Abstract
High-fidelity chromosome segregation requires that the sister chromatids produced during S phase also become paired during S phase. Ctf7p (Eco1p) is required to establish sister chromatid pairing specifically during DNA replication. However, Ctf7p also becomes active during G2/M in response to DNA damage. Ctf7p is a phosphoprotein and an in vitro target of Cdc28p cyclin-dependent kinase (CDK), suggesting one possible mechanism for regulating the essential function of Ctf7p. Here, we report a novel synthetic lethal interaction between ctf7 and cdc28. However, neither elevated CDC28 levels nor CDC28 Cak1p-bypass alleles rescue ctf7 cell phenotypes. Moreover, cells expressing Ctf7p mutated at all full- and partial-consensus CDK-phosphorylation sites exhibit robust cell growth. These and other results reveal that Ctf7p regulation is more complicated than previously envisioned and suggest that CDK acts in sister chromatid cohesion parallel to Ctf7p reactions.
CELL division is essential for embryonic growth, repair of damaged tissue, and the replacement of senescent cells. During S phase and long before a cell divides, chromosomes are replicated to produce identical sister chromatids. During mitosis, each sister chromatid orients to spindle pole microtubules opposite that of its sister chromatid, ensuring proper segregation of chromosomes into the newly forming daughter cells. In most vertebrate cells, chromosome replication and sister chromatid segregation are separated by extended periods of time and checkpoint functions. Thus, identifying the products of chromosome replication as sister chromatids during S phase and then maintaining that identity until anaphase are both fundamental facets of cell division and essential for progeny viability.
Early studies in yeast identified cohesin complexes (Mcd1p/Scc1p, Irr1p/Scc3p, Smc1p, and Smc3p) as the molecular glue that maintains sister chromatid pairing from S phase until anaphase onset (Strunnikov et al. 1993; Kurlandzka et al. 1995; Guacci et al. 1997; Michaelis et al. 1997; Toth et al. 1999). Biochemical and EM-based analyses revealed that a subset of cohesin subunits (Mcd1p, Smc1p, and Smc3p) form a huge triangular ring that likely encircles one or both chromatid fibers (Melby et al. 1998; Anderson et al. 2002; Haering et al. 2002; Gruber et al. 2003; Ivanov and Nasmyth 2005). Cohesin complexes are deposited along the chromosome length by a heterodimeric complex composed of Scc2p and Scc4p (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999; Ciosk et al. 2000; Laloraya et al. 2000; Glynn et al. 2004). Importantly, cohesins and their deposition onto sister chromatids are not sufficient for sister chromatid pairing. A third activity, termed establishment, is required to pair together cohesin-decorated sister chromatids. This function is provided by the essential and highly conserved Ctf7p (Ctf7p/Eco1p, EFO1/ESCO1, EFO2/ESCO2, DECO, ESO1) family of proteins (Skibbens et al. 1999; Toth et al. 1999; Tanaka et al. 2000; Bellows et al. 2003; Williams et al. 2003; Hou and Zou 2005; Vega et al. 2005). While the mechanism of cohesion establishment remains highly controversial, likely models are that Ctf7p catalyzes the pairing of cohesins associated with each sister chromatid or mediates cohesin dynamics during DNA replication (Skibbens et al. 2007).
In unperturbed cell cycles, Ctf7p functions exclusively during DNA replication after which Ctf7p becomes inactive (Skibbens et al. 1999; Toth et al. 1999). In response to DNA double-strand breaks during G2/M, however, Ctf7p-dependent sister chromatid pairing becomes reactivated. Thus, S phase pairing and G2/M re-pairing Ctf7p activities are tightly regulated (Strom et al. 2004, 2007; Unal et al. 2004, 2007). Prior studies revealed that Ctf7p is a phosphoprotein and an in vitro target of CDK activity (Ubersax et al. 2003), suggesting a mechanism for regulating Ctf7p. Here, we report on results from a synthetic lethal screen regarding novel interactions between CTF7, CAK1, and CDC28 and site-directed mutagenesis of Ctf7p phosphorylation consensus sites. These findings test the model that Ctf7p function is regulated through CDK activity.
MATERIALS AND METHODS
Media and cell growth:
Yeast and bacterial media, growth, sporulation, and transformation procedures were performed as described with minor modifications (Ito et al. 1983; Schiestl and Gietz 1989; Rose et al. 1990). Strains used in this study are listed in Table 1. Sectoring analyses to identify plasmid-dependent functions were based on previous genetic analyses and performed as previously described with minor modification (Koshland et al. 1985; Krantz and Holm 1990; Brands and Skibbens 2005).
TABLE 1.
Strains and mutations used in this study
| YAB1005 | MATα ade2 ade3 lys2 ura3 his3 leu2 ctf7∷HIS3 ctf7-203:LEU2 |
| YAB1010 | MATaade2 ade3 trp1 ura3 his3 leu2 ctf7∷HIS3 pAB1009 (CEN ADE3 URA3 CTF7) |
| YBS514 | MATaura3 lys2 ade2 his3 trp1 leu2 ctf7∷HIS3 ctf7-203:LEU2 (D168V, R199K, I231F, G259R) (Skibbens et al. 1999) |
| YLA1119 | MATaHIS3:LacI-GFP CLONAT:KAN:LacO:telomere IV PDS1-12MYC:TRP1 (Antoniacci and Skibbens 2006) |
| YBS1294 | YLA1119 containing cak1-277 |
| Mutations in full/partial consensus CDK/Cdc28p-phosphorylation Ctf7p residues | |
| YAB1200 | = YAB 1010 pAB1067 (CEN TRP1 ctf7-500 = S67A) |
| YAB1201 | = YAB 1010 pAB1068 (CEN TRP1 ctf7-502 = T94A) |
| YAB1202 | = YAB 1010 pAB1066 (CEN TRP1 ctf7-501 = S99A) |
| YAB1203 | = YAB 1010 pAB1069 (CEN TRP1 ctf7-503 = S105A) |
| YAB1204 | = YAB 1010 pAB1074 (CEN TRP1 ctf7-504 = S67A, T94A, S99A, S105A) |
Molecular methods and site-directed mutagenesis:
YAB1004 was generated by crossing YBS514 four times into the W303 background strain YCH128 (Hardy and Pautz 1996). YAB1028 was generated by transformation with pAB1009. pAB1009 was generated by inserting a 5.5-kb ADE3 locus-containing fragment into the SacI site of pRS316-CTF7 (Skibbens et al. 1999). All site-directed mutations were performed in pLC3 (CEN TRP1 CTF7 using high-fidelity Pfu DNA polymerase and the entire resulting CTF7 open reading frame sequenced in-house (ABI 310 DNA sequencer). Site-directed mutagenesis was performed using PCR and designed oligo pairs as previously described and following manufacturer instructions. DNA oligomers used in this study are available upon request.
CEN URA3 plasmids harboring various cak1 alleles (generous gifts from Ed Winter, Thomas Jefferson University), were digested with PvuI and the cak1-containing fragments ligated with CEN TRP1 fragments harvested from PvuI-digested pRS314.
