. Although both studies used a knockout line that originated in the same laboratory, the line was first described by Gu et al. in 1999 and has subsequently been maintained by others. The mice used by Felton et al. (2005) were obtained from Novartis (H. Perry, personal communication) while those used in this study were purchased from The Jackson Laboratory. Over time it is possible that differences may have been introduced into these independent colonies. The C57BL6/J control animals were also acquired from different sources: Harlan, UK in the Felton et al. (2005) study and The Jackson Laboratory in this study, which may further compound the differences. An alternative and possibly more likely explanation for these differences is the choice of prion strains. While Chandler/RML and ME7 are both mouse-adapted scrapie prion strains, they were independently derived and are distinct strains with differing biological and biochemical characteristics. Naturally occurring scrapie is associated with multiple strains (Bruce et al. 1991, 2002). Chandler/RML was derived from goat scrapie and subsequently passaged in sheep and CD-1 mice (Chandler 1961). In contrast, ME7 was derived from the spleen of a scrapie infected Suffolk sheep passaged in Swiss white mice and subsequently passaged in C57BL mice (Zlotnik and Rennie 1963; Dickinson et al. 1969). In inbred mice, ME7 and RML have different incubation times, show different patterns of neuropathology, and in vitro cells show differential susceptibility to both strains (Westaway et al. 1987; Bruce 1993, 2003; Klohn et al. 2003). Furthermore, biochemical analysis has established that ME7 and RML have different conformations of PrPSc (Thackray et al. 2007). Although Mcp1 is similarly upregulated in both ME7 and RML models it appears that its role in disease pathogenesis may be prion-strain specific.
Both pro- and anti-inflammatory molecules have been implicated in prion disease incubation time (Schultz et al. 2004; Thackray et al. 2004; Felton et al. 2005) and their results are all consistent with the hypothesis that inflammation may accelerate neuronal loss. However, our results demonstrate that the effects of individual molecules are likely to be prion-strain specific and therefore it may not be possible to extrapolate to human prion diseases where their role remains uncertain.
Acknowledgments
We thank Michelle Ahmet, Catherine O'Malley, and Caroline Powell for preparation of histological slides. We also thank David Key and team for animal care and Ray Young for preparation of figures. This work was supported by the Medical Research Council, United Kingdom.
References
- Asante, E. A., J. M. Linehan, M. Desbruslais, S. Joiner, I. Gowland et al., 2002. BSE prions propagate as either variant CJD-like or sporadic CJD-like prion strains in transgenic mice expressing human prion protein. EMBO J. 21 6358–6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, M. E., 1993. Scrapie strain variation and mutation. Br. Med. Bull. 49 822–838. [DOI] [PubMed] [Google Scholar]
- Bruce, M. E., 2003. TSE strain variation. Br. Med. Bull. 66 99–108. [DOI] [PubMed] [Google Scholar]
- Bruce, M. E., A. Boyle, S. Cousens, I. McConnell, J. Foster et al., 2002. Strain characterization of natural sheep scrapie and comparison with BSE. J. Gen. Virol. 83 695–704. [DOI] [PubMed] [Google Scholar]
- Bruce, M. E., I. McConnell, H. Fraser and A. G. Dickinson, 1991. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J. Gen. Virol. 72 595–603. [DOI] [PubMed] [Google Scholar]
- Carlson, G. A., C. Ebeling, M. Torchia, D. Westaway and S. B. Prusiner, 1993. Delimiting the location of the scrapie prion incubation time gene on chromosome 2 of the mouse. Genetics 133 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, G. A., P. A. Goodman, M. Lovett, B. A. Taylor, S. T. Marshall et al., 1988. Genetics and polymorphism of the mouse prion gene complex: control of scrapie incubation time. Mol. Cell. Biol. 8 5528–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, R. L., 1961. Encephalopathy in mice produced by inoculation with scrapie brain material. Lancet 1 1378–1379. [DOI] [PubMed] [Google Scholar]
- Collinge, J., 2001. Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci. 24 519–550. [DOI] [PubMed] [Google Scholar]
- Dickinson, A. G., V. M. Meikle and H. Fraser, 1969. Genetical control of the concentration of ME7 scrapie agent in the brain of mice. J. Comp. Pathol. 79 15–22. [DOI] [PubMed] [Google Scholar]
- Felton, L. M., C. Cunningham, E. L. Rankine, S. Waters, D. Boche et al., 2005. MCP-1 and murine prion disease: separation of early behavioural dysfunction from overt clinical disease. Neurobiol. Dis. 20 283–295. [DOI] [PubMed] [Google Scholar]
- Flint, J., and R. Mott, 2001. Finding the molecular basis of quantitative traits: successes and pitfalls. Nat. Rev. Genet. 2 437–445. [DOI] [PubMed] [Google Scholar]
