Abstract
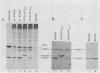
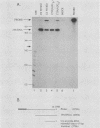
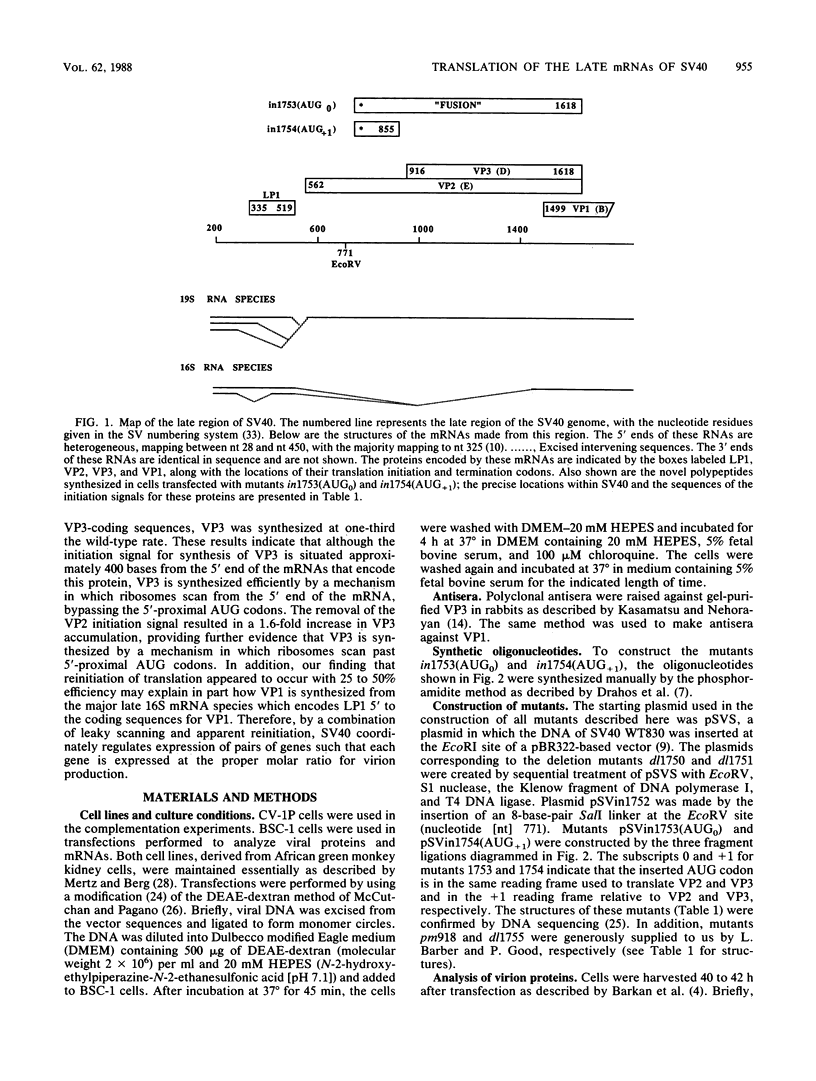
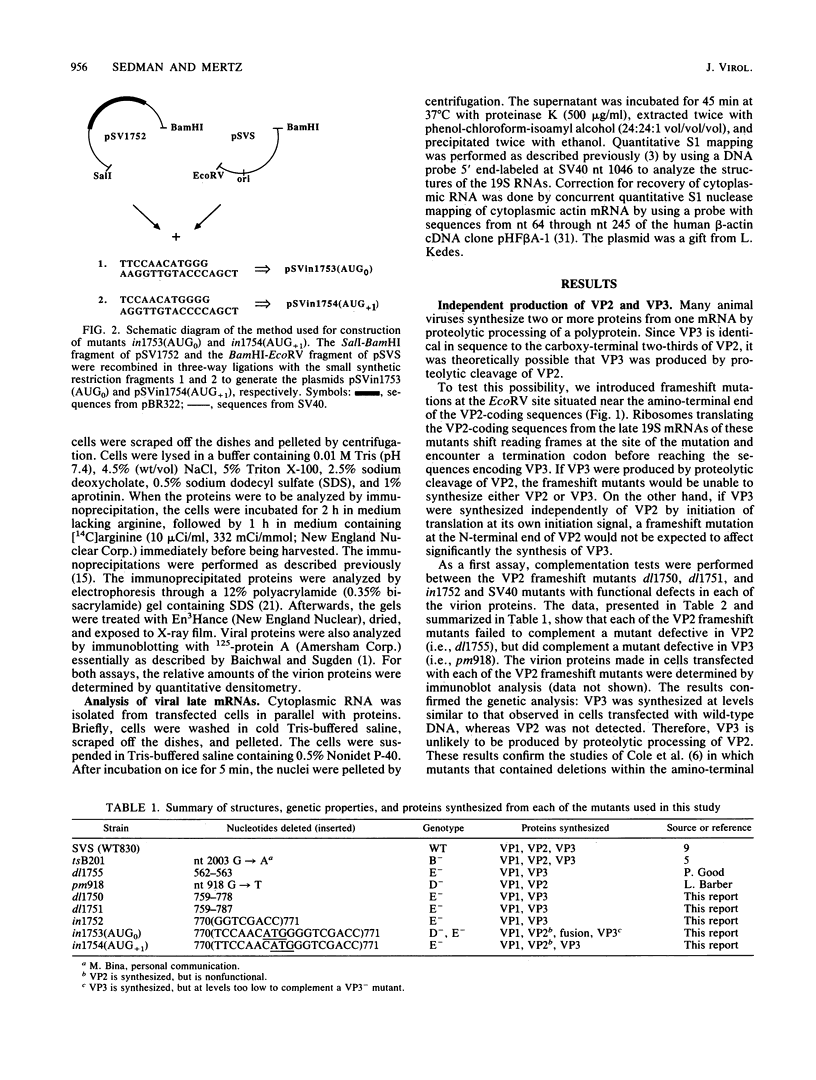
The late 19S RNAs of simian virus 40 (SV40) are functionally polycistronic, i.e., all encode both VP2 and VP3. The VP3-coding sequences are situated in the same reading frame as the VP2-coding sequences, within the carboxy-terminal two-thirds of the VP2-coding sequences. To test whether VP3 is produced by proteolytic processing of VP2, we introduced a variety of deletion and insertion mutations within the amino-terminal end of the VP2-coding sequences. Genetic and biochemical analysis of the proteins synthesized in cells transfected with these mutants indicated that VP2 and VP3 were synthesized independently of each other. A leaky scanning model for the synthesis of VP3 was tested by the insertion of a strong initiation signal (CCAACATGG) upstream of the VP3-coding sequences. When the signal was placed in the same reading frame as VP3, synthesis of VP3 was reduced by a factor of 10 to 20, whereas synthesis of the expected VP3-related fusion protein occurred at a rate similar to that observed for VP3 in cells transfected with wild-type SV40 DNA. Insertion of this strong initiation signal at the same site, but in a different reading frame, resulted in the synthesis of VP3 at one-third of the wild-type rate. Mutation of the VP2 initiator AUG resulted in a small but reproducible (1.6-fold) increase in VP3 accumulation. From these experiments we conclude that (i) VP3 is synthesized predominantly by independent initiation of translation via a leaky scanning mechanism, rather than by proteolytic processing of VP2 or direct internal initiation of translation; (ii) a strong initiation signal 5' of the VP3-coding sequences can significantly inhibit synthesis of VP3, but does not act as an absolute barrier to scanning ribosomes; (iii) approximately 70% of scanning ribosomes bypass the VP2 initiator AUG, which is present in a weak context (GGUCCAUGG), and initiate at the VP3 initiation signal located downstream; and (iv) reinitiation of translation appears to occur on the SV40 late 19S mRNAs at an efficiency of 25 to 50%.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bandyopadhyay P. K., Temin H. M. Expression from an internal AUG codon of herpes simplex thymidine kinase gene inserted in a retrovirus vector. Mol Cell Biol. 1984 Apr;4(4):743–748. doi: 10.1128/mcb.4.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Mertz J. E. The number of ribosomes on simian virus 40 late 16S mRNA is determined in part by the nucleotide sequence of its leader. Mol Cell Biol. 1984 Apr;4(4):813–816. doi: 10.1128/mcb.4.