Abstract
The erythroid membrane cytoskeletal protein 4.1 is the prototypical member of a genetically and topologically complex family that is generated by combinatorial alternative splicing pathways and is localized at diverse intracellular sites including the nucleus. To explore the molecular determinants for nuclear localization, we transfected COS-7 cells with epitope-tagged versions of natural red cell protein 4.1 (4.1R) isoforms as well as mutagenized and truncated derivatives. Two distant topological sorting signals were required for efficient nuclear import of the 4.1R80 isoform: a basic peptide, KKKRER, encoded by alternative exon 16 and acting as a weak core nuclear localization signal (4.1R NLS), and an acidic peptide, EED, encoded by alternative exon 5. 4.1R80 isoforms lacking either of these two exons showed decreased nuclear import. Fusion of various 4.1R80 constructs to the cytoplasmic reporter protein pyruvate kinase confirmed a requirement for both motifs for full NLS function. 4.1R80 was efficiently imported in the nuclei of digitonin-permeabilized COS-7 cells in the presence of recombinant Rch1 (human importin α2), importin β, and GTPase Ran. Quantitative analysis of protein–protein interactions using a resonant mirror detection technique showed that 4.1R80 bound to Rch1 in vitro with high affinity (KD = 30 nM). The affinity decreased at least 7- and 20-fold, respectively, if the EED motif in exon 5 or if 4.1R NLS in exon 16 was lacking or mutated, confirming that both motifs were required for efficient importin-mediated nuclear import of 4.1R80.
INTRODUCTION
Protein 4.1 was first identified in the erythrocyte plasma membrane. It links the spectrin–actin cytoskeleton to specific transmembrane proteins (Anderson and Lovrien, 1984; Pasternack et al., 1985; Jöns and Drenckhahn, 1992; Hemming et al., 1994; Marfatia et al., 1995). In nucleated cells, 4.1-immunoreactive proteins are present not only in the plasma membrane but also in the nucleus and in the centrosome (Correas, 1991; Chasis et al., 1993; De Carcer et al., 1995; Shimizu et al., 1996; Krauss et al., 1997a,b; Lallena and Correas, 1997; Luque et al., 1998). In addition, 4.1 may interact with microtubules and stress fibers (Cohen et al., 1982; Correas and Avila, 1988). However, the precise identity of 4.1 isoforms present in these various subcellular structures and identification of their binding partners and putative functional roles remain to be defined.
The wide cellular distribution of 4.1 may reflect the coexistence in the cell of various isoforms of the protein generated by alternative splicing and/or by expression of multiple 4.1 genes (Granger and Lazarides, 1985; Anderson et al., 1988; Chasis et al., 1993; Gascard et al. 1998). The best characterized 4.1 gene, designated red cell protein 4.1 (4.1R) to reflect its abundant expression in red cells, also encodes multiple distinct isoforms in nucleated cells via complex alternative pre-mRNA splicing pathways (Ngai et al., 1987; Conboy et al., 1988, 1991; Tang et al., 1990; Chasis et al., 1993; Gascard et al., 1998). In addition to splicing of numerous exons throughout the coding sequence, variable use of two alternative start codons leads to the synthesis of 4.1R isoforms of ∼135 kDa (initiated at upstream AUG-1) and 80 kDa (initiated at downstream AUG-2). The major erythroid 80-kDa isoform, 4.1R80, is composed of four chymotryptic fragments: a 30-kDa N-terminal domain, a 16-kDa domain, a 10-kDa domain, and a 24 kDa C-terminal domain (Leto and Marchesi, 1984). The 30-kDa domain contains binding sites for the transmembrane proteins glycophorin C, and band 3 and is thus referred to as the membrane binding domain (Anderson and Lovrien, 1984; Pasternack et al., 1985; Jöns and Drenckhahn, 1992; Hemming et al., 1994; Marfatia et al., 1995). It also binds calmodulin (Tanaka et al., 1991). The 10-kDa domain contains the spectrin–actin binding (SAB) domain (Correas et al., 1986; Schischmanoff et al., 1995). The C-terminal domain interacts with the nuclear mitotic apparatus protein (NuMA) and with elongation factor 1α (Mattagajasingh et al., 1996). No function has been attributed thus far to the 16-kDa domain.
Recent studies have shed some new insights into potential functions of 4.1R in the nucleus. 4.1R colocalizes with spliceosome assembly factors such as SC35 and may thus be involved in splicing regulation (De Carcer et al., 1995; Lallena and Correas, 1997; Lallena et al., 1998). In addition, 4.1R may play a key role in mitotic events, because it undergoes dramatic redistribution during the cell cycle (Krauss et al., 1997a) and because it binds to NuMA (Mattagajasingh et al., 1996), another key protein in mitotic events. 4.1R is present in the nucleus and in the centrosome during interphase, whereas it is also detected in the mitotic spindle during mitosis, in perichromatin during telophase, and in the midbody during cytokinesis.
Large proteins, such as 4.1R, cannot freely diffuse into the nucleus. They are transported to nuclear pore complexes by specialized shuttle proteins, which bind to specific domains of the transported protein called nuclear localization signals (NLSs; for review, see Görlich and Mattaj, 1996; Görlich, 1998). Typical NLSs, characterized by either a single or a bipartite cluster of basic residues, bind to the importin α/β heterodimer. Importin α provides the NLS binding site, whereas importin β mediates docking to the nuclear pore complex. The transfer of the trimeric NLS substrate–importin α/β complex through the nuclear pore is energy dependent and requires GTP hydrolysis by GTPase Ran. However, the recent identification of two potent NLSs with novel sequences (Siomi and Dreyfuss, 1995; Pollard et al., 1996; Michael et al., 1997) and the characterization of numerous types and isoforms of shuttle proteins (Miyamoto et al., 1997; for review, see Görlich, 1998; Wozniak et al., 1998) have revealed additional complexity and diversity in nuclear import machinery. In some instances, other regions, either immediately flanking the NLSs or more distant from them, may override or activate the NLSs (Rihs and Peters, 1989; Rihs et al., 1991; Hong and Engler, 1991; van Zee et al., 1991; Zhou et al., 1991; Gao and Knipe, 1992; Gashler et al., 1993; Jans and Jans, 1994; Schmolke et al., 1995; Douglas and Quinlan, 1996; Knuehl et al., 1996).
As a first step toward defining the potential function(s) of nuclear 4.1R proteins, we report here mapping of 4.1R domains required for its efficient nuclear import. This has been achieved by comparing the expression pattern of three major 80-kDa 4.1R isoforms after transfection into COS-7 cells and of mutants generated from these isoforms. We show that two domains of 4.1R80 are involved in efficient nuclear import of this protein. In addition, we establish that 4.1R80 is translocated to the nucleus in vitro by direct interaction with Rch1, an α subunit of the human nuclear shuttle complex importin.
MATERIALS AND METHODS
Preparation of 4.1R
4.1R was purified from human erythrocytes as described by Tyler et al. (1979) with minor modifications.
Erythroid Progenitor Isolation
Erythroblasts, isolated from human bone marrow as previously described (Gascard et al., 1998), were 95% pure, mostly at the early polychromatophilic stage as shown by May–Grunmald–Giemsa staining.
RNA Preparation and DNA Cloning
Full-length 4.1R cDNAs were cloned by reverse transcriptase-PCR of erythroblast total RNA as previously described (Gascard et al., 1998). 4.1R cDNAs were tagged at their C terminus with either SV40 large T antigen KT3 epitope or influenza viral hemagglutinin (HA) epitope before insertion into mammalian expression vectors pSV2neo (provided by Dr. P. Yaswen, Lawrence Berkeley National Laboratory) or pCDNA3 (Invitrogen, La Jolla, CA) as previously described (Gascard et al., 1998). HA epitope-tagged human generally expressed protein 4.1 (4.1G) was cloned into pCDNA3 as previously described (Parra et al., 1998). Mouse neuron-specific protein 4.1 (4.1N) was cloned into a cytomegalovirus promoter-driven mammalian expression vector as previously described (Walensky, Blackshaw, Liao, Watkins, Weier, Parra, Huganir, Conboy, Mohandas, and Snyder, unpublished data). Chicken muscle pyruvate kinase (PK) cDNA, tagged at its N-terminus with a c-myc epitope and cloned into pCDNA3 (Siomi and Dreyfuss, 1995; Pollard et al., 1996), was provided by Dr. G. Dreyfuss (Howard Hughes Medical Institute, University of Pennsylvania, Philadelphia, PA). To generate PK–4.1R fusion constructs, various 4.1R domains were amplified with a 5′ PCR primer that created a KpnI site and a 3′ PCR primer including specific sequences complementary to 4.1R followed by a stop codon and a NotI site. 4.1R PCR products were inserted at the C terminus end of PK using KpnI and NotI restriction sites.
DNA Mutagenesis
Site-directed mutagenesis of 4.1R clones was performed using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions. Truncations of various domains of 4.1R clones were performed using either the Seamless kit (Stratagene) or the splice overlap extension method (Horton et al., 1989).
Cell Culture and Transfections
NIH/3T3 cells were obtained from American Type Culture Collection (Rockville, MD), COS-7 cells were provided by Dr. C. Collins (Lawrence Berkeley National Laboratory), and HeLa cells were provided by Dr. P. Yaswen (Lawrence Berkeley National Laboratory). Cells were transiently transfected by lipofection as previously described with minor modifications (Krauss et al., 1997a; Gascard et al., 1998). Briefly, cells grown on coverslips for 24 h were incubated for 8–14 h in 1 ml of Opti-MEM I medium containing 6 μl of LipofectAMINE (Life Technologies, Gaithersburg, MD) and 2.5 μg of cDNAs encoding 4.1N, HA epitope-tagged 4.1G, various KT3 or HA epitope-tagged 4.1R isoforms, or c-myc epitope-tagged PK fusion proteins. After transfection, cells were incubated in growth medium for an additional 34–40 h.