Cell and colony morphological analyses and cohesion assays:
Yeast cells harvested for phenotypic analyses were fixed in 3.7% formaldeyde for 30 min, washed, and the cell wall removed using zymolyase prior to adhering the cells to slides pretreated with poly-l-lysine. Cells were then permeabilized and flattened by immersion for 4 min in −20° methanol. Mounting mix containing DAPI to allow for DNA visualization was used to adhere coverslips to the glass slides. All images were acquired using a Nikon Eclipse 800 equipped with either a 100× Plan Apo 1.4 objective for cells or 10× Plan 0.25 objective for colonies and captured using a CoolSnap fx (Photometrics) cooled CCD camera operated by IPLab software (version 3.5.2).
Images of sectored colonies were acquired using a Nikon SMZ 1500 microscope equipped with a HR Plan Apo 1× WD 54 lens. Optivar settings of ∼1.5 were used to achieve a 0.55 cm field of view applied to all sector colony assay images. Colony images were captured using a Nikon DXM 1200 CCD camera and ACT1 image software.
Cohesion defects were assessed by crossing cak1-277 allele into our telomere proximal cohesion assay strain YLA1119 (LacI-GFP:HIS3, LacO:KAN, and PDS1-13MYC:TRP) previously described (Antoniacci and Skibbens 2006). The resulting diploids were sporulated, dissected, and independent isolates of progeny containing the appropriate markers identified. In parallel, cak1-277 was incorporated into a centromere-proximal cohesion assay strain YBS1042 (TetO:URA3, TetR-GFP:LEU2, and PDS1-13MYC:TRP1) using a similar strategy (Kenna and Skibbens 2003). Cohesion assay diploid strains homozygous for either wild-type CDC28 (AK1429), or cdc28-B119 (AK1439), or cdc28-B28 (AK1445) alleles were obtained as a generous gift from Ana Kitazono and Steven Kron (Kitazono et al. 2003). For cdc28 mutant strains, the lac operator is integrated on chromosome III at the LEU3 locus (Kitazono et al. 2003). Cohesion assays were performed as described previously with the following modifications (Kenna and Skibbens 2003; Antoniacci and Skibbens 2006). Cells synchronized in late G1 or early S phase were preshifted to 37° during the final 30–45 min of the incubation. For cak1 mutant cells, preanaphase cells were obtained by supplementing fresh medium with nocodazole. In contrast, cdc28-B119 and cdc28-B28 cells are deficient in spindle checkpoint function but they will arrest prior to anaphase onset in response to DNA damage (Kitazono et al. 2003 and this study). Thus, synchronized cdc28 mutant cells were released into fresh medium supplemented with zeocin (100 μg/ml) to induce double-strand breaks and produce a G2/M arrest. Pds1 is not epitope tagged in these cells, requiring analyses to be based on the 2C DNA bias resulting from zeocin exposure and in comparison to wild-type cells. All results represent data tallied from at least two separate experiments in which 100 cells were counted for each strain in each experiment.
RESULTS
ctf7-203 synthetic lethality screen identifies a novel cak1 allele:
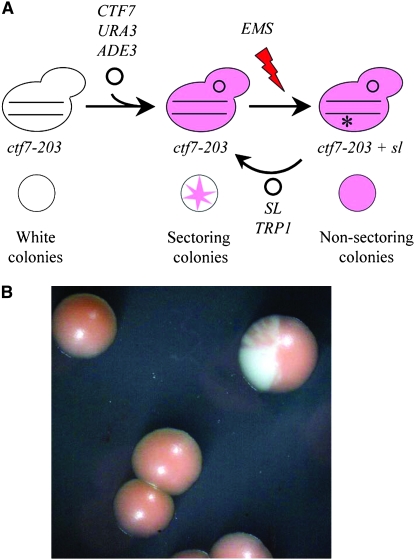
To identify novel cohesion establishment factors, we performed a synthetic lethal screen based on the conditional ctf7-203 mutant strain. An ade2, ade3 temperature-sensitive ctf7-203 strain, which gives rise to white colonies, was transformed with a CTF7 URA3 ADE3 plasmid. At temperatures permissive for ctf7-203p function, transformants remain viable despite plasmid loss and give rise to red/white sectored colonies. Transformed cells were mutagenized to a 20% survival rate using ethyl methylsulfonate (EMS). Cells harboring mutated genes synthetically lethal with ctf7-203 are obligated to maintain the CTF7 URA3 ADE3 plasmid and readily detected as giving rise to nonsectored red colonies (Figure 1). From a population of ∼60,000 randomly mutagenized cells, we identified five nonsectoring strains that depended on plasmid-borne CTF7 for viability. One of the nonsectoring strains was temperature sensitive, suggesting that the new mutation occurred within an essential gene. CEN TRP1 genomic library-based complementation of both temperature sensitivity at 37° and nonsectoring at 23° identified four genomic inserts that contained CTF7 and three inserts that contained CAK1 (Figure 1). Deletion and subcloning analyses confirmed that wild-type CAK1 fully rescues the synthetic lethal interaction, documenting a genetic interaction between CTF7 and CAK1. Sequence analyses identified a single G277D mutation within the cak1 amino acid sequence of the synthetic lethal strain. From here on, we refer to this allele as cak1-277.
Figure 1.—
ctf7-203-based synthetic lethal screen identifies a novel cak1 allele. (A) Schematic of synthetic lethal screen: ade2 ade3 ctf7-203 cells (top) produce white colonies (bottom). When transformed with CTF7 URA3 ADE3, cells give rise to red/white sectored colonies on nonselective 23° medium. Random EMS mutation of genes (asterisk) that are synthetically lethal (sl) with ctf7-203 oblige cells to retain the CTF7 URA3 ADE3 plasmid, producing nonsectored red colonies. Transformation of a genomic library (TRP1 SL rescuing plasmid) identified CAK1 as allowing for loss of the CTF7 URA3 ADE3 plasmid. (B) Micrograph shows transformation of synthetic lethal strain with CAK1 genomic clone results in a sectored red and white colony.
cak1-277 cells exhibit bud growth, polarity, and nuclear division defects:
We first considered the possibility that we had identified a novel sister chromatid pairing role for Cak1p. However, an alignment between numerous kinases (including Cak1p, Cdc28p, Csk1p, and Fus1p) revealed that glycine 277 in Cak1p is invariably conserved through evolution (Chun and Goebl 1997) and thus not likely to uncover a unique Cak1p function. Indeed, phenotypic analyses of cak1-277 mutant cells identified bud polarity defects, multiple and elongated bud structures, and chromosome segregation phenotypes previously documented in other cak1 mutant cells (supplemental Figure 1) (Kaldis et al. 1996; Thuret et al. 1996; Chun and Goebl 1997; Sutton and Freiman 1997).