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Welch R. C., Mertz J. E. Missense mutations in the VP1 gene of simian virus 40 that compensate for defects caused by deletions in the viral agnogene. J Virol. 1987 Oct;61(10):3190–3198. doi: 10.1128/jvi.61.10.3190-3198.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J. Y., Martin R. G. Complementation analysis of simian virus 40 mutants. J Virol. 1974 May;13(5):1101–1109. doi: 10.1128/jvi.13.5.1101-1109.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Landers T., Goff S. P., Manteuil-Brutlag S., Berg P. Physical and genetic characterization of deletion mutants of simian virus 40 constructed in vitro. J Virol. 1977 Oct;24(1):277–294. doi: 10.1128/jvi.24.1.277-294.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L. K., Hohn T. Initiation of translation of the cauliflower mosaic virus genome from a polycistronic mRNA: evidence from deletion mutagenesis. EMBO J. 1984 Dec 1;3(12):2731–2736. doi: 10.1002/j.1460-2075.1984.tb02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drahos D., Galluppi G. R., Caruthers M., Szybalski W. Synthesis of the nutL DNA segments and analysis of antitermination and termination functions in coliphage lambda. Gene. 1982 Jun;18(3):343–354. doi: 10.1016/0378-1119(82)90173-1. [DOI] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Good P. J., Welch R. C., Barkan A., Somasekhar M. B., Mertz J. E. Both VP2 and VP3 are synthesized from each of the alternative spliced late 19S RNA species of simian virus 40. J Virol. 1988 Mar;62(3):944–953. doi: 10.1128/jvi.62.3.944-953.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S., Mellstrom K., Kosik E., Tamanoi F., Brugge J. Mutation of a termination codon affects src initiation. Mol Cell Biol. 1984 Sep;4(9):1738–1746. doi: 10.1128/mcb.4.9.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen H., Schümperli D., Rosenberg M. Affecting gene expression by altering the length and sequence of the 5' leader. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7698–7702. doi: 10.1073/pnas.81.24.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamatsu H., Nehorayan A. Intracellular localization of viral polypeptides during simian virus 40 infection. J Virol. 1979 Nov;32(2):648–660. doi: 10.1128/jvi.32.2.648-660.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- Kozak M. Bifunctional messenger RNAs in eukaryotes. Cell. 1986 Nov 21;47(4):481–483. doi: 10.1016/0092-8674(86)90609-4. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Translation of insulin-related polypeptides from messenger RNAs with tandemly reiterated copies of the ribosome binding site. Cell. 1983 Oct;34(3):971–978. doi: 10.1016/0092-8674(83)90554-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu C. C., Simonsen C. C., Levinson A. D. Initiation of translation at internal AUG codons in mammalian cells. Nature. 1984 May 3;309(5963):82–85. doi: 10.1038/309082a0. [DOI] [PubMed] [Google Scholar]