Immunofluorescence Microscopy
Samples were processed for immunofluorescence microscopy as previously described with minor modifications (Krauss et al., 1997a). All steps were performed at room temperature. Forty-eight hours after transfection, cells were fixed with 3% paraformaldehyde for 30 min and permeabilized with 0.5% Triton X-100 for 10 min. After blocking in 10% (vol/vol) goat serum for 1 h, samples were probed either with an anti-KT3 tag monoclonal antibody (provided by Dr. B. Schwer, Cornell University Medical College, New York, NY) or with an affinity-purified anti-HA tag polyclonal antibody (Zymed Laboratories, South San Francisco, CA) diluted 1:10 and 1:300, respectively. Cells transfected with PK constructs were processed as described above using an anti-c-myc tag monoclonal antibody (Boehringer Mannheim, Indianapolis, IN) diluted to 5 μg/ml. Because the c-myc tag was located at the N terminus of the fusion protein and to ensure that PK–4.1R fusion proteins were fully translated products, in some experiments samples were processed with polyclonal 4.1R antibodies specific for the C-terminal region of the fusion constructs in addition to the anti-c-myc antibody. 4.1R antibodies against either synthetic peptide 10-1 or peptide 24-3, sequences encoded by exons 16 and 21, respectively (Krauss et al., 1997a), were used at 10 μg/ml. After incubation with primary antibodies, samples were incubated for 1 h either with anti-mouse immunoglobulin G (IgG) conjugated to Texas Red or anti-rabbit IgG conjugated to FITC (Molecular Probes, Eugene, OR), diluted 1:2000 and 1:5000 respectively. Coverslips were mounted using Vectorshield containing DAPI as a nuclear DNA staining probe (Vector Laboratories, Burlingame, CA). Microscopic analysis of the samples and image processing were performed as previously described (Krauss et al., 1997a). The distribution pattern for each 4.1R construct was expressed as mean ± SD of the percentage of cells displaying any of five nucleocytoplasmic distribution patterns in three independent transfections (200–220 cells per transfection). The individual percentages did not deviate from the mean value by >20%. The significance of the differences observed between the distribution of various 4.1R80 constructs was tested using the χ2 statistic.
Preparation of Recombinant Proteins for In Vitro Nuclear Import Assay
cDNAs encoding C-terminal HA-tagged 4.1R80 and N-terminal c-myc-tagged PK and their derived clones were inserted into pET30a(+) vector (Novagen, Madison, WI). Rch1 and importin β cloned into pQE60 vector and GTPase Ran cloned into pQE32 vector (Qiagen, Hilden, Germany) were provided by Dr. Dirk Görlich (Zentrum fur Molekulare Biologie der Universitat Heidelberg, Heidelberg, Germany). The vectors described above enabled bacterial expression of 6x histidine-tagged proteins. In addition to the histidine tag, pET30a(+) vector encoded a N-terminal S tag epitope, which was used for detection of 4.1R80 in in vitro nuclear import assays described below. Histidine-tagged proteins were purified according to instructions of the manufacturer with minor modifications (Novagen). Expression of recombinant proteins was induced in the presence of 1 mM isopropyl-1-thio-β-d-galactopyranoside for 3 h at 37°C, except for Ran and importin β, which were induced for 4 h at 25°C. After induction, cells were spun down, frozen at −70°C, thawed out quickly, and lysed by sonication for 1 min in binding buffer (Novagen) supplemented with 6 M urea. Lysate supernatants, dialyzed against binding buffer to renature proteins, were incubated with a Ni2+ matrix for 30 min. Matrix was washed with 30 vol of binding buffer and 20 vol of washing buffer (Novagen) and eluted with 3 vol of elution buffer (Novagen) diluted 1:5 (final imidazole concentration, 200 mM). Proteins were dialyzed against either PBS and 0.05% Tween 20, if used in resonant mirror detection experiments, or 110 mM potassium acetate, 20 mM HEPES, pH 7.3, and 2 mM magnesium acetate (import buffer), if used in in vitro nuclear import assays. The purity of proteins was assessed by SDS-PAGE (7% gel). Protein concentration was measured using DC protein assay reagent (Bio-Rad Laboratories, Hercules, CA).
In Vitro Nuclear Import Assay
The in vitro nuclear import assay was carried out as previously described (Adam et al., 1990; Knuehl et al., 1996) with minor modifications. Briefly, subconfluent COS-7 cells grown on coverslips were permeabilized for 5 min at room temperature in import buffer containing 50 μg/ml high-grade digitonin (Calbiochem-Novabiochem, La Jolla, CA). In one case, cells were preincubated for 15 min at room temperature with import buffer containing 100 μg/ml wheat germ agglutinin (WGA; Sigma, St. Louis, MO) before performing the nuclear import assay. The assay was initiated by flipping coverslips over 50-μl drops of import buffer supplemented with 0.5 mM GTP, an ATP regeneration system (0.5 mM ATP, 20 U/ml creatine phosphokinase, and 5 mM creatine phosphate; Sigma) and recombinant histidine-tagged proteins, including 50 μg/ml N-terminal c myc-tagged PK (either wild type or fused with the SV40 NLS, PKKKRKV) or C-terminal HA-tagged 4.1R80 (either wild type or lacking exon 16 [4.1R80 ΔE16] or bearing a mutation KKK→AAA in NLS [4.1R80 mutKKK]), 6 μg/ml Rch1, 10 μg/ml importin β, and 50 μg/ml Ran. In some samples, select components of the incubation medium were omitted: GTP, an ATP regeneration system, Rch1, or importin β. The assay was carried out for 30 min in a humidified chamber at 30°C (except for one sample at 4°C). Coverslips were fixed for 20 min in import buffer containing 3% paraformaldehyde and permeabilized for 10 min in import buffer containing 0.5% Triton X-100. Samples were blocked and incubated with primary and secondary antibodies as described in Immunofluorescence Microscopy. Primary antibody was a polyclonal antibody raised against S tag (Santa Cruz Biotechnology, Santa Cruz, CA) used at 5 μg/ml, and the secondary antibody was the anti-rabbit IgG coupled to FITC described above. In one control sample, the primary S tag antibody was pre-exhausted with the control peptide (1:100 [mol/mol] antibody:peptide) for 1 h at room temperature before use.
Preparation of Recombinant GST Fusion Proteins Used for Binding Affinity Measurements
cDNAs encoding the recombinant 30-kDa domain of 4.1R (r30 kDa) or the polypeptide encoded by exon 5 of the 30-kDa domain were cloned into pGEX-KG vector (Guan and Dixon, 1991). GST fusion proteins were purified from bacterial lysates by affinity column chromatography using glutathione-Sepharose 4B as previously described (Discher et al., 1993; Schischmanoff et al., 1995). Purified proteins were cleaved with thrombin. r30 kDa was further purified by affinity column chromatography using glutathione-Sepharose 4B to remove cleaved GST. The purity of proteins was assessed by SDS-PAGE (15% gel). The protein concentrations were determined using the following relationship: protein concentration (mg/ml) = 1.45 A280 − 0.74 A260.
Measurement of Binding Affinities
Protein–protein interactions were studied using the resonant mirror detection method (Watts and Lowe, 1994; George et al., 1995) of the IAsys system (Affinity Sensors, Cambridge, United Kingdom). All experimental procedures were carried out at 25°C with constant stirring. Aminosilane or carboxymethyldextran (CMD) cuvettes (Affinity Sensors) were activated with either 2 mM bis-(sulfosuccinimidyl) suberate or 0.2 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and 0.05 M N-hydroxysuccinimide (Pierce, Rockford, IL), respectively. Recombinant Rch1 resuspended in PBS and 0.05% Tween 20 at a concentration of 0.1 mg/ml was immobilized for 30 min on either aminosilane cuvettes or CMD cuvettes preincubated in immobilization buffer (50 mM sodium acetate, pH 4.4). PBS containing 2 mg/ml BSA (aminosilane cuvettes), or 1 M ethanolamine (CMD cuvettes) was added to reduce nonspecific binding. Importin β, various recombinant constructs of 4.1R80 or PK, and 4.1R80 purified from human red cells were probed with the Rch1-coated cuvettes. Cuvettes immobilized with BSA or recombinant GST were used as negative controls. Two dissociation constants (KD from kinetic analysis [K(D)kin] and KD from Scatchard analysis) were determined using the results from the binding assay as previously described (Nunomura et al., 1997). The KD from Scatchard analysis, derived under a variety of experimental conditions, closely matched the corresponding K(D)kin values calculated. Consequently, only K(D)kin values are presented in Results. At least two cuvettes were used to determine various binding constants, and the derived values differed by <10% between the two measurements.
RESULTS
Alternatively Spliced Isoforms of Protein 4.1R Exhibit Differential Subcellular Localization
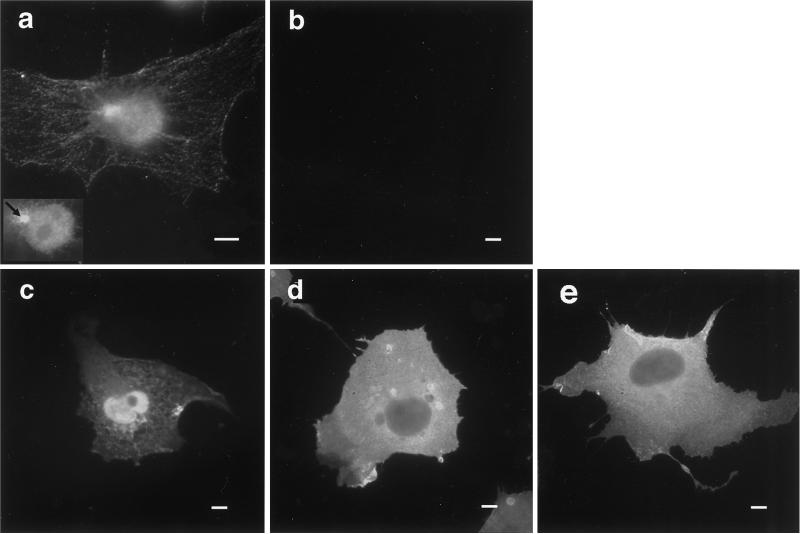
Endogenous protein 4.1R visualized by indirect immunofluorescence exhibits a complex expression pattern in nontransfected COS-7 cells (Figure 1a). Prominent nuclear staining in a punctate pattern, decoration of a cytoplasmic filament network, and colocalization with a perinuclear structure corresponding to the centrosome (Figure 1a, inset, arrow) were observed. Because nucleated cells express multiple 4.1 polypeptides via alternative pre-mRNA splicing (Granger and Lazarides, 1985; Anderson et al., 1988; Chasis et al., 1993; Gascard et al., 1998), we hypothesized that this immunofluorescence pattern represents a composite of several specifically compartmentalized 4.1R isoforms. To clearly localize individual isoforms, 4.1R cDNAs of known structure were linked to an HA epitope tag, transfected into COS-7 cells, and visualized with an antibody directed against the HA tag. Figure 1c shows that the major erythroid form of 80-kDa protein 4.1R, 4.1R80, containing the complete SAB domain including the peptide encoded by alternative exon 16, was strongly expressed in the nucleus as previously reported (Krauss et al., 1997a; Gascard et al., 1998; Luque et al., 1998). Eighty-four percent of transfected cells exhibited strong nuclear staining, as well as a variable cytoplasmic staining (Table 1). In contrast, as previously reported (Gascard et al., 1998; Luque et al., 1998), expression of a 4.1R isoform lacking the 21 amino acids encoded by exon 16 (4.1R80ΔE16 isoform) was mostly cytoplasmic (Figure 1d), with only 28% of the cells showing strong nuclear staining (Table 1). Staining was specific because it was not observed in cells transfected with the expression vector alone (Figure 1b). Additional controls showed that the differential localization was neither cell line nor epitope tag dependent, because similar results were obtained in NIH/3T3 and HeLa cells and with 4.1R isoforms tagged with the KT3 epitope (our unpublished results). This finding demonstrated unambiguously that alternative splicing generates 4.1R isoforms with distinct subcellular localization. Moreover, these data imply an important role for exon 16, which encodes part of the SAB domain, in 4.1R nuclear localization (Gascard et al., 1998; Luque et al., 1998).
