Abstract
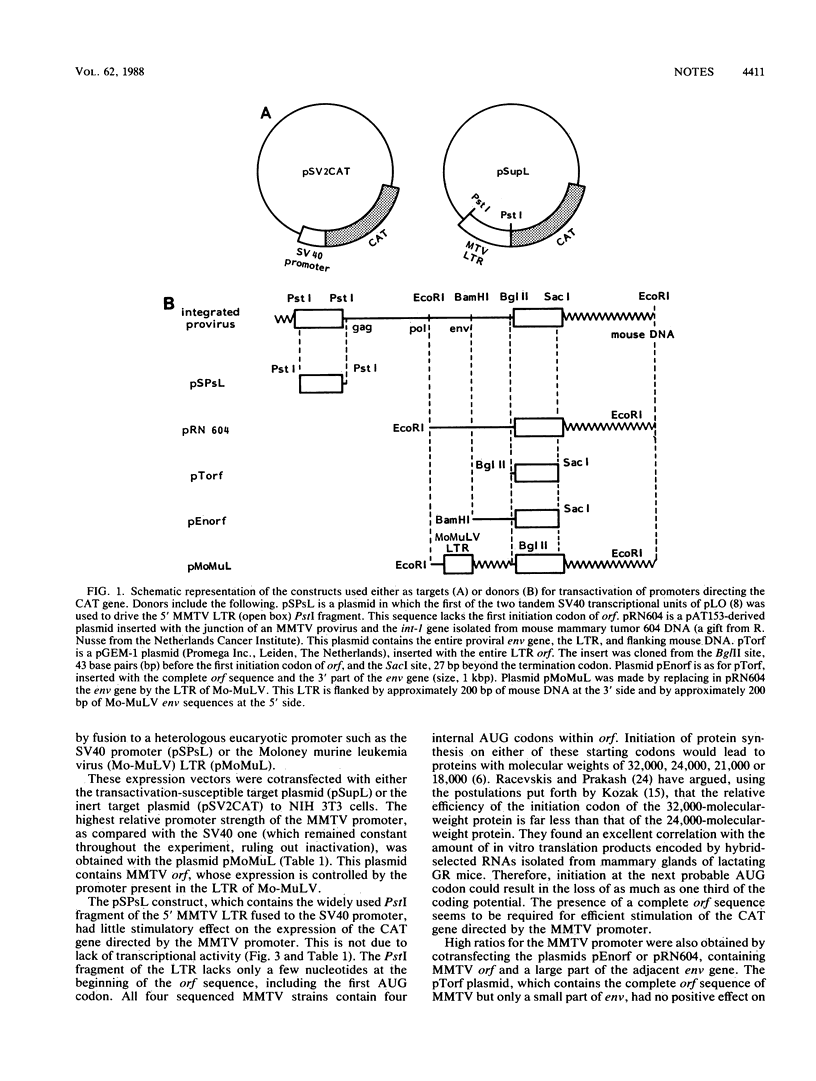
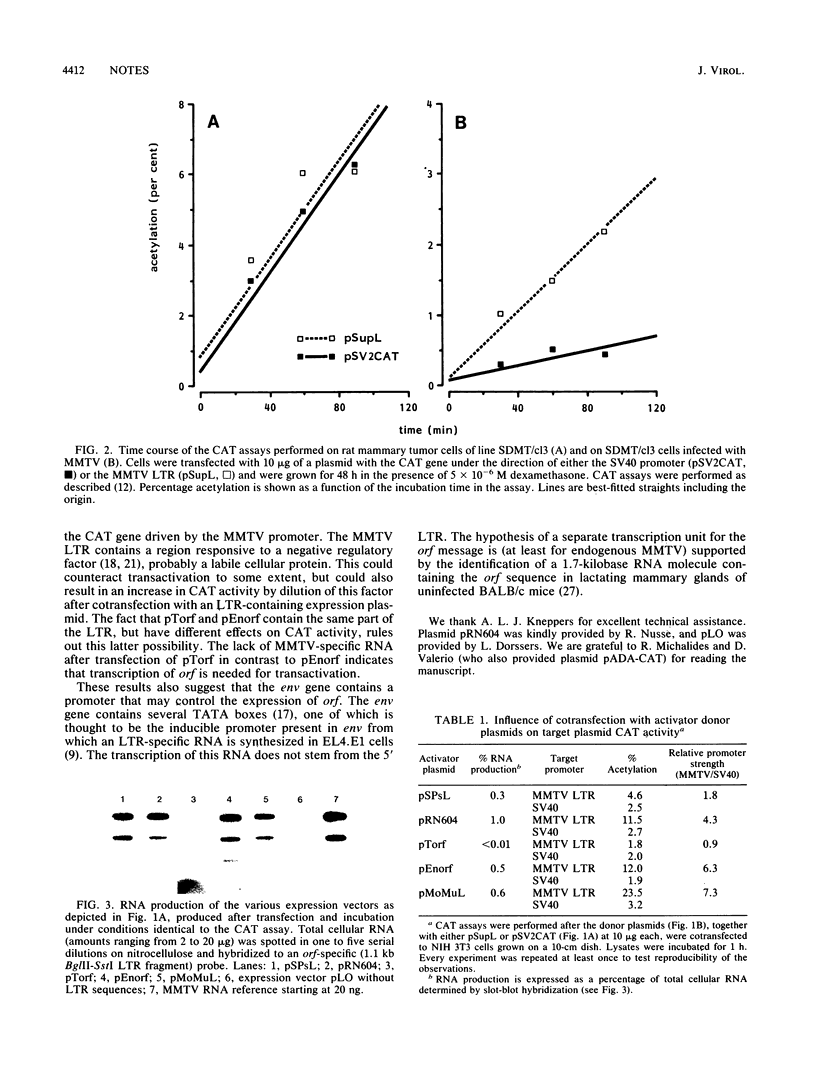
The procaryotic chloramphenicol acetyltransferase (CAT) gene controlled by the murine mammary tumor virus (MMTV) promoter shows reduced activity in a rat mammary tumor cell line after infection with MMTV but to a considerably lesser extent than the CAT gene controlled by a heterologous promoter, indicating trans-acting regulation of promoter activity by MMTV. Cotransfection of vectors capable of expressing RNA from the 3' open reading frame (orf) of MMTV with the CAT-MMTV construct resulted in enhanced CAT activity, suggesting that orf carries a transactivating potential. Since transactivation was also found with a vector containing only orf and part of the viral env gene, it was concluded that a separate transcriptional unit exists for the orf message.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arya S. K., Guo C., Josephs S. F., Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science. 1985 Jul 5;229(4708):69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- Dickson C., Peters G. Protein-coding potential of mouse mammary tumor virus genome RNA as examined by in vitro translation. J Virol. 1981 Jan;37(1):36–47. doi: 10.1128/jvi.37.1.36-47.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson C., Smith R., Peters G. In vitro synthesis of polypeptides encoded by the long terminal repeat region of mouse mammary tumour virus DNA. Nature. 1981 Jun 11;291(5815):511–513. doi: 10.1038/291511a0. [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Fleurdelys B., Hager G. L. Further evidence for the protein coding potential of the mouse mammary tumor virus long terminal repeat: nucleotide sequence of an endogenous proviral long terminal repeat. J Virol. 1983 Mar;45(3):941–949. doi: 10.1128/jvi.45.3.941-949.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A., Huang A. L., Hager G. L. Regulatory and coding potential of the mouse mammary tumor virus long terminal redundancy. J Virol. 1981 Jan;37(1):226–238. doi: 10.1128/jvi.37.1.226-238.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorssers L., Burger H., Bot F., Delwel R., Geurts van Kessel A. H., Löwenberg B., Wagemaker G. Characterization of a human multilineage-colony-stimulating factor cDNA clone identified by a conserved noncoding sequence in mouse interleukin-3. Gene. 1987;55(1):115–124. doi: 10.1016/0378-1119(87)90254-x. [DOI] [PubMed] [Google Scholar]