We next tested whether ctf7-203 synthetic lethality could be recapitulated using other cak1 alleles (Wagner et al. 1997). The results show that ctf7 cak1 cells that contain cak1-17 plasmid required the CEN URA3 CTF7 plasmid for viability and thus gave rise to red nonsectored colonies. ctf7 cak1 cells transformed with either cak1-22 or cak1-K31R provided only for infrequent CEN URA3 CTF7 loss, consistent with prior studies that cak1-K31R and cak1-17 mutations exhibit increased levels of activity relative to cak1-22 (Wagner et al. 1997). These observations reveal that ctf7-203 interactions with cak1-277 are not unique to this allele.
CDC28-dependent bypass of cak1 phenotypes but not ctf7 phenotypes:
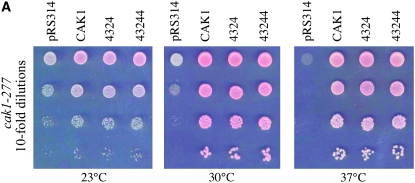
Cak1p phosphorylates many substrates including Kin28p, Bur1p, and Ctk1p, but the only essential role for Cak1p kinase is to activate Cdc28p (Espinoza et al. 1996, 1998; Cross and Levine 1998; Yao and Prelich 2002; Ostapenko and Solomon 2005; Ganem et al. 2006). If the synthetic lethality that we observed between ctf7 and cak1 was in fact due to diminished CDK activation, we reasoned that Cak1p-bypass CDC28 alleles (Cross and Levine 1998) should rescue both cak1-277 temperature sensitivity and ctf7 cak1 synthetic lethality. To test the first of these predictions, wild-type CAK1-, CDC28-bypass alleles (4324 and 43244) and vector alone were transformed into cak1-277 cells. Independent isolates for each strain were then plated on rich medium at permissive and restrictive temperatures. As expected, CAK1 fully rescued cak1-277 mutant cell temperature sensitivity while vector alone did not (Figure 2A). CDC28-bypass alleles fully rescued the conditional growth of the cak1-277 strain, documenting that cak1-277p is conditionally defective in CDK activation (Figure 2A).
Figure 2.—
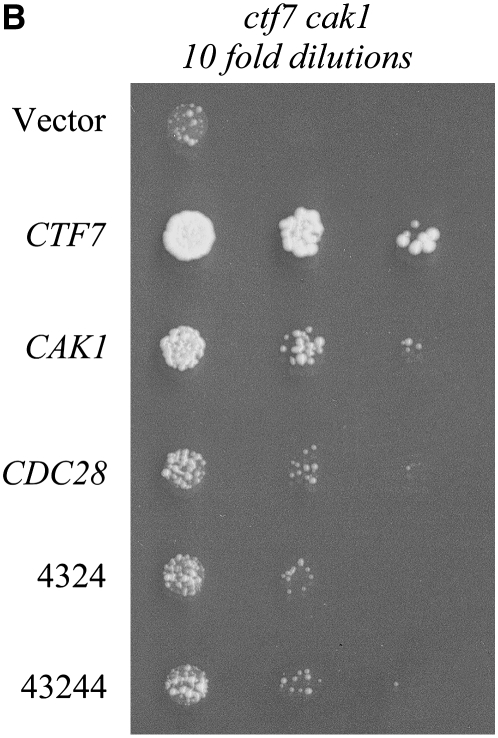
(A) CDC28-bypass alleles suppress cak1-277 temperature sensitivity. Tenfold serial dilutions of cak1-277 mutant cells containing empty vector (pRS314), a CAK1 genomic clone, or CDC28-bypass alleles (4324 and 43244) were spotted on selective medium plates and incubated at 23°, 30°, or 37°. (B) CDC28-bypass alleles partly suppress double-mutant ctf7-203 cak1-277 synthetic lethality. Tenfold serial dilutions of ctf7-203 cak1-277 double mutants harboring CEN CTF7 URA3 and transformed with either CEN TRP1 vector alone or vector containing CTF7-, CAK1-, CDC28-, or CDC28-bypass alleles (4324 or 43244) grown on medium containing 5-FOA at 23°.
To confirm this interpretation, we tested whether ctf7 cak1 synthetic lethality is based on reduced Cdc28p activation. cak1 ctf7 double-mutant cells harboring CEN CTF7 URA3 plasmid were transformed with CEN TRP1 plasmids containing either CTF7, CAK1, CDC28, or CDC28 CAK1-bypass alleles (4324 and 43244). Independent isolates of the transformants were placed on medium supplemented with 5′ fluoro-orotic acid (5-FOA) to counterselect cells harboring the CEN CTF7 URA3 plasmid (Boeke et al. 1987). Both CAK1 and CTF7, but not vector alone, supported cell growth at 23° (Figure 2B). CTF7 provided for more robust cell growth, relative to CAK1, because the ctf7-203 allele is compromised for growth and chromosome transmission fidelity even at 23° (Skibbens et al. 1999). CDC28-bypass alleles (4324 and 43244) also supported cak1 ctf7 double-mutant cell viability, although not to the extent of either CTF7 or CAK1 (Figure 2B). Even a CEN-based increase of wild-type CDC28 provided for partial rescue of cak1 ctf7 double-mutant cell growth. While these findings do not exclude a minor role for Cak1p in Ctf7p function, the data reveal that ctf7 cak1 synthetic lethality is largely due to diminished Cdc28p function.
cdc28 ctf7 double-mutant cells are inviable:
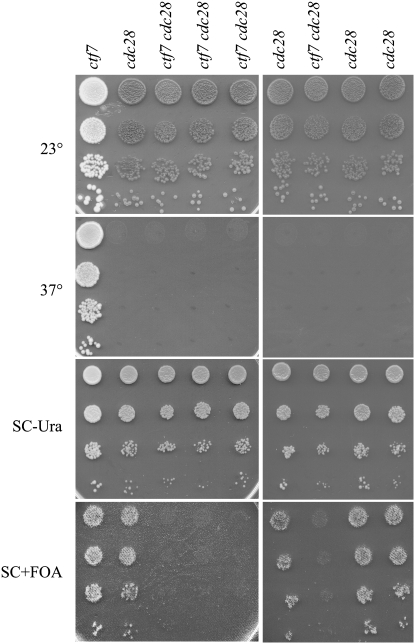
To directly test the model that ctf7 mutant cells are inviable in the presence of diminished CDK function, a reporter strain (CTF7∷HIS3, ctf7-203-LEU2, ade2, ade3 harboring a CEN CTF7 URA3 plasmid) was mated to two different cdc28 mutant strains (cdc28-cte2-HIS3 or cdc28-cte8-HIS3). These cdc28 alleles were of particular interest because they exhibit defects in genetic stability, consistent with a model that Cdc28p functions in chromosome segregation (Kitazono and Kron 2002). The resulting diploids were sporulated and eight His+ Leu+ Ura+ strains that also exhibited temperature sensitivity at 37° (attributable to cdc28) selected for further analyses (Figure 3). Tenfold serial dilutions of the parental ctf7-203 strain and all eight spores were plated on uracil-deficient medium (to maintain the CEN CTF7 URA3 plasmid) and then replica plated two times onto medium plates supplemented with 5-FOA to counterselect cells harboring the CEN URA3 CTF7 plasmid. If ctf7 and cdc28 alleles are indeed synthetically lethal, approximately half of these strains should remain 5-FOA resistant while the other half should be 5-FOA sensitive and thus inviable. Indeed, four of the eight strains exhibited robust growth on 5-FOA plates identical to the ctf7-203 parent strain (Figure 3). The remaining four strains were unable to survive exposure to medium containing 5-FOA. These findings reveal that these four strains contain both cdc28 and ctf7 alleles and that this combination is lethal (Figure 3).