Figure 1.
Expression of endogenous protein 4.1R in COS-7 cells and of HA epitope-tagged 4.1R isoforms in transfected COS-7 cells. Nontransfected COS-7 cells (a) or COS-7 cells transfected with empty expression vector (b), HA epitope-tagged 4.1R80 (c), HA epitope-tagged 4.1R80ΔE16 (d), or HA epitope-tagged 4.1R80mutKKK (e) were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for immunofluorescence using, as primary antibody, either the polyclonal protein 4.1R 24-3 antibody (Krauss et al., 1997a) to probe nontransfected cells or a polyclonal anti-HA-tag antibody to probe transfected cells. An anti-rabbit IgG coupled to FITC was used in all samples as secondary antibody. Because of COS-7 cell thickness, the nucleus and cytoplasmic network cannot be in focus in the same field. (a, inset) Focused closeup of nuclear and centrosome staining of the cell displayed in this panel. Bar, 10 μm.
Table 1.
Subcellular distribution of various 4.1R80 isoforms expressed in transfected COS-7 cells
| % of cells with strong nuclear staining
|
% of cells with weak or no nuclear staining
|
||||
|---|---|---|---|---|---|
| N | N > C | N = C | N < C | C | |
| 4.1R80 | |||||
| Wild type | 1 ± 2 | 43 ± 8 | 40 ± 3 | 7 ± 5 | 9 ± 1 |
| ΔE16 | 1 ± 0 | 9 ± 3 | 18 ± 4 | 13 ± 6 | 59 ± 2 |
| mutKKK | 1 ± 1 | 3 ± 1 | 17 ± 2 | 17 ± 6 | 62 ± 7 |
| ΔE5 | 2 ± 1 | 34 ± 3 | 5 ± 1 | 5 ± 1 | 54 ± 2 |
| mutEED | 1 ± 1 | 33 ± 6 | 16 ± 5 | 7 ± 3 | 43 ± 8 |
HA epitope-tagged 4.1R80 isoforms were expressed in transfected COS-7 cells as described in MATERIALS AND METHODS. The distribution pattern of each isoform was assessed in 200–220 cells randomly chosen in three independent experiments. Results are expressed as percentage of cells (mean ± SD) showing one of the five following distribution patterns: N, strong staining, exclusively nuclear; N > C, strong nuclear staining with variable cytoplasmic staining; N = C, strong staining equally distributed in nucleus and cytoplasm; N < C, strong cytoplasmic staining with weak nuclear expression; C, strong staining, exclusively cytoplasmic. The distribution patterns of 4.1R80 isoforms lacking either exon 16 or 5 or bearing mutations in these exons were significantly different from that of wild-type 4.1R80 (p < 10−6 for all constructs using the χ2 statistic).
Exon 16 Contains an NLS (4.1R NLS) That Is Necessary for 4.1R Nuclear Localization
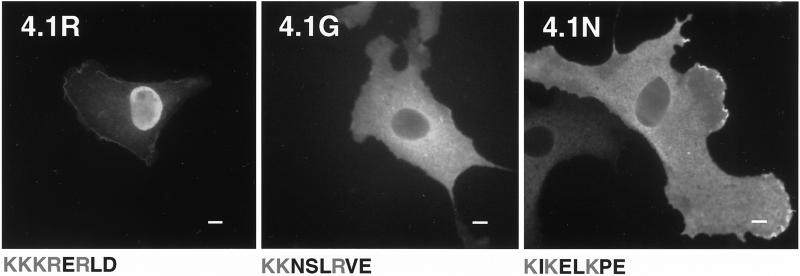
By amino acid sequence homology with other NLS-containing proteins, Correas and Avila (1988) proposed that 4.1R contains a putative NLS characterized by a cluster of basic residues, Lys-Lys-Lys-Arg-Glu-Arg (KKKRER). This motif is located at the beginning of the SAB domain, at the junction of sequences encoded by exon 13 (first lysine) and exon 16 (the remaining five amino acids). Luque et al. (1998) reported that extensive mutation of this motif results in nuclear exclusion of 4.1R. To gain further insights into the involvement of 4.1R NLS in 4.1R80 nuclear targeting, we studied the effects of limited mutations of the KKKRER cassette. Mutation of one lysine (KKK→KAK) in the putative NLS did not affect nuclear localization compared with wild-type protein (our unpublished results). By contrast, mutation of the three lysines (KKK→AAA) in NLS (isoform 4.1R80 mutKKK) induced a dramatic decrease in 4.1R80 nuclear localization (Figure 1e). Indeed, only 21% of the nuclei showed strong staining (Table 1). Furthermore, 4.1R was the only member of the recently characterized protein 4.1 family to show nuclear expression after transfection (Figure 2). Indeed, in contrast with human 4.1R80, which showed strong nuclear expression, human 4.1G and mouse 4.1N were totally excluded from the nucleus in interphase COS cells (Figure 2). Interestingly, as shown in Figure 2, a comparison of the aligned primary amino acid sequences of the three gene products revealed that the 4.1R NLS (KKKRER) was not conserved in either 4.1G (KKNSLR) or in 4.1N (KIKELK), consistent with the requirement of the 4.1R NLS for 4.1R nuclear import. However, Luque et al. (1998) showed that fusion of a polypeptide encompassing the 4.1R NLS to the cytoplasmic reporter protein β-galactosidase failed to promote nuclear targeting of the protein. This finding implied that the KKKRER motif was a weak core NLS, which required other domains of 4.1R to mediate efficient nuclear import.
Figure 2.
Comparison of cellular distribution of various proteins of the 4.1 protein family in transfected COS-7 cells. COS-7 cells were transfected with HA-tagged 4.1R80, HA-tagged 4.1G, or 4.1N. Cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for immunofluorescence using, as primary antibody, either a polyclonal anti-HA tag antibody (4.1R80 and 4.1G) or a polyclonal 4.1N antibody (Walensky et al., unpublished data). An anti-rabbit IgG coupled to FITC was used in all samples as secondary antibody. The 4.1R NLS and the corresponding aligned primary amino acid sequences in 4.1G and 4.1N are displayed. The basic residues are shown in gray. Note that because of poor conservation of the 4.1N sequence within the corresponding region of 4.1R and 4.1G, the 4.1N sequence displayed results from the best alignment achievable (Walensky et al., unpublished data). Bar, 10 μm.
Sequences Upstream of 4.1R NLS Are Required for Efficient Nuclear Localization of 4.1R
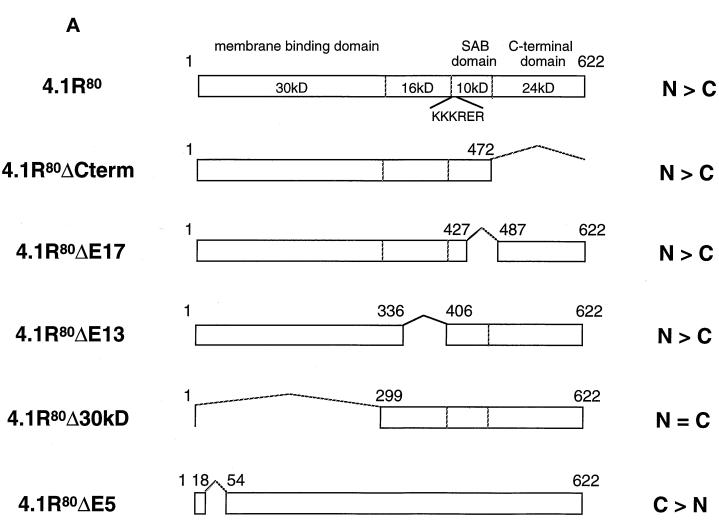
To characterize the minimal regions reconstituting a functional 4.1R NLS, we designed a series of 4.1R deletion constructs to test the requirement for other domains in specifying 4.1R nuclear localization (Figure 3A).