- Elliott J. F., Pohajdak B., Talbot D. J., Shaw J., Paetkau V. Phorbol diester-inducible, cyclosporine-suppressible transcription from a novel promoter within the mouse mammary tumor virus env gene. J Virol. 1988 Apr;62(4):1373–1380. doi: 10.1128/jvi.62.4.1373-1380.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasel N., Pearson K., Buetti E., Diggelmann H. The region of mouse mammary tumor virus DNA containing the long terminal repeat includes a long coding sequence and signals for hormonally regulated transcription. EMBO J. 1982;1(1):3–7. doi: 10.1002/j.1460-2075.1982.tb01115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallahan D., Callahan R. Mammary tumorigenesis in feral mice: identification of a new int locus in mouse mammary tumor virus (Czech II)-induced mammary tumors. J Virol. 1987 Jan;61(1):66–74. doi: 10.1128/jvi.61.1.66-74.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard D. K., Schlom J. Isolation of host-range variants of mouse mammary tumor viruses that efficiently infect cells in vitro. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5718–5722. doi: 10.1073/pnas.75.11.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy N., Knedlitschek G., Groner B., Hynes N. E., Herrlich P., Michalides R., van Ooyen A. J. Long terminal repeats of endogenous mouse mammary tumour virus contain a long open reading frame which extends into adjacent sequences. Nature. 1982 Feb 18;295(5850):622–624. doi: 10.1038/295622a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequencing of an apparent proviral copy of env mRNA defines determinants of expression of the mouse mammary tumor virus env gene. J Virol. 1983 Sep;47(3):495–504. doi: 10.1128/jvi.47.3.495-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley K. L., Toohey M. G., Peterson D. O. Transcriptional repression of a hormone-responsive promoter. Nucleic Acids Res. 1987 Sep 11;15(17):6973–6989. doi: 10.1093/nar/15.17.