Figure 3.—
ctf7 cdc28 double-mutant strains are inviable. Tenfold dilutions of ctf7 parental cells and eight spores obtained from ctf7 cdc28 heterozygous diploids. All spores harbor CEN CTF7 URA3 plasmid (growth on SC −Ura medium) but are inviable on rich medium placed at 37°, indicating that each spore contains the temperature-sensitive cdc28 allele. Replica plating onto medium supplemented with 5-FOA counterselects for cells containing the URA3-based plasmid. See text for details.
Mutational analyses of candidate Ctf7p phosphorylation sites:
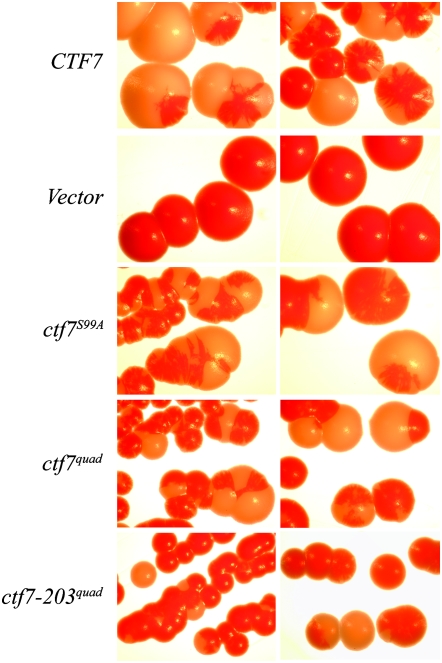
ATP-analog-specific Cdc28p kinase reactions identified Ctf7p as an in vitro substrate for Cdc28p phosphorylation (Ubersax et al. 2003). Ctf7p is a small protein of 281 amino acids and contains only one full-CDK-consensus phosphorylation site (S99) and three partial-consensus sites (S67, T94, and S105). If Cdc28p phosphorylates Ctf7p to regulate cohesion establishment, then these candidate phosphorylation residues should be critical for cell viability. To test this model, we used PCR-based site-directed mutagenesis to replace each of these residues with alanine (herein referred to as ctf7phos− constructs). The mutated regions were cloned into pRS314 (CEN TRP1 plasmid) and resequenced to ensure that no other mutations were generated. The resulting plasmids were then transformed into a ctf7 null ade2 ade3 strain in which viability was maintained by a CEN ADE3 URA3 CTF7 plasmid. As controls, we included in our analyses CEN TRP1 vector alone and vector harboring wild-type CTF7. Independent isolates of the resulting transformants were placed on rich nonselective medium and assayed for plasmid loss. As expected, ctf7 null ade2 ade3 cells harboring CEN URA3 ADE3 CTF7 and transformed with a CEN TRP vector gave rise to solid red colonies while cells transformed with CEN TRP CTF7 produced highly sectored red/white colonies (Figure 4). Importantly, cells transformed with any of the four single candidate ctf7phos− alleles also produced highly sectored red/white colonies (Figure 4). We then generated a quadruple ctf7phos− mutant (S99A, S67A, T94A, and S105A herein termed a “quad” mutant). When transformed into the reporter assay, this construct also gave rise to highly sectored red/white colonies (Figure 4). Thus, Ctf7p mutated at any or all consensus CDK phosphorylation residues is competent to provide for Ctf7p function at 23°.
Figure 4.—
CTF7 mutated at candidate CDK phosphorylation sites maintain cell viability. ctf7 null ade2 ade3 strains harboring CEN ADE3 URA3 CTF7 and CEN TRP1 plasmids: the latter of which contains wild-type CTF7, CTF7 mutated at either the full-consensus CDK phosphorylation site (ctf7S99A), all candidate CDK phosphorylation sites (ctf7S99A S67A T94A S105A or quad mutant), the ctf7-203 allele in which the quad mutations were also introduced (ctf7-203quad), or vector alone (Vector). Ability of various ctf7 constructs to support cell growth is observed as white and red sectored colonies on nonselective rich medium held at 23°. Two independent isolates for each strain are shown. Colony size differences reflect only cell crowding and local nutrient conditions, not growth defects associated with ctf7 alleles. Each image in this and subsequent colony sectoring figures corresponds to ∼0.55 cm.
We next decided to test whether conserved serine/threonine residues were crucial for Ctf7p function. From sequence analyses, we identified five additional serine and threonine residues (T76, S213, T228, S252, and T255) in budding yeast that are highly conserved in fission yeast, Drosophila, mouse, and human cells (Bellows et al. 2003). We mutated each of these five residues to alanine using PCR-based site-directed mutagenesis and confirmed each construct (ctf7conserved S/T−) by sequencing. Upon transformation into our reporter strain, all five ctf7conserved S/T− constructs supported robust cell growth at 23° (supplemental Figure S2).
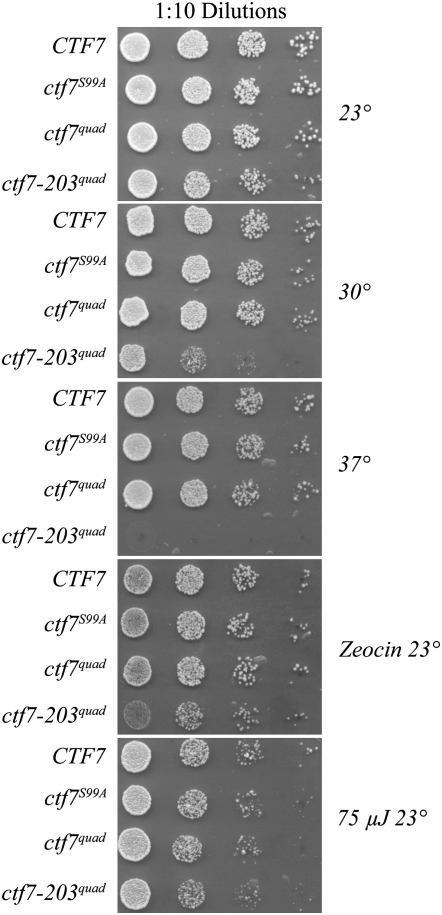
Recent studies revealed that Ctf7p activity is required during G2/M in response to DNA damage (Strom et al. 2007; Unal et al. 2007). Thus, we decided to test if any of the five ctf7phos− or five ctf7conserved S/T− constructs could support cell viability in the presence of various challenges such as elevated temperatures or exposure to genotoxic agents. To obtain cells that expressed ctf7phos− or ctf7conserved S/T− as the sole source of Ctf7p function, we isolated Trp+ Ura− cells that gave rise to solid white colonies (Figure 4 and supplemental Figure 2). Tenfold serial dilutions of the four single ctf7phos− strains, the quad ctf7phos− mutant, and the five different ctf7conserved S/T− mutant strains were then plated onto rich medium and maintained at 23°, 30°, or 37°. In parallel, duplicate samples were exposed to 75 μJ of UV irradiation or challenged with DNA double-strand breaks by incubating the cultures for 60 min in medium containing 100 μg/ml zeocin. The results show that each of the single and quad ctf7phos− alleles provided for robust growth identical to wild-type CTF7 regardless of the type of challenge (Figure 5). Only ctf7-203 mutant cells exhibited temperature and zeocin sensitivity, providing an internal control that further supported the finding that ctf7phos− strains are not genotoxic sensitive. In parallel, each of the 5 ctf7conserved S/T− constructs provided for robust growth, regardless of temperature and genotoxic challenges (supplemental Figure S3).