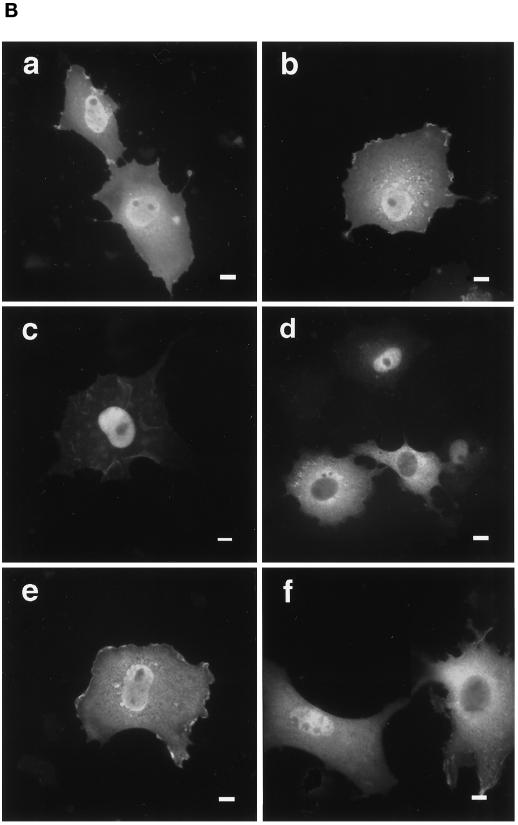
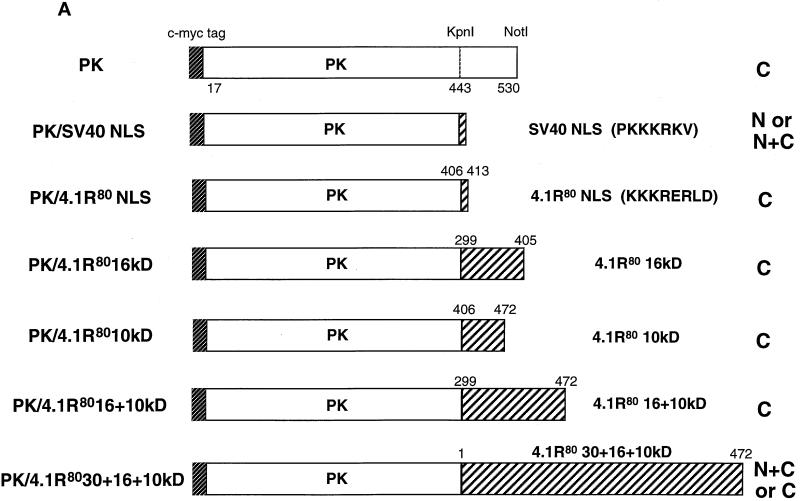
Figure 3.
Effects of truncation or point mutation of protein 4.1R domains flanking 4.1R NLS on protein 4.1R nuclear import. (A) Map of protein 4.1R constructs used to investigate nuclear localization. The four chymotryptic fragments of 4.1R80 isoform are shown at the top (30-kDa membrane binding domain, 16-kDa domain, 10-kDa SAB domain [containing the weak core NLS KKKRER], and 24-kDa domain). The protein is 622 amino acids in length. The numbers displayed refer to the amino acids present in each construct. The predominant distribution pattern is displayed for each construct and is based on examination of at least 600 cells per construct: N > C, predominant nuclear staining; C > N, predominant cytoplasmic staining; N = C, equivalent nuclear and cytoplasmic staining. (B) COS-7 cells were transfected with various HA-tagged mutants of 4.1R80. Cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and then processed for immunofluorescence using an affinity-purified polyclonal antibody against the HA tag as primary antibody and anti-rabbit IgG coupled to FITC as secondary antibody. Cell imaging was performed on samples analyzed by conventional microscopy. Cells were transfected with the following: (a) 4.1R80ΔC-term; (b) 4.1R80ΔE17; (c) 4.1R80ΔE13; (d) 4.1R80ΔE5 (the protein distribution pattern of cells expressing 4.1R80Δ30-kDa domain isoform is similar to that of 4.1R80ΔE5 isoform); (e) 4.1R80mut KH(X)7KR exon5; (f) 4.1R80mut EED exon 5. Bar, 10 μm.
The potential involvement of downstream sequences was ruled out because 4.1R80ΔC-term (Figure 3A), bearing a C-terminal deletion of residues 473–622, was predominantly nuclear (Figure 3B, a). Similarly, a smaller deletion within the SAB domain (4.1R80ΔE17; Figure 3A), corresponding to amino acids 428–486 encoded by exon 17 that have been shown to be critical for efficient spectrin–actin binding (Discher et al., 1995; Schischmanoff et al., 1995), also had no effect on 4.1R80 nuclear distribution (Figure 3B, b). Thus, nuclear import and nuclear localization can be functionally dissociated from spectrin–actin binding activity and from the reported NuMA binding domain within the C-terminus (Mattagajasingh et al., 1996).
The region upstream of the 4.1R NLS, encoded by constitutive exon 13, includes a basic dipeptide that could be part of an unusually spaced bipartite NLS (Zhu et al., 1995) with the structure KK(X)16KKKRERLD. To explore this possibility, construct 4.1R80ΔE13 (Figure 3A) was made by deletion of amino acids 337–405. This construct still exhibited nuclear localization (Figure 3B, c), indicating that this immediate upstream region was also not required for 4.1R nuclear import.
The contribution of the far upstream region comprising the N-terminal 30-kDa domain was evaluated via additional deletion constructs. Construct 4.1R80Δ30 kDa (Figure 3A), lacking amino acids 2–298 including the entire 30-kDa domain, was still permissive for nuclear localization. However, because the resulting protein is only ∼35 kDa in size, it is possible that its nuclear localization might have occurred by passive diffusion rather than by active import pathway. To explore this possibility, the effect of a smaller deletion in the 30-kDa domain was also tested. 4.1R80ΔE5 (Figure 3A), a natural 4.1R isoform lacking amino acids 19–53 because of alternative splicing of exon 5 (Gascard et al., 1998), exhibited a reduced nuclear inclusion (Figure 3B, d). Only 41% of 4.1R80ΔE5-transfected COS-7 cells showed strong nuclear staining (Table 1), compared with 84% of 4.1R80-transfected cells.
Point mutations were made to further define the nature of determinants of nuclear import within exon 5. Mutations of two doublets of basic residues, KH(X)7KR→IL(X)7IL, which might have acted as a redundant NLS, failed to reproduce the effects of exon 5 deletion (Figure 3B, e). In contrast, mutation of a cluster of negatively charged residues EED→AAA resulted in a marked decrease in 4.1R80 nuclear expression (Figure 3B, f). Only 51% of cells transfected with this mutant showed strong nuclear staining (Table 1). Similar acidic motifs have been previously reported to affect NLS function in other proteins (Jans and Jans, 1994; Hübner et al., 1997; Xiao et al., 1997). Taken together, these results suggest that two determinants are required for optimal 4.1R nuclear localization: a basic peptide KKKRER encoded within exon 16, which resembles canonical NLSs, and an upstream acidic peptide EED encoded within exon 5.
Both Weak Core 4.1R NLS and 4.1R 30-kDa Domain Are Required for Efficient Nuclear Import of Cytoplasmic Reporter Protein PK
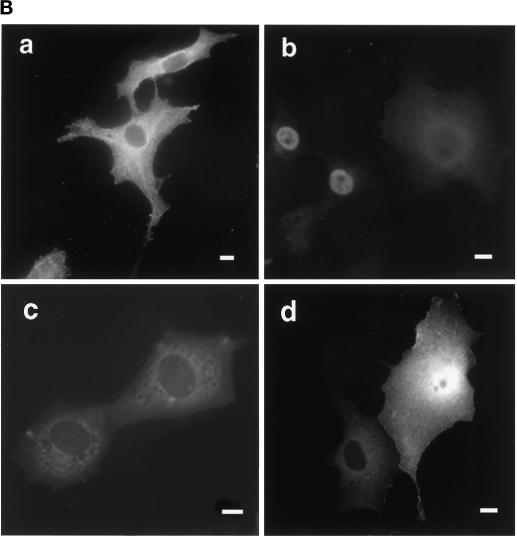
To confirm independently that a functional 4.1R NLS required two distinct motifs, located in exons 5 and 16, respectively, we tested the ability of various 4.1R constructs to promote nuclear import of chicken muscle PK (Lonberg and Gilbert, 1983), a heterologous reporter protein exclusively expressed in the cytoplasm (Figure 4, A and B, a). Although only 5% of cells transfected with PK exhibited nuclear staining, PK fused to the canonical SV40 NLS, PKKKRKV (Figure 4A; Kalderon et al., 1984), displayed nuclear expression in 75% of transfected cells (Figure 4B, b). In contrast, the weak core 4.1R NLS fused to PK (PK/4.1R80KKKRERLD) failed to target the protein to the nucleus (Figure 4, A and B, c), confirming a previous report using β-galactosidase as a reporter protein (Luque et al., 1998). Expansion of the 4.1R NLS cassette to include the entire SAB domain (PK/4.1R8010 kDa, containing amino acids 406–472), or the SAB domain plus some additional upstream sequence (PK/4.1R8016 + 10 kDa, containing amino acids 299–472), also failed to promote nuclear import of PK (Figure 4A). This fulfilled our prediction that the weak core 4.1R NLS requires other domains, in addition to the SAB domain, for efficient nuclear import. Only when the fusion protein included the 4.1R80 30-kDa domain (PK/4.1R8030 + 16 + 10 kDa, containing amino acids 1–472) was targeting of PK to the nucleus partially reconstituted (Figure 4A). In this case 45% of transfected cells exhibited nuclear localization (Figure 4B, d). These data further confirmed our finding that both the weak core NLS and a motif located within the 30-kDa domain were required for efficient nuclear targeting of 4.1R.
Figure 4.