- Freter, R. R., J. A. Alberta, K. K. Lam and C. D. Stiles, 1995. A new platelet-derived growth factor-regulated genomic element which binds a serine/threonine phosphoprotein mediates induction of the slow immediate-early gene MCP-1. Mol. Cell. Biol. 15 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese, A., D. R. Brown, M. H. Groschup, C. Feldmann, I. Haist et al., 1998. Role of microglia in neuronal cell death in prion disease. Brain Pathol. 8 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, L., B. Rutledge, J. Fiorillo, C. Ernst, I. Grewal et al., 1997. In vivo properties of monocyte chemoattractant protein-1. J. Leukoc. Biol. 62 577–580. [DOI] [PubMed] [Google Scholar]
- Gu, L., S. C. Tseng and B. J. Rollins, 1999. Monocyte chemoattractant protein-1. Chem. Immunol. 72 7–29. [DOI] [PubMed] [Google Scholar]
- Hughes, P. M., P. R. Allegrini, M. Rudin, V. H. Perry, A. K. Mir et al., 2002. Monocyte chemoattractant protein-1 deficiency is protective in a murine stroke model. J. Cereb. Blood Flow Metab. 22 308–317. [DOI] [PubMed] [Google Scholar]
- Kim, C. G., S. L. Swendeman, K. M. Barnhart and M. Sheffery, 1990. Promoter elements and erythroid cell nuclear factors that regulate alpha-globin gene transcription in vitro. Mol. Cell. Biol. 10 5958–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klohn, P., L. Stoltze, E. Flechsig, M. Enari and C. Weissmann, 2003. A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc. Natl. Acad. Sci USA 100 11666–11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, S., O. N. Onwuazor, J. Beck, G. Mallinson, M. Farrall et al., 2001. Identification of multiple quantitative trait loci linked to prion disease incubation period in mice. Proc. Natl. Acad. Sci. USA 98 6279–6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, S. E., and J. Collinge, 2005. Genetic susceptibility to prion diseases in humans and mice. Curr. Genomics 6 1–12. [Google Scholar]
- Lloyd, S. E., J. B. Uphill, P. V. Targonski, E. M. Fisher and J. Collinge, 2002. Identification of genetic loci affecting mouse-adapted bovine spongiform encephalopathy incubation time in mice. Neurogenetics 4 77–81. [DOI] [PubMed] [Google Scholar]
- Lu, B., B. J. Rutledge, L. Gu, J. Fiorillo, N. W. Lukacs et al., 1998. Abnormalities in monocyte recruitment and cytokine expression in monocyte chemoattractant protein 1-deficient mice. J. Exp. Med. 187 601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, T. F., 2001. Quantitative trait loci in Drosophila. Nat. Rev. Genet. 2 11–20. [DOI] [PubMed] [Google Scholar]
- Manolakou, K., J. Beaton, I. McConnell, C. Farquar, J. Manson et al., 2001. Genetic and environmental factors modify bovine spongiform encephalopathy incubation period in mice. Proc. Natl. Acad Sci USA 98 7402–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R. C., J. Hope, P. A. McBride, I. McConnell, J. Selfridge et al., 1998. Mice with gene targetted prion protein alterations show that Prnp, Sinc and Prni are congruent. Nat. Genet. 18 118–125. [DOI] [PubMed] [Google Scholar]
- Moreno, C. R., F. Lantier, I. Lantier, P. Sarradin and J. M. Elsen, 2003. Detection of new quantitative trait loci for susceptibility to transmissible spongiform encephalopathies in mice. Genetics 165 2085–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, V. H., C. Cunningham and D. Boche, 2002. Atypical inflammation in the central nervous system in prion disease. Curr. Opin. Neurol. 15 349–354. [DOI] [PubMed] [Google Scholar]
- Schultz, J., A. Schwarz, S. Neidhold, M. Burwinkel, C. Riemer et al., 2004. Role of interleukin-1 in prion disease-associated astrocyte activation. Am. J. Pathol. 165 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson, D. A., K. Chiotti, C. Ebeling, D. Groth, S. J. Dearmond et al., 2000. Quantitative trait loci affecting prion incubation time in mice. Genomics 69 47–53. [DOI] [PubMed] [Google Scholar]
- Teuscher, C., R. J. Butterfield, R. Z. Ma, J. F. Zachary, R. W. Doerge et al., 1999. Sequence polymorphisms in the chemokines Scya1 (TCA-3), Scya2 (monocyte chemoattractant protein (MCP)-1), and Scya12 (MCP-5) are candidates for eae7, a locus controlling susceptibility to monophasic remitting/nonrelapsing experimental allergic encephalomyelitis. J. Immunol. 163 2262–2266. [PubMed] [Google Scholar]
- Thackray, A. M., L. Hopkins, M. A. Klein and R. Bujdoso, 2007. Mouse-adapted ovine scrapie prion strains are characterized by different conformers of PrPSc. J. Virol. 81 12119–12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray, A. M., A. N. McKenzie, M. A. Klein, A. Lauder and R. Bujdoso, 2004. Accelerated prion disease in the absence of interleukin-10. J. Virol. 78 13697–13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway, D., P. A. Goodman, C. A. Mirenda, M. P. McKinley, G. A. Carlson et al., 1987. Distinct prion proteins in short and long scrapie incubation period mice. Cell 51 651–662. [DOI] [PubMed] [Google Scholar]
- Yalcin, B., S. A. Willis-Owen, J. Fullerton, A. Meesaq, R. M. Deacon et al., 2004. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat. Genet. 36 1197–1202. [DOI] [PubMed] [Google Scholar]
- Zlotnik, I., and J. C. Rennie, 1963. Further observations on the experimental transmission of scrapie from sheep and goats to laboratory mice. J. Comp. Pathol. 73 150–162. [DOI] [PubMed] [Google Scholar]