Figure 5.—
Cells harboring mutations in candidate Ctf7p phosphorylation residues as the sole source of Ctf7p function exhibit robust growth not only at 23° but also despite exposure to increased temperatures (30° and 37°) and to genotoxic agents (zeocin and 75 μJ of ultraviolet irradiation). Tenfold serial dilutions of each strain were plated onto rich medium and treated as indicated. See text for details.
Analyses of ctf7 alleles in combination with cak1-277:
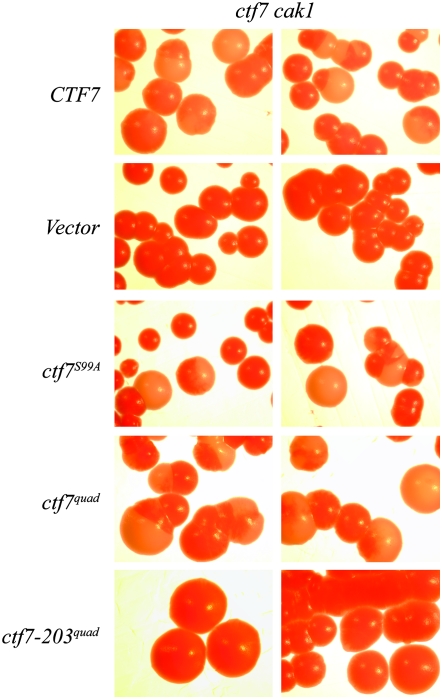
The synthetic lethality between ctf7 and cdc28 (and cak1) suggested that we might uncover phenotypes associated with either our ctf7phos− or ctf7conserved S/T− alleles if placed within the context of the original cak1 allele. To test this possibility, each construct was transformed into the original synthetic lethal strain (ade2 ade3 ctf7-203 cak1-277 harboring CEN URA3 ADE3 CTF7 plasmid). Independent isolates of each were then plated to rich medium at 23° to allow for plasmid loss. As expected, cak1 ctf7 double-mutant cells transformed with CEN TRP1 vector alone were unable to lose the reporter plasmid and gave rise only to solid red colonies. In contrast, cells transformed with CEN TRP1 CTF7 gave rise to highly sectored red and white colonies (Figure 6). Each of the ctf7 mutant constructs tested (S99A, the quad allele, and all five conserved serine/threonine point mutation constructs) provided for robust cell viability despite the presence of cak1-277 as the sole source of Cak1p function (Figure 6 and supplemental Figure S4). We included in our analyses a CEN TRP ctf7-203 plasmid further engineered to contain all four quad mutations. Despite the additional copy of the ctf7-203quad allele, these cells required the CEN CTF7 URA3 ADE3 reporter plasmid for viability (Figure 6).
Figure 6.—
CTF7 mutated at all candidate CDK phosphorylation residues are competent to support cell viability in the presence of cak1 mutations. See text for details.
Testing cak1 and cdc28 mutant cells for cohesion defects:
Synthetic lethality between ctf7 and both cak1 and cdc28 raised the possibility that cells harboring mutations in either CAK1 or CDC28 might exhibit sister chromatid-pairing defects. To test the first of these possibilities, cak1-277 mutant cells were crossed into a telomere cohesion assay strain (Antoniacci and Skibbens 2006) in which LacO arrays are integrated proximal to the telomere of chromosome IV and detected via GFP-tagged LacI-GFP expression. We inactivated cak1-277p starting both before and after START by first synchronizing log phase wild-type and cak1-277 mutant strains in G1 (α-factor) or early S phase (hydroxyurea) in 23° media. All cultures were shifted to 37° during the final 30–45 min of synchronization. The resulting cultures were washed and placed in 37° fresh media supplemented with nocodazole to arrest cells prior to anaphase onset. Parallel cell samples were then assessed for DNA content, cell morphology, Pds1p content (an inhibitor of anaphase onset) (Cohen-Fix et al. 1996), and disposition of sister chromatid loci via GFP. Both wild-type and cak1 mutant cells exposed to nocodazole arrested as large budded cells with Pds1p coincident with DAPI staining (supplemental Figure 5). Results from numerous assays reveal that preanaphase cak1 mutant cells retained a single GFP focus at levels indistinguishable from wild-type cells, regardless of whether the mutant cells were released from G1 or early S phase synchronization (supplemental Figure 5).
Our telomere-based cohesion assay cannot rule out a model in which cak1 mutant cells contain sister chromatids that are separated except at their telomeres. To address this issue, we repeated our assay but this time placed the cak1-277 allele into a strain in which the TetO arrays are integrated within 40 kb of the centromere of chromosome V and visualized by TetR-GFP expression. As before, log phase wild-type and cak1 mutant cells were synchronized in G1 using α-factor at the permissive temperature, then washed and released into fresh medium supplemented with nocodazole, and maintained at the restrictive temperature. Three independent cak1 isolates were tested for sister chromatid pairing defects in cells that retained Pds1p staining (supplemental Figure 5). Each of the cak1 isolates exhibited precocious sister chromatid dissociation levels identical to that of wild-type cells.