Constructs used to identify protein 4.1R domains necessary for targeting cytoplasmic PK to the nucleus. (A) The top construct corresponds to PK lacking its first 16 amino acids, which are replaced by a c-myc epitope tag (Siomi and Dreyfuss, 1995). A construct consisting of PK fused with a KpnI–NotI fragment encoding SV40 NLS (PKKKRKV; Kalderon et al., 1984) was used as a positive control. The PK clone was used to generate fusion proteins with KpnI–NotI fragments encoding various 4.1R domains. Numbers displayed above dashed boxes refer to the 4.1R80 amino acids present in each KpnI–NotI fragment. The predominant distribution pattern is displayed for each construct and is based on examination of at least 600 cells per construct. N, strong nuclear staining with no or weak cytoplasmic staining; C, strong cytoplasmic staining with no nuclear staining; N+C, variable nuclear staining and strong cytoplasmic staining. (B) COS-7 cells transfected with the various c-myc epitope-tagged PK fusion proteins were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100, and processed for immunofluorescence using a monoclonal anti-c-myc epitope tag antibody as primary antibody and anti-mouse IgG coupled to Texas Red as secondary antibody. Cells were transfected with the following: (a) PK; (b) PK/SV40 NLS; (c) PK/4.1R80KKKRERLD (including amino acids 406–413); (d) PK/4.1R8030 + 16 + 10 kDa (including amino acids 1–472). The image b shows a representative population of the two major patterns observed: strong nuclear and weak cytoplasmic staining or similar nuclear and cytoplasmic staining accounting for 45 and 30% of transfected cells, respectively, 25% of the cells showing no nuclear staining. The image in d shows a representative population of the two major patterns observed: nuclear and cytoplasmic staining or only cytoplasmic staining accounting for 45 and 55% of transfected cells, respectively. An identical pattern was observed if sequences of the 10-kDa domain encoded by exon 17 were deleted (our unpublished results). Nuclear localization was not observed for several fusion constructs containing either the 16-kDa domain (PK/4.1R8016 kDa, including amino acids 299–406) or the NLS with additional flanking sequences (PK/4.1R8010 kDa, including amino acids 406–472; PK/4.1R8016 + 10 kDa, including amino acids 299–472). Bar, 10 μm.
Nuclear Import of 4.1R Is Mediated by Direct Interaction of 4.1R with Rch1, the Human Importin α2
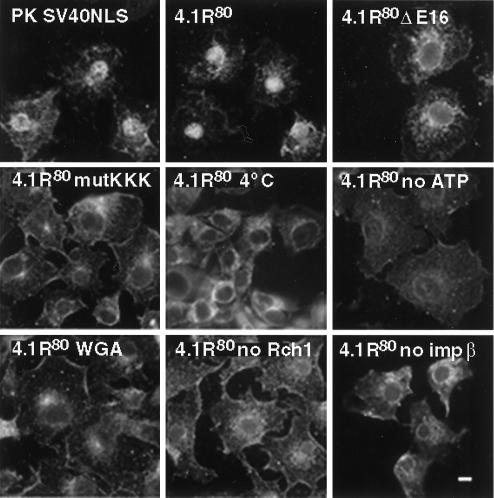
Given the complexity of the motifs involved in efficient nuclear import of 4.1R80, it became clear that defining the 4.1R80 nuclear transport pathway was critical for deciphering the molecular mechanism of 4.1R80 intracellular trafficking. Indeed, it was important to determine whether the weak core 4.1R NLS mediates direct interaction with the nuclear import machinery or, alternatively, whether 4.1R80 would be translocated to the nucleus via a piggyback effect. In the latter case, 4.1R80 might interact with an adaptor protein bearing a strong NLS, which would in turn bind to importin α (Zacksenhaus et al., 1993; Kambach and Mattaj, 1994; Mizuno et al., 1996; Jans et al., 1997). To address this issue, we carried out an in vitro import assay (Adam et al., 1990; Knuehl et al., 1996) using digitonin-permeabilized COS-7 cells. As a positive control, PK fused to SV40 NLS showed nuclear translocation (Figure 5), whereas wild-type PK was restricted to the cytoplasm (our unpublished results). Addition of recombinant 4.1R80 to permeabilized cells in the presence of Rch1, importin β, Ran, GTP, and an ATP regeneration system resulted in efficient nuclear translocation of 4.1R80 (Figure 5). In contrast, 4.1R80ΔE16 or 4.1R80mutKKK failed to be efficiently transported to the nucleus (Figure 5). 4.1R80 nuclear transport was temperature and ATP dependent: incubation at either 4 or 30°C in the absence of GTP and an ATP regeneration system resulted in inhibition of 4.1R80 nuclear translocation (Figure 5). 4.1R80 transport was also inhibited if cells were preincubated with WGA (Figure 5), a lectin known to obstruct nuclear pore complexes by interaction with nucleoporins (Radu et al., 1994). Finally, 4.1R80 transport was impaired when either Rch1 or importin β was omitted in the assay, clearly establishing that 4.1R80 nuclear transport was importin-mediated (Figure 5).
Figure 5.
In vitro nuclear import assay of PK and 4.1R80 in permeabilized COS-7 cells. Subconfluent COS-7 cells grown on coverslips were permeabilized in import buffer containing 50 μg/ml digitonin and washed twice in import buffer. Cells were then incubated with recombinant import substrates together with GTP, an ATP regeneration system, and recombinant importin α2 (Rch1), importin β, and GTPase Ran. Import substrates include PK/SV40NLS, 4.1R80, 4.1R80ΔE16, and 4.1R80mutKKK (first four panels). Control experiments showed that import was blocked by incubation at 4°C, by preincubation with WGA, by omission of GTP and the ATP regeneration system, or by omission of Rch1 or importin β (remaining panels). Cells were fixed in 3% paraformaldehyde and permeabilized in 0.5% Triton X-100. Samples were then processed for immunofluorescence using an affinity-purified polyclonal antibody against the S tag as primary antibody and anti-rabbit IgG coupled to FITC as secondary antibody. Specificity of detection was assessed using samples in which primary S-tag antibody was either omitted or pre-exhausted with control peptide. Cell imaging was performed on samples analyzed by conventional microscopy. The pattern of cells probed with wild-type PK was similar to that of 4.1R80 mutants. Bar, 10 μm.
Both Exon 16 Weak Core 4.1R NLS and Exon 5-encoded Peptide Interact with Rch1
To confirm a direct interaction of 4.1R80 with Rch1, both proteins, purified as recombinant histidine-tagged proteins, were incubated in vitro, and their binding affinities were characterized by resonant mirror detection using an IAsys machine. As shown in Table 2, recombinant 4.1R80 bound to Rch1 with high affinity (KD = 30 nM). 4.1R80, purified from human red cells, bound to Rch1 with a similar affinity (our unpublished results). In contrast, the binding affinity was significantly reduced in isoforms or mutants with alteration in either exon 16- or exon 5-encoded peptides (Table 2). Isoform 4.1R80ΔE16 and mutant 4.1R80mutKKK exhibited the most dramatic reductions, with KD values of 833 and 592 nM, respectively. This result confirmed a direct interaction between the basic KKKRER motif and Rch1 but also suggested the existence of additional lower-affinity interactions involving another region of 4.1R, such as exon 5. To test whether the N-terminal peptide encoded by exon 5 was also interacting with Rch1, additional binding studies were performed. As shown in Table 2, a decreased affinity was observed for isoform 4.1R80ΔE5 and mutant 4.1R80mutEED, with KD values of 327 and 221 nM, respectively. The interaction between exon 5 and Rch1 was further explored in shorter polypeptides representing the membrane binding domain. Thus, intact r30 kDa had an affinity of 239 nM, whereas r30 kDa lacking exon 5 (r30 kDaΔE5) had no measurable binding. Binding was also observed between Rch1 and exon 5-encoded peptide (KD = 102 nM), confirming that this motif represented a second interaction domain for nuclear import. Additional controls showed that importin β and PK fused to SV40 NLS (PKKKRKV) interacted strongly with Rch1, with KD values of 69 and 56 nM, respectively, whereas PK or GST alone exhibited no detectable binding (Table 2). Finally, none of the PK or 4.1R80 constructs bound with high affinity to importin β (our unpublished results). Taken together, these data support the theory that sequences encoded by exons 16 and 5 both participate in the interaction of 4.1R80 with Rch1.
Table 2.
Binding analysis of various 4.1R80 and pyruvate kinase constructs and of importin β to human importin α2 Rch 1
| Protein | ka (1/M·s) | kd (1/s) | K(D)kin (nM) |
|---|---|---|---|
| 4.1R80 | |||
| Wild type | 7.7 ± 0.5 × 104 | 2.3 ± 0.1 × 10−3 | 30 |
| ΔE16 | 1.2 ± 0.1 × 104 | 1.0 ± 0.3 × 10−2 | 833 |
| mutKKK | 1.4 ± 0.2 × 104 | 8.3 ± 0.5 × 10−3 | 592 |
| ΔE5 | 2.2 ± 0.2 × 104 | 7.2 ± 0.7 × 10−3 | 327 |
| mutEED | 2.2 ± 0.3 × 104 | 4.9 ± 0.8 × 10−3 | 221 |
| r30kDa | |||
| Full length | 2.3 ± 0.2 × 104 | 5.5 ± 0.5 × 10−3 | 239 |
| ΔE5 | No binding | No binding | No binding |
| GST + E5 | 8.4 ± 0.2 × 104 | 8.6 ± 0.5 × 10−3 | 102 |
| Controls | |||
| Importin β | 1.2 ± 0.1 × 105 | 8.3 ± 0.2 × 10−3 | 69 |
| PK + SV40 NLS | 9.7 ± 0.8 × 104 | 5.4 ± 0.1 × 10−3 | 56 |
| PK | No binding | No binding | No binding |
| GST | No binding | No binding | No binding |
Recombinant histidine-tagged or GST fusion proteins were expressed and purified as described in MATERIALS AND METHODS. The binding assay was carried out in PBS containing 0.05% Tween 20. From the binding curves obtained by resonant mirror detection method, ka, kd, and K(D)kin were determined using the software package FAST-Fit.
DISCUSSION
In this study we have identified naturally occurring 4.1R isoforms that can either be imported efficiently into the nucleus or be restricted predominantly to the cytoplasm in transfected cells. Two stretches of charged amino acids, located in the 10-kDa SAB and 30-kDa membrane binding domain, appear to play an important role in nuclear localization of 4.1R. Our findings strongly support the hypothesis that individual isoforms of 4.1R exhibit different subcellular localizations based on the expression of transport signals and/or protein interaction domains encoded by alternative exons. By such mechanisms, cells could simultaneously express multiple isoforms of 4.1R and sort them to various intracellular compartments. This is a feature shared by other nuclear and centrosomal proteins, such as ninein (Bouckson-Castaing et al., 1996) and NuMA (Gueth-Hallonet et al., 1996). In addition to identifying the domains in 4.1R responsible for protein nuclear localization, we also defined the mechanism of 4.1R nuclear import by showing a direct interaction of 4.1R with human importin α2 Rch1.