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982 Nov;31(1):99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Nusse R., van Ooyen A., Cox D., Fung Y. K., Varmus H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature. 1984 Jan 12;307(5947):131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- Ostrowski M. C., Huang A. L., Kessel M., Wolford R. G., Hager G. L. Modulation of enhancer activity by the hormone responsive regulatory element from mouse mammary tumor virus. EMBO J. 1984 Aug;3(8):1891–1899. doi: 10.1002/j.1460-2075.1984.tb02064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G., Brookes S., Smith R., Dickson C. Tumorigenesis by mouse mammary tumor virus: evidence for a common region for provirus integration in mammary tumors. Cell. 1983 Jun;33(2):369–377. doi: 10.1016/0092-8674(83)90418-x. [DOI] [PubMed] [Google Scholar]
- Peters G., Smith R., Brookes S., Dickson C. Conservation of protein coding potential in the long terminal repeats of exogenous and endogenous mouse mammary tumor viruses. J Virol. 1982 Jun;42(3):880–888. doi: 10.1128/jvi.42.3.880-888.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racevskis J., Prakash O. Proteins encoded by the long terminal repeat region of mouse mammary tumor virus: identification by hybrid-selected translation. J Virol. 1984 Sep;51(3):604–610. doi: 10.1128/jvi.51.3.604-610.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Sodroski J. G., Willems L., Kettmann R., Campbell K., Zaya R., Burny A., Haseltine W. A. The 3' region of bovine leukemia virus genome encodes a trans-activator protein. EMBO J. 1986 Oct;5(10):2585–2589. doi: 10.1002/j.1460-2075.1986.tb04538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodroski J. G., Rosen C. A., Haseltine W. A. Trans-acting transcriptional activation of the long terminal repeat of human T lymphotropic viruses in infected cells. Science. 1984 Jul 27;225(4660):381–385. doi: 10.1126/science.6330891. [DOI] [PubMed] [Google Scholar]
- van Ooyen A. J., Michalides R. J., Nusse R. Structural analysis of a 1.7-kilobase mouse mammary tumor virus-specific RNA. J Virol. 1983 May;46(2):362–370. doi: 10.1128/jvi.46.2.362-370.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]