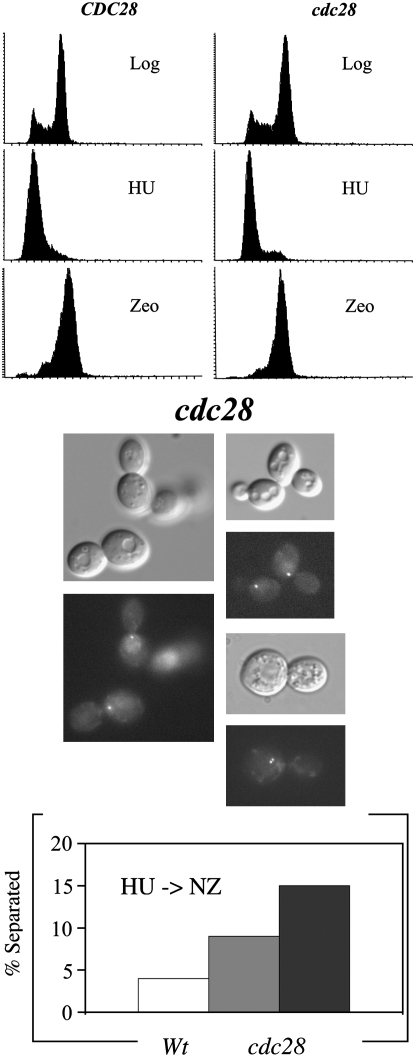
One limitation of the above analyses is that the assay procedure includes a cell cycle arrest that may allow for Cdc28p to accumulate and thus obscure a cohesion defect. To directly test for Cdc28p function in cohesion, we obtained cohesion assay diploid strains homozygous for either wild-type CDC28, or cdc28-B119, or cdc28-B28 alleles but heterozygous for the GFP-tagged loci (Kitazono et al. 2003). cdc28-B119 and cdc28-B28 cells are deficient in spindle checkpoint function but arrest prior to anaphase onset in response to DNA damage (Kitazono et al. 2003 and this study). Log phase wild-type and both cdc28 mutant strains were synchronized in early S phase using hydroxyurea and then released into rich medium at 37°. Twenty to 30 min postrelease, the media was supplemented with zeocin (100 μg/ml final concentration) to induce postreplicative double-strand breaks. In response to continued exposure to zeocin, wild-type cells arrested tightly in G2/M phase with very low levels (4%) of sister chromatid separation, indicative of a preanaphase arrest (Figure 7). In contrast, both cdc28 mutant strains exhibited precocious sister chromatid separation: the weaker of the two alleles (cdc28-B119) exhibited a small but significant level (9%) of separation whereas the stronger of the two alleles (cdc28-B28) exhibited a more robust cohesion defect (15%)—roughly four times the level observed in wild-type cells. This latter defect level is similar to that reported for many DNA replication cohesion mutant strains (Kenna and Skibbens 2003; Warren et al. 2004; Moldovan et al. 2006).
Figure 7.—
cdc28 mutant cells exhibit modest cohesion defects. (Top) Flow cytometer profiles reveal DNA contents of wild-type and cdc28 cells during log growth (log), synchronized in early S phase (HU), and then released to the restrictive temperature in the presence of zeocin (Zeo) to obtain preanaphase cells. (Middle) Micrograph pairs of cdc28 mutant cells show cell morphology above and sister chromatid loci immediately below. (Bottom) Quantification of cohesion defects in zeocin-treated wild-type cells and cdc28 mutant cells. Analyses for wild type (open bars) and both cdc28-B119 (shaded bar) and cdc28-B28 (solid bar) are shown.
DISCUSSION
How cells ensure the pairing of only sister chromatids while precluding the catastrophic pairing of nonsister chromatids remains one of the least understood and controversial mechanisms in the field of chromosome segregation (Skibbens 2008). Regardless of these models, the data clearly show that Ctf7p sister chromatid pairing reactions are tightly regulated in unperturbed cell cycles and inducible in response to specific challenges such as DNA double-strand breaks (Skibbens et al. 1999; Milutinovich et al. 2007; Strom et al. 2007; Unal et al. 2007).
One plausible mechanism of regulating cohesion establishment is predicated on the identification of Ctf7p as a phosphoprotein and potential Cdc28p substrate in vitro (Ubersax et al. 2003). Here, we uncovered a synthetic lethal interaction between ctf7-203 and a novel CAK1 allele from a nonbiased genetic screen. The essential function of Cak1p is to activate Cdc28p-CDK (Cross and Levine 1998). On the basis of this, we tested and subsequently found that ctf7 is synthetically lethal when combined with cdc28 alleles. That the interaction of interest occurs between CTF7 and CDC28 (and not between CTF7 and CAK1) is made clear by the findings that (1) cak1-277 conditional lethality is completely rescued by CDC28-CAK1-bypass hypermorphs, (2) both CDC28 hypermorphs and increased dosage of wild-type CDC28 partially rescue cak1 ctf7 synthetic lethality, and (3) cdc28 mutant cells exhibit cohesion defects. In contrast, no cohesion defect was observed in cak1 mutant cells. These results rule out an essential cohesion role for Cak1p. However, we cannot exclude a model in which Cak1p supplies some indirect role in cohesion, especially given the observation that CAK1 suppresses ctf7 cak1 lethality more efficiently than CDC28-Cak1p-bypass alleles (Figure 2).
A priori, these interactions raise numerous possibilities: (1) Cdc28p directly regulates Ctf7p, (2) Ctf7p and Cak1p coregulate Cdc28p, and (3) Cdc28p promotes cohesion in parallel to but distinct from Ctf7p reactions. The results presented here fail to support the first two of these models. First, CDC28-bypass alleles rescues cak1-277 cell temperature sensitivity but do not bypass Ctf7p nor suppress ctf7 cell phenotypes. Second, cells that express ctf7phos− mutant constructs exhibit robust growth at all temperatures tested and despite genotoxic challenges. Third, cells expressing either ctf7phos− or ctf7conserved S/T− mutant constructs failed to recapitulate the synthetic lethal interaction when combined with cak1. We also note that ctf7-203, the allele that genetically interacts with both cak1 and cdc28, is wild type at all candidate phosphorylation residues. There is also a paucity of evidence to support a model that Ctf7p affects Cdc28p-dependent cell cycle regulation. ctf7 phenotypes minimally overlap with the numerous and pleiotropic effects observed in either cak1 or cdc28 mutant cells (Kaldis et al. 1996; Thuret et al. 1996; Chun and Goebl 1997; Sutton and Freiman 1997; Skibbens et al. 1999; Toth et al. 1999).
A more likely scenario is that Cdc28p affects sister chromatid pairing independent of Ctf7p. While speculative, this model is not without basis. Of the ∼40 CDK in vitro substrates identified by Morgan and colleagues (Ubersax et al. 2003), several promote efficient sister chromatid pairing or regulate cohesion dynamics. In addition to Ctf7p, these include ORCs, Kar3p, Slk19, Cdc5p, and Pds1p (Ciosk et al. 1998; Mayer et al. 2004; Yu and Koshland 2005; Zhang et al. 2006; Shimada and Gasser 2007). How might diminished CDK function alter cohesion to the point of lethality when combined with mutations in CTF7? Using one of the CDK substrates listed above as an example, it is well established that Cdc28p phosphorylates origin of recognition complex (ORC) components and Cdc6p (Duncker et al. 1999; Sanchez et al. 1999; Nguyen et al. 2001; Weinreich et al. 2001; Cook et al. 2002; Ubersax et al. 2003). There is now strong evidence that ORCs participate in cohesin-independent sister chromatid cohesion (Dillin and Rine 1998; Suter et al. 2004; Shimada and Gasser 2007). That ORCs act in parallel to but separate from Ctf7p/cohesin is supported by findings that both orc2 smc1 and orc2 ctf7 double-mutant cells produce additive cohesion defects, compared to single-mutant cells (Shimada and Gasser 2007). Additive cohesion defects are consistent with the notion that separate sister chromatid pairing reactions function in a loci-specific manner (Sullivan et al. 2004; Chang et al. 2005; Antoniacci and Skibbens 2006; Shimada and Gasser 2007). In combination, these results indicate that ORC's cohesion function is likely to be regulated by Cdc28p and act in parallel to Ctf7p/cohesin-based activities. These studies provide for a unique starting point from which to study CDK-dependent modes of cohesion. Our study further suggests that the field must pursue alternate models regarding Ctf7p regulation.