Detailed characterization of the requirements for 4.1R80 nuclear localization indicates that the peptide KKKRER, encoded by the junction between exons 13 and 16, may be classified as a weak core NLS (Görlich and Mattaj, 1996). This peptide is necessary for nuclear import of 4.1R80, as shown by the predominantly cytoplasmic localization of 4.1R80 mutKKK mutant and of 4.1G and 4.1N, two recently identified proteins sharing very high sequence homology with 4.1R but lacking a motif similar to the 4.1R NLS (Parra et al., 1998; Walensky et al., unpublished data). However, this motif is not sufficient to target PK to the nucleus, consistent with other observations that the function of weak core NLS motifs requires other regions of the protein, either in proximity to the NLS or more distant from it (Rihs and Peters, 1989; Rihs et al., 1991; Hong and Engler, 1991; van Zee et al., 1991; Zhou et al., 1991; Gao and Knipe, 1992; Gashler et al., 1993; Jans and Jans, 1994; Schmolke et al., 1995; Douglas and Quinlan, 1996; Knuehl et al., 1996). Our study shows that this is also the case for protein 4.1R NLS, because the 30-kDa domain appears essential for optimal NLS function. A 35-amino acid deletion within the 30-kDa domain, corresponding to the natural isoform 4.1R80ΔE5 (Gascard et al., 1998), resulted in a marked decrease in nuclear localization in COS-7 cells. Although constructs lacking the 30-kDa domain were imported relatively efficiently, this is likely attributable to diffusion of the truncated 4.1R isoform (with a predicted molecular mass of ∼35 kDa) into the nucleus independent of the nuclear import pathway (Gao and Knipe, 1992). Among the PK–4.1R fusion proteins, the most efficient nuclear translocation occurred in constructs containing both 30- and 16-kDa domains in addition to the NLS.
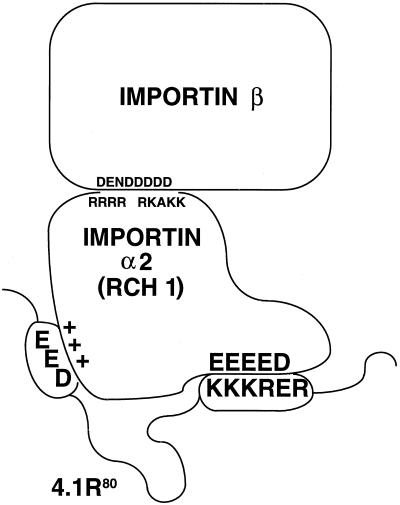
Several mechanisms may explain the requirement for the upstream domains for optimal 4.1R80 nuclear localization. First, specific amino acid residues in other regions might contribute directly to NLS function by increasing the affinity of 4.1R80 for importin α. This hypothesis is consistent with the observations that the absence of exon 5-encoded peptide or the mutation of the EED motif in exon 5 results in decreased binding affinities of 4.1R80 for Rch1. The involvement of the EED motif, in addition to the KKKRER motif, in NLS function leads us to suggest the model shown in Figure 6. In this model, we propose that the basic KKKRER peptide in the SAB domain and the N-terminal acidic EED peptide interact with specific charged residues of Rch 1, and that both interactions are required for highest affinity binding. A similar model was proposed in recent studies of the SV40 large-T antigen NLS, in which it was shown that binding affinity of the canonical NLS for importin α/β complex, as well as kinetics of SV40 NLS nuclear import, were significantly improved by negatively charged residues located immediately upstream of the basic NLS (Jans and Jans, 1994; Hübner et al., 1997; Xiao et al., 1997). In that model, the NLS and clusters of negatively charged residues of the SV40 large-T antigen interact with clusters of negatively charged and positively charged residues of importin α, respectively (Xiao et al., 1997). In the case of 4.1R80, however, the negatively charged EED motif is located much further upstream of the NLS. Precedence for influence of distant regions on function of an NLS motif has been reported previously (van Zee et al., 1991; Gao and Knipe, 1992). Second, we cannot rule out that the 30-kDa domain contains additional conformational determinants that contribute to the function of the downstream NLS. Third, it is possible that motifs distant from the basic NLS may anchor 4.1R in the nucleus by interacting with nuclear proteins in a similar manner that the A/B domain of retinoblastoma gene product p110RB1 interacts with E1A (Zacksenhaus et al., 1993). Although 4.1R NLS is located within the SAB domain, this particular interaction cannot be solely responsible for 4.1R nuclear anchoring, because deletion of essential SAB sequences encoded by exon 17 did not reduce nuclear localization. In addition, the reported interaction between 4.1R C-terminal sequences and NuMA also appears dispensable for nuclear localization, because C-terminal sequences could be deleted without effect.
Figure 6.
Schematic representation of the interaction between 4.1R and Rch1. We propose a model similar to that of Xiao et al. (1997) for the interaction of 4.1R with Rch1. In that model, the positively charged 4.1R NLS KKKRER in exon 16 and the negatively charged motif EED in exon 5 of 4.1R interact with clusters of negatively charged and positively charged residues of Rch1, respectively. Amino acids responsible for electrostatic interactions between Rch1 and importin β are also displayed.
Interestingly, the isoform exhibiting the strongest and most frequent nuclear expression in these studies, 4.1R80, is identical to the major 4.1 isoform found in the erythrocyte cytoskeleton. This observation exemplifies how a particular 4.1R isoform may have a broad distribution pattern within the cell, perhaps depending on the cell cycle and on specifically localized interacting partners competing for 4.1R binding. Thus, 4.1R80 is a component of the membrane cytoskeleton in red cells, which lack a nucleus but contain a spectrin–actin skeleton, which serves as a high-affinity target for 4.1R binding; in nucleated cells, the 4.1R80 isoform localizes not only to the plasma membrane but also to the nucleus and possibly to the centrosome (Gascard et al., 1998). Interestingly, the EED motif in exon 5 of 4.1R80, which interacts with Rch1 to promote 4.1R80 nuclear import, has been previously shown to be involved in 4.1R80 binding to the transmembrane protein band 3 (Jöns and Drenckhahn, 1992). The fact that the same motif in 4.1R80 may interact with different cellular proteins may shed light on the complex cellular distribution pattern of 4.1R80. There is compelling evidence that cytoskeletal proteins can reside in multiple cellular compartments (for review, see Smalheiser, 1996). Besides the increasing evidence for nuclear 4.1, the cytoskeletal proteins actin and tau and various actin-binding proteins have also been identified in the nucleus (Ankenbauer et al., 1989; Rimm and Pollard, 1989; Milankov and De Boni, 1993; Sahlas et al., 1993; Wang et al., 1993; Amankwah and De Boni, 1994; Parfenov et al., 1995).
In the present study, we present evidence that 4.1R nuclear import is mediated by the high-affinity NLS receptor Rch1/importin β. Our study confirms a previous report showing that Rch1, the α2 subunit of human importin, functions efficiently in binding NLS substrates (Moroianu et al., 1995). The binding affinities of NLS-containing proteins for importin α presented in our study are in accordance with those measured by others using an ELISA (Efthymiadis et al., 1997; Hübner et al., 1997; Xiao et al., 1997). These studies reported KD values of ∼10 nM for binding of optimal SV40 NLS substrates to importin α/β complex, with decreased affinities (by a factor of 4–10) for binding to importin α alone. From these data, we expected KD values ranging from 40 to 100 nM for binding of NLS substrates to Rch1, a range consistent with 4.1R80 and PK SV40NLS binding affinities reported here (30 and 56 nM, respectively). The specificity of our results is supported by the fact that 1) proteins lacking NLS (such as PK and 4.1R80ΔE16) or bearing a mutant NLS (such as 4.1R80mutKKK) do not bind or bind only very weakly to Rch1, and 2) all proteins tested, except Rch 1, bind very weakly to importin β, confirming that importin α is the only subunit to bind SV40-like NLS substrates.
These new data extend previous observations reporting nuclear localization of 4.1R (Krauss et al., 1997a; Luque et al., 1998; Gascard et al., 1998) by demonstrating that a critical feature facilitating nuclear import of 4.1R80 is the simultaneous presence of an acidic motif, EED, found in alternative exon 5 and an NLS-like basic peptide, KKKRER, encoded by alternative exon 16 (Correas and Avila, 1988). Understanding the structure and function of nuclear 4.1R is complicated by the extraordinary diversity of 4.1 isoforms expressed in the cell. Both alternative splicing of the prototypical 4.1R gene (Gascard et al., 1998) as well as expression of multiple distinct protein 4.1 gene(s) (Parra et al., 1998; Walensky et al., unpublished data) contribute to the repertoire of cellular 4.1 proteins. Thus, although the studies reported here have defined at least one nuclear isoform, the observation that multiple 4.1 polypeptides are detected in nuclear protein extracts (Correas, 1991; De Carcer et al., 1995) indicates that there are likely to be others. For example, we recently reported that 4.1R135 isoforms, initiated at AUG-1 and containing a longer N-terminal domain, may also be localized to the nucleus if they contain exon 16 and thus the weak core NLS (Gascard et al., 1998).
Our data confirm previous studies reporting a complex distribution pattern of protein 4.1R isoforms overexpressed in transfected COS cells (Gascard et al., 1998; Luque et al., 1998). The causes for such heterogeneity may result from several mechanisms. First, the 4.1R80 isoform has been shown to be targeted to various cellular compartments (Gascard et al., 1998). Second, 4.1R undergoes various posttranslational modifications, including phosphorylation (Cohen and Gascard, 1992), which may result in differential cellular sorting of the protein. This hypothesis is supported by the observation that activation of Ca2+-dependent protein kinases in keratinocytes results in redistribution of endogenous 4.1R to the periphery of the cells (Shimizu et al., 1996). Finally, nonspecific nuclear trapping of the protein after mitosis may explain the nuclear expression of isoforms lacking an optimal 4.1R NLS, such as 4.1R80ΔE16 and 4.1R80ΔE5, in a significant population of transfected cells as reported here and in earlier reports (Gascard et al., 1998; Luque et al., 1998).
Finally, it is important to note that the alternative splicing of NLS-containing exon 16 is tissue specific (Tang et al., 1988, 1990; Conboy et al., 1991) and developmentally regulated (Chasis et al., 1993). Thus nuclear protein 4.1R may play a key role in a subset of tissues and in specific steps of cell maturation or embryonic development. The identification of the transduction pathways responsible for protein 4.1R redistribution during cell division and their importance in the overall biology of the cell will be the focus of our future studies.