- Lomedico P. T., McAndrew S. J. Eukaryotic ribosomes can recognize preproinsulin initiation codons irrespective of their position relative to the 5' end of mRNA. Nature. 1982 Sep 16;299(5880):221–226. doi: 10.1038/299221a0. [DOI] [PubMed] [Google Scholar]
- Luthman H., Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983 Mar 11;11(5):1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCutchan J. H., Pagano J. S. Enchancement of the infectivity of simian virus 40 deoxyribonucleic acid with diethylaminoethyl-dextran. J Natl Cancer Inst. 1968 Aug;41(2):351–357. [PubMed] [Google Scholar]
- Mertz J. E. A detailed genetic analysis of the late complementation groups of simian virus 40. Virology. 1984 Jan 15;132(1):173–185. doi: 10.1016/0042-6822(84)90101-6. [DOI] [PubMed] [Google Scholar]
- Mertz J. E., Berg P. Defective simian virus 40 genomes: isolation and growth of individual clones. Virology. 1974 Nov;62(1):112–124. doi: 10.1016/0042-6822(74)90307-9. [DOI] [PubMed] [Google Scholar]
- Peabody D. S., Berg P. Termination-reinitiation occurs in the translation of mammalian cell mRNAs. Mol Cell Biol. 1986 Jul;6(7):2695–2703. doi: 10.1128/mcb.6.7.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody D. S., Subramani S., Berg P. Effect of upstream reading frames on translation efficiency in simian virus 40 recombinants. Mol Cell Biol. 1986 Jul;6(7):2704–2711. doi: 10.1128/mcb.6.7.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponte P., Ng S. Y., Engel J., Gunning P., Kedes L. Evolutionary conservation in the untranslated regions of actin mRNAs: DNA sequence of a human beta-actin cDNA. Nucleic Acids Res. 1984 Feb 10;12(3):1687–1696. doi: 10.1093/nar/12.3.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodia S. S., Cleveland D. W., Sollner-Webb B. A combination of RNase H and S1 nuclease circumvents an artefact inherent to conventional S1 analysis of RNA splicing. Nucleic Acids Res. 1987 Mar 11;15(5):1995–2011. doi: 10.1093/nar/15.5.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T. C., Hirano A. Structure and function of bicistronic RNA encoding the phosphoprotein and matrix protein of measles virus. J Virol. 1987 Feb;61(2):584–589. doi: 10.1128/jvi.61.2.584-589.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]