At present it remains unclear why cak1 mutant cells exhibit negligible cohesion defects. One possibility is that the cohesion assay includes a cell cycle delay that allows for an accumulation of CDK. If true, this suggests that noncohesin-based mechanisms of cohesion maintenance (possibly via ORCs) require active CDK through G2/M. We further note that cak1-277 is not synthetically lethal when combined with a cohesin mutant: sporulation of a heterozygous cak1-277 mcd1-1 diploid strain generated the expected frequency of viable double-mutant haploid cells (A. Brands and R. V. Skibbens, unpublished data).
Acknowledgments
The authors thank Ed Winter, David Morgan, Ana Kitazono, and Stephen Kron for generously sharing of reagents and expertise and Skibbens lab members for comments throughout the course of these experiments. The authors are also indebted to Becton-Dickenson for the gift of a FACSCantoII cytometer to Lehigh University. This study was initiated under support by the National Science Foundation to R.V.S. (MCB-0212323) and completed under funding by the Susan G. Komen for the Cure Foundation (BCTR0707708). Any opinions, findings, and conclusions or recommendations expressed in this study are those of the author(s) and do not necessarily reflect the views of the National Science Foundation nor of the Susan G. Komen Foundation.
References
- Anderson, D. E., A. Losada, H. P. Erickson and T. Hirano, 2002. Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 156 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniacci, L. M., and R. V. Skibbens, 2006. Sister-chromatid telomere cohesion is nonredundant and resists both spindle forces and telomere motility. Curr. Biol. 16 902–906. [DOI] [PubMed] [Google Scholar]
- Bellows, A. M., M. A. Kenna, L. Cassimeris and R. V. Skibbens, 2003. Human EFO1p exhibits acetyltransferase activity and is a unique combination of linker histone and Ctf7p/Eco1p chromatid cohesion establishment domains. Nucleic Acids Res. 31 6334–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat, Y., and N. Kleckner, 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98 249–259. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., J. Trueheart, G. Natsoulis and G. R. Fink, 1987. 5-Flouoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154 164–175. [DOI] [PubMed] [Google Scholar]
- Brands, A., and R. V. Skibbens, 2005. Ctf7p/Eco1p exhibits acetyltransferase activity: But does it matter? Curr. Biol. 15 50–51. [DOI] [PubMed] [Google Scholar]
- Chang, C-R., C-S. Wu, Y. Hom and M. R. Gartenberg, 2005. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 19 3031–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, K. T., and M. G. Goebl, 1997. Mutational analysis of Cak1p, an essential protein kinase that regulates cell cycle progression. Mol. Gen. Genet. 256 365–375. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchanko, M. Mann et al., 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93 1067–1076. [DOI] [PubMed] [Google Scholar]
- Ciosk, R., M. Shirayama, A. Shevchenko, T. Tanaka, A. Toth et al., 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4. Mol. Cell 5 243–254. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix, O., J. M. Peters, M. W. Kirschner and D. Koshland, 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10 3081–3093. [DOI] [PubMed] [Google Scholar]
- Cook, J. G., C-H. Park, T. W. Burke, G. Leone, J. DeGregori et al., 2002. Analysis of Cdc6 function in the assembly of mammalian pre-replication complexes. Proc. Natl. Acad. Sci. USA 99 1347–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, F. R., and K. Levine, 1998. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol. Cell. Biol. 18 2923–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin, A., and J. Rine, 1998. Roles for ORC in M phase and S phase. Science 279 1733–1737. [DOI] [PubMed] [Google Scholar]
- Duncker, B. P., P. Pasero, D. Braguglia, P. Heun, M. Weinreich et al., 1999. Cyclin B-cdk kinase stimulates ORC- and Cdc6-independent steps of semiconservative plasmid replication in yeast nuclear extracts. Mol. Cell. Biol. 19 1226–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza, F. H., A. Farrell, H. Erdjument-Bromage, P. Tempst and D. O. Morgan, 1996. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science 273 1714–1717. [DOI] [PubMed] [Google Scholar]
- Espinoza, F. H., A. Farrell, J. L. Nourse, H. M. Chamberlin, O. Gileadi et al., 1998. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol. Cell. Biol. 18 6365–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem, C., C. Miled, C. Facca, J-G. Valay, G. Labesse et al., 2006. Kinase Cak1 functionally interacts with the PAF1 complex and phophatase Ssu72 via kinase Ctk1 and Bur1. Mol. Genet. Genomics 275 136–147. [DOI] [PubMed] [Google Scholar]
- Glynn, E. F., P. C. Megee, H-G. Yu, C. Mistrot, E. Unal et al., 2004. Genome-wide mapping of the cohesin complex in yeast Saccharomyces cereivisiae. PLoS Biol. 2 e259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, S., C. H. Haering and K. Nasmyth, 2003. Chromosomal cohesin forms a ring. Cell 112 765–777. [DOI] [PubMed] [Google Scholar]
- Guacci, V., D. Koshland and A. Strunnikov, 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering, C. H., J. Lowe, A. Hochwagen and K. Nasmyth, 2002. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9 773–788. [DOI] [PubMed] [Google Scholar]
- Hardy, C. F., and A. Pautz, 1996. A novel role for Cdc5p in DNA replication. Mol. Cell. Biol. 16 6775–6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, F., and H. Zou, 2005. Two human orthologues of Eco1/Ctf7p acetyltransferases are both required for proper sister-chromatid cohesion. Mol. Biol. Cell 16 3908–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., Y. Jukuda, K. Murata and A. Kimura, 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, D., and K. Nasmyth, 2005. A topological interaction between cohesin rings and a circular minichromosome. Cell 122 849–860. [DOI] [PubMed] [Google Scholar]
- Kaldis, P., A. Sutton and M. J. Solomon, 1996. The Cdk-activating kinase (CAK) from budding yeast. Cell 86 553–564. [DOI] [PubMed] [Google Scholar]
- Kenna, M. A., and R. V. Skibbens, 2003. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 23 2999–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazono, A., and S. Kron, 2002. An essential function of yeast cyclin-dependent kinase Cdc28 maintains chromosome stability. J. Biol. Chem. 277 48627–48634. [DOI] [PubMed] [Google Scholar]
- Kitazono, A., D. A. Garza and S. J. Kron, 2003. Mutations in the yeast cyclin-dependent kinase Cdc28 reveal a role in the spindle assembly checkpoint. Mol. Genet. Genomics 269 672–684. [DOI] [PubMed] [Google Scholar]
- Koshland, D., J. C. Kent and L. H. Hartwell, 1985. Genetic analysis of the mitotic transmission of minichromosomes. Cell 40 393–403. [DOI] [PubMed] [Google Scholar]