ACKNOWLEDGMENTS
We thank Dr. I. Auffray and Dr. L. Coulombel (Institut National de la Santé et de la Recherche Médicale U362, Villejuif, France) for isolating burst-forming unit erythroid–derived erythroblasts and purifying total RNA from these cells. We also thank Dr. G. Dreyfuss and Dr. H. Siomi (Howard Hughes Medical Institute, University of Pennsylvania, Philadelphia, PA) for providing us with the c-myc epitope-tagged PK construct, Dr. D. Görlich for providing us with the histidine epitope-tagged Rch1, importin β, and small GTPase Ran constructs, and Dr. B. Schwer (Cornell University Medical College, New York, NY) for providing us with the anti-KT3 tag antibody. We thank Dr. S. Snyder (The Johns Hopkins School of Medicine, Baltimore) for providing us with mouse 4.1N cDNA and mouse 4.1N antibody. We are very grateful to Dr. D. Callahan and K. Benson (Lawrence Berkeley National Laboratory, Berkeley, CA) for their invaluable help in cell imaging and to Linda Geniesse and Derek Clark for their help in preparation of the artwork. This work was supported by National Institutes of Health grant DK-32094 to (N.M.).
Abbreviations used:
- CMD
carboxymethyldextran
- 4.1G
generally expressed protein 4.1
- 4.1N
neuron-specific protein 4.1
- 4.1R
red cell protein 4.1
- HA
hemagglutinin
- IgG
immunoglobulin G
- K(D)kin
KD from kinetic analysis
- NLS
nuclear localization signal
- NuMA
nuclear mitotic apparatus protein
- PK
pyruvate kinase
- r30 kDa
recombinant 30-kDa membrane binding domain of protein 4.1R
- SAB
spectrin–actin binding
- WGA
wheat germ agglutinin
REFERENCES
- Adam SA, Sterne Marr R, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amankwah KS, De Boni U. Ultrastructural localization of filamentous actin within neuronal interphase nuclei in situ. Exp Cell Res. 1994;210:315–325. doi: 10.1006/excr.1994.1044. [DOI] [PubMed] [Google Scholar]
- Anderson RA, Correas I, Mazzucco C, Castle JD, Marchesi VT. Tissue-specific analogues of erythrocyte protein 4.1 retain functional domains. J Cell Biochem. 1988;37:269–284. doi: 10.1002/jcb.240370303. [DOI] [PubMed] [Google Scholar]
- Anderson RA, Lovrien RE. Glycophorin is linked by band 4.1 protein to the human erythrocyte membrane skeleton. Nature. 1984;307:655–658. doi: 10.1038/307655a0. [DOI] [PubMed] [Google Scholar]
- Ankenbauer T, Kleinschmidt JA, Walsh MJ, Weiner OH, Franke WW. Identification of a widespread nuclear actin binding protein. Nature. 1989;342:822–825. doi: 10.1038/342822a0. [DOI] [PubMed] [Google Scholar]
- Bouckson-Castaing V, Moudjou M, Ferguson DJ, Muchlow S, Belkaid Y, Milon G, Crocker PR. Molecular characterization of ninein, a new coiled-coil protein of the centrosome. J Cell Sci. 1996;109:179–190. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]
- Chasis JA, Coulombel L, Conboy J, McGee S, Andrews K, Kan YW, Mohandas N. Differentiation-associated switches in protein 4.1 expression. J Clin Invest. 1993;91:329–338. doi: 10.1172/JCI116189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CM, Foley SF, Korsgren C. A protein immunologically related to erythrocyte band 4.1 is found on stress fibers on non erythroid cells. Nature. 1982;299:648–650. doi: 10.1038/299648a0. [DOI] [PubMed] [Google Scholar]
- Cohen CM, Gascard P. Regulation and posttranslational modification of erythrocyte membrane and membrane-skeletal proteins. Semin Hematol. 1992;29:244–292. [PubMed] [Google Scholar]
- Conboy JG, Chan JY, Chasis JA, Kan YW, Mohandas N. Tissue-and development-specific alternative RNA splicing regulates expression of multiple isoforms of erythroid membrane protein 4.1. J Biol Chem. 1991;266:8273–8280. [PubMed] [Google Scholar]
- Conboy JG, Chan J, Mohandas N, Kan YW. Multiple protein 4.1 isoforms produced by alternative splicing in human erythroid cells. Proc Natl Acad Sci USA. 1988;85:9062–9065. doi: 10.1073/pnas.85.23.9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correas I. Characterization of isoforms of protein 4.1 present in the nucleus. Biochem J. 1991;279:581–585. doi: 10.1042/bj2790581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correas I, Avila J. Erythrocyte protein 4.1 associates with tubulin. Biochem J. 1988;255:217–221. [PMC free article] [PubMed] [Google Scholar]
- Correas I, Leto TL, Speicher DW, Marchesi VT. Identification of the functional site of erythrocyte protein 4.1 involved in spectrin-actin associations. J Biol Chem. 1986;261:3310–3315. [PubMed] [Google Scholar]
- De Carcer G, Lallena M-J, Correas I. Protein 4.1 is a component of the nuclear matrix of mammalian cells. Biochem J. 1995;312:871–877. doi: 10.1042/bj3120871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D, Parra M, Conboy JG, Mohandas N. Mechanochemistry of the alternatively spliced spectrin-actin binding domain in membrane skeletal protein 4.1. J Biol Chem. 1993;268:7186–7195. [PubMed] [Google Scholar]
- Discher DE, Winardi R, Schischmanoff PO, Parra M, Conboy JG, Mohandas N. Mechanochemistry of protein 4.1’s spectrin-actin-binding domain: ternary complex interactions, membrane binding, network integration, structural strengthening. J Cell Biol. 1995;130:897–907. doi: 10.1083/jcb.130.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Quinlan MP. Structural limitations of the Ad5 E1A 12s nuclear localization signal. Virology. 1996;220:339–349. doi: 10.1006/viro.1996.0322. [DOI] [PubMed] [Google Scholar]
- Efthymiadis A, Shao H, Hübner S, Jans DA. Kinetic characterization of the human retinoblastoma protein bipartite nuclear localization sequence (NLS) in vivo and in vitro. A comparison with the SV40 large T-antigen NLS. J Biol Chem. 1997;272:22134–22139. doi: 10.1074/jbc.272.35.22134. [DOI] [PubMed] [Google Scholar]
- Gao M, Knipe DM. Distal protein sequences can affect the function of a nuclear localization signal. Mol Cell Biol. 1992;12:1330–1339. doi: 10.1128/mcb.12.3.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascard P, Lee G, Coulombel L, Auffray I, Lum M, Parra M, Conboy JG, Mohandas N, Chasis JA. Characterization of multiple isoforms of protein 4.1R expressed during erythroid terminal differentiation. Blood. 1998;92:4404–4414. [PubMed] [Google Scholar]
- Gashler AL, Swaminathan S, Sukhatme VP. A novel repression module, an extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol Cell Biol. 1993;13:4556–4571. doi: 10.1128/mcb.13.8.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George AJ, French RR, Glennie MJ. Measurement of kinetic binding constants of a panel of antisaponin antibodies using a resonant mirror biosensor. J Immunol Methods. 1995;183:51–63. doi: 10.1016/0022-1759(95)00031-5. [DOI] [PubMed] [Google Scholar]
- Görlich D. Transport into and out of the nucleus [review] EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Mattaj IW. Nucleocytoplasmic transport [review] Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Granger BL, Lazarides E. Appearance of new variants of membrane skeletal protein 4.1 during terminal differentiation of avian erythroid and lenticular cells. Nature. 1985;313:238–241. doi: 10.1038/313238a0. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. J Biol Chem. 1991;266:17026–17030. [PubMed] [Google Scholar]
- Gueth-Hallonet C, Weber K, Osborn M. NuMa: a bipartite nuclear location signal and other functional properties of the tail domain. Exp Cell Res. 1996;225:207–218. doi: 10.1006/excr.1996.0171. [DOI] [PubMed] [Google Scholar]
- Hemming NJ, Anstee DJ, Staricoff MA, Tanner MJA, Mohandas N. Identification of the membrane attachment sites for protein 4.1 in the human erythrocyte. J Biol Chem. 1994;270:5360–5366. doi: 10.1074/jbc.270.10.5360. [DOI] [PubMed] [Google Scholar]
- Hong JS, Engler J. The amino-terminus of the adenovirus fiber protein encodes the nuclear localization signal. Virology. 1991;185:758–767. doi: 10.1016/0042-6822(91)90547-o. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Hübner S, Xiao CY, Jans DA. The protein kinase CK2 site (Ser111/112) enhances recognition of the SV40 large T-antigen nuclear localization sequence by importin. J Biol Chem. 1997;272:17191–17195. doi: 10.1074/jbc.272.27.17191. [DOI] [PubMed] [Google Scholar]
- Jans DA, Briggs LJ, Gustin SE, Jans P, Ford S, Young IG. The cytokine interleukin-5 (IL-5) effects cotransport of its receptor subunits to the nucleus in vitro. FEBS Lett. 1997;410:368–372. doi: 10.1016/s0014-5793(97)00622-4. [DOI] [PubMed] [Google Scholar]
- Jans DA, Jans P. Negative charge at the casein kinase II site flanking the nuclear localization signal of the SV40 large T-antigen is mechanistically important for enhanced nuclear import. Oncogene. 1994;9:2961–2968. [PubMed] [Google Scholar]
- Jöns T, Drenckhahn D. Identification of the binding interface involved in linkage of cytoskeletal protein 4.1 to the erythrocyte anion exchanger. EMBO J. 1992;11:2863–2867. doi: 10.1002/j.1460-2075.1992.tb05354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kambach C, Mattaj IW. Nuclear transport of the U2 snRNP-specific U2B" protein is mediated by both direct and indirect signaling mechanisms. J Cell Sci. 1994;107:1807–1816. doi: 10.1242/jcs.107.7.1807. [DOI] [PubMed] [Google Scholar]
- Knuehl C, Seelig A, Brecht B, Henklein P, Kloetzel PM. Functional analysis of eukaryotic 20S proteasome nuclear localization signal. Exp Cell Res. 1996;225:67–74. doi: 10.1006/excr.1996.0157. [DOI] [PubMed] [Google Scholar]