- Krantz, J. E., and C. Holm, 1990. Cloning by function: an alternate approach for identifying yeast homologs of genes from other organisms. Proc. Natl. Acad. Sci. USA 87 6629–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurlandzka, A., J. Rytka, R. Gromadka and M. Murawski, 1995. A new essential gene located on Saccharomyces cerevisiae chromosome IX. Yeast 11 885–890. [DOI] [PubMed] [Google Scholar]
- Laloraya, S., V. Guacci and D. Koshland, 2000. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M. L., I. Pot, M. Chang, H. Xu, V. Aneliunas et al., 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell 15 1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee, P. C., C. Mistrot, V. Guacci and D. Koshland, 1999. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell 4 445–450. [DOI] [PubMed] [Google Scholar]
- Melby, T. E., C. N. Ciampaglio, G. Briscoe and H. P. Erickson, 1998. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 142 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis, C., R. Ciosk and K. Nasmyth, 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91 35–45. [DOI] [PubMed] [Google Scholar]
- Milutinovich, M., E. Unal, C. Ward, R. V. Skibbens and D. Koshland, 2007. A multi-step pathway for the establishment of sister chromatid cohesion. PLoS Genet. 3 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan, G-L., B. Pfander and S. Jentsch, 2006. PCNA controls establishment of sister chromatid cohesion during S-phase. Mol. Cell 23 723–732. [DOI] [PubMed] [Google Scholar]
- Nguyen, V. Q., C. Co and J. J. Li, 2001. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411 1068–1073. [DOI] [PubMed] [Google Scholar]
- Ostapenko, D., and M. J. Solomon, 2005. Phosphorylation by Cak1 regulates the C-terminal domain kinase Ctk1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 25 3906–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., F. Winston and P. Hieter, 1990. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanchez, M., A. Calzada and A. Bueno, 1999. Functionally homologous DNA replication genes in fission and budding yeast. J. Cell Sci. 112 2381–2390. [DOI] [PubMed] [Google Scholar]
- Schiestl, R. H., and R. D. Gietz, 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16 339–346. [DOI] [PubMed] [Google Scholar]
- Shimada, K., and S. M. Gasser, 2007. The origin recognition complex functions in sister chromatids cohesion in Saccharomyces cerevisiae. Cell 128 85–99. [DOI] [PubMed] [Google Scholar]
- Skibbens, R. V., L. B. Corson, D. Koshland and P. Hieter, 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens, R. V., M. Maradeo and L. Eastman, 2007. Fork it over: the cohesion establishment factor Ctf7p and DNA replication. J. Cell Sci. 120 2471–2477. [DOI] [PubMed] [Google Scholar]
- Skibbens, R. V., 2008. Mechanisms of sister chromatid pairing. Int. Rev. Cell Mol. Biol. 269 283–339. [DOI] [PubMed] [Google Scholar]
- Strom, L., H. B. Lindroos, K. Shirahige and C. Sjogren, 2004. Postreplicative recruitment of cohesin to double-strand breaks in required for DNA repair. Mol. Cell 16 1003–1015. [DOI] [PubMed] [Google Scholar]
- Strom, L., C. Karlsson, H. B. Lindroos, S. Wedahl, Y. Katou et al., 2007. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317 242–245. [DOI] [PubMed] [Google Scholar]
- Strunnikov, A. V., V. L. Larionov and D. Koshland, 1993. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell Biol. 123 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, M., T. Higuchi, V. L. Katis and F. Uhlmann, 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase. Cell 117 471–482. [DOI] [PubMed] [Google Scholar]
- Sutton, A., and R. Freiman, 1997. The Cak1p protein kinase is required at G1/S and G2/M in the budding yeast cell cycle. Genetics 147 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter, B., A. Tong, M. Chang, L. Yu, G. W. Brown et al., 2004. The origin recognition complex links replication, sister chromatid cohesion, and transcriptional silencing in Saccharomyces cerevisiae. Genetics 167 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T., M. P. Cosma, K. Wirth and K. Nasmyth, 1999. Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98 847–858. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., T. Yonekawa, Y. Kawasaki, K. Furuya, M. Iwasaki et al., 2000. Fission yeast Eso1p is required for establishment cohesion during S phase. Mol. Cell. Biol. 20 3459–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuret, J-Y., J-G. Valay, G. Faye and C. Mann, 1996. Civ1 (CAK In Vivo), a novel Cdk-activating kinase. Cell 86 565–576. [DOI] [PubMed] [Google Scholar]
- Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer et al., 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 13 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax, J. A., E. L. Woodbury, P. N. Quang, M. Paraz, J. D. Blethrow et al., 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425 859–864. [DOI] [PubMed] [Google Scholar]
- Unal, E., A. Arbel-Eden, U. Sattler, R. Shroff, M. Lichten et al., 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16 991–1002. [DOI] [PubMed] [Google Scholar]
- Unal, E., J. M. Heidinger-Pauli and D. Koshland, 2007. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317 245–248. [DOI] [PubMed] [Google Scholar]
- Vega, H., Q. Waisfisz, M. Gordillo, N. Sakai, I. Yanagihara et al., 2005. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat.Genet. 37 468–470. [DOI] [PubMed] [Google Scholar]
- Yu, H. G., and D. Koshland, 2005. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell 123 397–407. [DOI] [PubMed] [Google Scholar]
- Wagner, M., M. Pierce and E. Winter, 1997. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 16 1305–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, C. D., D. M. Eckley, M. S. Lee, J. S. Hanna, A. Hughes et al., 2004. S-phase checkpoint genes safeguard high-fidelity sister chromatid cohesion. Mol. Biol. Cell 15 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich, M., C. Liang, H. H. Chen and B. Stillman, 2001. Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc. Natl. Acad. Sci. USA 98 11211–11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B. C., C. M. Garrett-Engele, Z. Li, E. V. Williams, E. D. Rosenman et al., 2003. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr. Biol. 13 2025–2036. [DOI] [PubMed] [Google Scholar]
- Yao, S., and G. Prelich, 2002. Activation of the Bur1-Bur2- cyclin-dependent kinase by Cak1. Mol. Cell. Biol. 22 6750–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, T., H. H. Lim, C. S. Cheng and U. Surana, 2006. Deficiency of centromere-associated protein Slk19 causes premature nuclear migration and loss of centromeric elasticity. J. Cell Sci. 119 519–631. [DOI] [PubMed] [Google Scholar]