- Krauss SW, Chasis JA, Larabell CA, Lockett S, Gascard P, Blaschke R, Mohandas N. Structural protein 4.1 in the nucleus of human cells: dynamic rearrangements during cell division. J Cell Biol. 1997a;137:275–289. doi: 10.1083/jcb.137.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss SW, Chasis JA, Rogers C, Mohandas N, Krockmalnic G, Penman S. Structural protein 4.1 is located in mammalian centrosomes. Proc Natl Acad Sci USA. 1997b;94:7297–7302. doi: 10.1073/pnas.94.14.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallena M-J, Correas I. Transcription-dependent redistribution of nuclear protein 4.1 to SC35-enriched nuclear domains. J Cell Sci. 1997;110:239–247. doi: 10.1242/jcs.110.2.239. [DOI] [PubMed] [Google Scholar]
- Lallena M-J, Martinez C, Valcarcel J, Correas I. Functional association of nuclear protein 4.1 with premRNA splicing factors. J Cell Sci. 1998;111:1963–1971. doi: 10.1242/jcs.111.14.1963. [DOI] [PubMed] [Google Scholar]
- Leto TL, Marchesi VT. A structural model of human erythrocyte protein 4.1. J Biol Chem. 1984;259:4603–4608. [PubMed] [Google Scholar]
- Lonberg N, Gilbert W. Primary structure of chicken muscle pyruvate kinase mRNA. Proc Natl Acad Sci USA. 1983;80:3661–3665. doi: 10.1073/pnas.80.12.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque CM, Lallena M-J, Alonso MA, Correas I. An alternative domain determines nuclear localization in multifunctional protein 4.1. J Biol Chem. 1998;273:11643–11649. doi: 10.1074/jbc.273.19.11643. [DOI] [PubMed] [Google Scholar]
- Marfatia SM, Lue RA, Branton D, Chishti AH. Identification of the protein 4.1 binding interface on glycophorin C and p55, a homologue of the Drosophila discs-large tumor suppressor protein. J Biol Chem. 1995;270:715–719. doi: 10.1074/jbc.270.2.715. [DOI] [PubMed] [Google Scholar]
- Mattagajasingh SN, Huang SC, Benz EJ. Direct evidence for a nuclear localization and function of protein 4.1 in the nucleus: in vivo association with mitotic apparatus proteins. Blood. 1996;88,suppl 1:276a. [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milankov K, De Boni U. Cytochemical localization of actin and myosin aggregates in interphase nuclei in situ. Exp Cell Res. 1993;209:189–199. doi: 10.1006/excr.1993.1301. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Imamoto N, Sekimoto T, Tachibana T, Seki T, Tada S, Enomoto T, Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Okamoto T, Yokoi M, Izumi M, Kobayashi A, Hachiya T, Tamai K, Inoue T, Hanaoka F. Identification of the nuclear localization signal of mouse DNA primase: nuclear transport of p46 subunit is facilitated by interaction with p54 subunit. J Cell Sci. 1996;109:2627–2636. doi: 10.1242/jcs.109.11.2627. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin α1β and α2β heterodimers: α1 or α2 subunit binds nuclear localization signal and β subunit interacts with peptide repeat-containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngai J, Stack JH, Moon RT, Lazarides E. Regulated expression of multiple chicken erythroid membrane skeletal protein 4.1 variants is governed by differential RNA processing and translational control. Proc Natl Acad Sci USA. 1987;84:4432–4436. doi: 10.1073/pnas.84.13.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunomura W, Takakuwa Y, Tokimitsu R, Krauss SW, Kawashima M, Mohandas N. Regulation of CD44-protein 4.1 interaction by Ca2+ and calmodulin. J Biol Chem. 1997;272:30322–30328. doi: 10.1074/jbc.272.48.30322. [DOI] [PubMed] [Google Scholar]
- Parfenov V, Davis D, Pochukalina G, Sample C, Bugaeva E, Murti K. Nuclear actin filaments and their topological changes in frog oocytes. Exp Cell Res. 1995;217:385–394. doi: 10.1006/excr.1995.1101. [DOI] [PubMed] [Google Scholar]
- Parra M, Gascard P, Walensky LD, Snyder SH, Mohandas N, Conboy JG. Cloning and characterization of 4.1G (EPB41L2), a new member of the skeletal protein 4.1 (EPB41) gene family. Genomics. 1998;49:298–306. doi: 10.1006/geno.1998.5265. [DOI] [PubMed] [Google Scholar]
- Pasternack GR, Anderson RA, Leto TL, Marchesi VT. Interactions between protein 4.1 and band 3. J Biol Chem. 1985;260:3676–3683. [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Radu A, Blobel G, Wozniak RW. Nup107 is a novel nuclear pore complex protein that contains a leucine zipper. J Biol Chem. 1994;269:17600–17605. [PubMed] [Google Scholar]
- Rihs HP, Jans DA, Fan H, Peters R. The rate of nuclear cytoplasmic protein transport is determined by the casein kinase II site flanking the nuclear localization sequence of the SV40 T-antigen. EMBO J. 1991;10:633–639. doi: 10.1002/j.1460-2075.1991.tb07991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihs HP, Peters R. Nuclear transport kinetics depend on phosphorylation site containing sequences flanking the karyophilic signal of the SV40 T-antigen. EMBO J. 1989;8:1479–1484. doi: 10.1002/j.1460-2075.1989.tb03531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimm DL, Pollard TD. Purification and characterization of an Acanthamoeba nuclear actin binding protein. J Cell Biol. 1989;109:585–591. doi: 10.1083/jcb.109.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlas DJ, Milankov K, Park PC, De Boni U. Distribution of snRNPs, splicing factor SC-35 and actin in interphase nuclei: immunocytochemical evidence for differential distribution during changes in functional states. J Cell Sci. 1993;105:347–357. doi: 10.1242/jcs.105.2.347. [DOI] [PubMed] [Google Scholar]
- Schischmanoff PO, Winardi R, Discher DE, Parra MK, Bicknese SE, Witkowska HE, Conboy JG, Mohandas N. Defining of the minimal domain of protein 4.1 involved in spectrin-actin binding. J Biol Chem. 1995;270:21243–21250. doi: 10.1074/jbc.270.36.21243. [DOI] [PubMed] [Google Scholar]
- Schmolke S, Drescher P, Jahn G, Plachter B. Nuclear targeting of the tegument protein pp65 (UL83) of human cytomegalovirus: an unusual bipartite nuclear localization signal functions with other portions of the protein to mediate its efficient nuclear transport. J Virol. 1995;69:1071–1078. doi: 10.1128/jvi.69.2.1071-1078.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Takakuwa Y, Koizumi H, Ishibashi T, Okhawara A. Calcium-dependent peripheral localization of 4.1-like proteins and fodrin in cultured human keratinocytes. Biol Cell. 1996;86:19–26. [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR. Proteins in unexpected locations. Mol Biol Cell. 1996;7:1003–1014. doi: 10.1091/mbc.7.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Kadowaki K, Lazarides E, Sobue K. Ca2+-dependent regulation of the spectrin/actin interaction by calmodulin and protein 4.1. J Biol Chem. 1991;266:1134–1140. [PubMed] [Google Scholar]
- Tang TK, Leto TL, Correas I, Alonso MA, Marchesi VT, Benz EJ., Jr Selective expression of an erythroid-specific isoform of protein 4.1. Proc Natl Acad Sci USA. 1988;85:3713–3717. doi: 10.1073/pnas.85.11.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TK, Qin Z, Leto T, Marchesi VT, Benz EJ. Heterogeneity of mRNA and protein products arising from the protein 4.1 gene in erythroid and non erythroid tissues. J Cell Biol. 1990;110:617–624. doi: 10.1083/jcb.110.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JM, Hargreaves WR, Branton D. Purification of two spectrin-binding proteins: biochemical and electron microscopic evidence for site-specific reassociation between spectrin and bands 2.1 and 4.1. Proc Natl Acad Sci USA. 1979;76:5192–5196. doi: 10.1073/pnas.76.10.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zee K, Appel F, Fanning E. A hydrophobic protein sequence can override a nuclear localization signal independently of protein context. Mol Cell Biol. 1991;11:5137–5146. doi: 10.1128/mcb.11.10.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Loomis P, Zinkowski R, Binder L. A novel tau transcript in cultured human neuroblastoma cells expressing nuclear tau. J Cell Biol. 1993;121:257–267. doi: 10.1083/jcb.121.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts HJ, Lowe CR. Real-time detection and quantification of DNA hybridization by an optical biosensor. Anal Chem. 1994;66:2465–2470. doi: 10.1021/ac00119a013. [DOI] [PubMed] [Google Scholar]
- Wozniak RW, Rout MP, Aitchison JD. Karyopherins and kissing cousins [review] Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- Xiao C-Y, Hübner S, Jans DA. SV40 large tumor antigen nuclear import is regulated by the double-stranded DNA-dependent protein kinase site (Ser120) flanking the nuclear localization sequence. J Biol Chem. 1997;272:22191–22198. doi: 10.1074/jbc.272.35.22191. [DOI] [PubMed] [Google Scholar]
- Zacksenhaus E, Bremner R, Phillips RA, Gallie BL. A bipartite nuclear localization signal in the retinoblastoma gene product and its importance for biological activity. Mol Cell Biol. 1993;13:4588–4599. doi: 10.1128/mcb.13.8.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Doorbar J, Sun XY, Crawford LV, McLean CS, Frazer IH. Identification of the nuclear localization signal of human papillomavirus type 16 L1 protein. Virology. 1991;185:625–632. doi: 10.1016/0042-6822(91)90533-h. [DOI] [PubMed] [Google Scholar]
- Zhu X, Chang K-H, He D, Mancini MA, Brinkley WR, Lee W-H. The C terminus of mitosin is essential for its nuclear localization, centromere/kinetochore targeting, and dimerization. J Biol Chem. 1995;270:19545–19550. doi: 10.1074/jbc.270.33.19545. [DOI] [PubMed] [Google Scholar]