Abstract
In free-spawning invertebrates sperm–egg incompatibility is a barrier to mating between species, and divergence of gamete recognition proteins (GRPs) can result in reproductive isolation. Of interest are processes that create reproductive protein diversity within species, because intraspecific variants are potentially involved in mate choice and early speciation. Sperm acrosomes of the Pacific oyster Crassostrea gigas contain the protein bindin that bonds sperm to egg during fertilization. Oyster bindin is a single-copy gene encoding a diversity of protein variants. Oyster bindins have a conserved N-terminal region followed by one to five tandem fucose-binding lectin (F-lectin) domains. These repeats have diversified by positive selection at eight sites clustered on the F-lectin's fucose binding face. Additional bindin variants result from recombination in an intron in each F-lectin repeat. Males also express alternatively spliced bindin cDNAs with one to five repeats, but typically translate only one or two isoforms into protein. Thus, positive selection, alternative splicing, and recombination can create thousands of bindin variants within C. gigas. Models of sexual conflict predict high male diversity when females are diverse and sexual conflict is strong. The amount of intraspecific polymorphism in male GRPs may be a consequence of the relative efficiency of local (molecular recognition) and global (electrical, cortical, and physical) polyspermy blocks that operate during fertilization.
Keywords: combinatorial diversity, F-type lectin, fertilization, polymorphism, sperm-egg binding
Selection shapes molecular variation in at least two ways: by modifying the rate at which mutations are fixed or eliminated and by favoring gene architectures and mutational mechanisms that increase protein diversity. These two responses to selection are evident in many biological recognition proteins including those of host–parasite interaction, self-incompatibility, and development (1–3). These modular gene architectures allow combinatorial mechanisms like recombination and alternative splicing to produce hundreds or thousand of protein variants from a single gene. The complexity of these diversity-generating mechanisms suggests that they are evolved responses to sustained selection favoring protein diversity.
Gamete-recognition proteins (GRPs) mediating sperm–egg recognition and fusion are foremost examples of selection causing rapid protein evolution, perhaps because of a sexual conflict between sperm and egg over optimal fertilization rate (4). Because GRP divergence could initiate reproductive isolation between incipient species, studying mechanisms that create intraspecific variation is key to understanding how GRPs and the selective pressures that cause their divergence contribute to speciation. The amount of polymorphism observed in GRPs varies considerably (5). Abalone sperm lysin monomorphism (6) is contrasted by considerable polymorphism within and between populations in mussel sperm lysin-M7 (7, 8) and sea urchin sperm bindin (9, 10), yet these taxa are all free-spawning invertebrates. GRP polymorphism is minor compared with the combinatorial diversity that recombination and alternative splicing create in other biological recognition genes.
Oyster sperm have ring-shaped acrosome vesicles that exocytose to expose an insoluble protein that bonds sperm to microvilli in the egg vitelline envelope (11). The insoluble acrosomal rings (ARs) contain oyster bindin, a protein that agglutinates unfertilized eggs. Glycopeptides derived from egg surfaces block this agglutination, suggesting that oyster bindin has lectin affinity for saccharide residues (12). Sperm ARs from the Pacific oyster Crassostrea gigas contain bindin proteins of 35, 48, 63, 75, and 88 kDa with one or two size variants per male. These proteins are products of a single gene that is subjected to recombination and alternative splicing, creating, potentially, thousands of bindin variants. Models of sexual conflict indicate that the maintenance of high male diversity in a GRP requires female diversity and strong sexual conflict (13). Oyster eggs have a weak global block to polyspermy (14–17). We propose that the burden of preventing polyspermy is placed on molecular recognition mediated by bindin, increasing the relative intensity of sexual conflict at fertilization and the potential for polymorphism in oyster bindin.
Results
Bindin Size Variation.
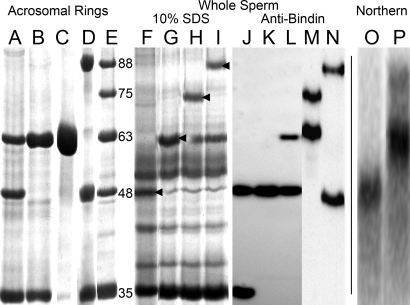
ARs isolated from individual males and separated by SDS/PAGE yield bindin proteins of five molecular masses: 35, 48, 63, 75, and 88 kDa (Fig. 1 A–E). The bindin bands are evident in 10% SDS solubilized whole sperm, indicating the different bindins are distinct proteins and not proteolytic products (Fig. 1 F–I). Peptide sequencing showed that residues 1–16 of the 48-, 63-, and 88-kDa bands are identical. Western blots, probed with a rabbit antibody raised against the 63-kDa bindin, react with all five size variants with one or two bindins per male (Fig. 1 J–N). Isolated ARs also contain a 34-kDa protein that is solubilized in 0.6 M KCl, leaving the AR intact (Fig. 1C). Peptide sequences identified this protein as molluscan sperm nuclear basic protein (18).
Fig. 1.
Size variation in oyster bindin proteins and mRNA transcripts. (A–D) AR isolates from individual males (50 μg protein per lane, Coomassie-stained). (C) The removal of the 34-kDa sperm nuclear basic protein from sample B with 0.6 M KCl. (E) AR isolates pooled from several males with 35-, 48-, 63-, 75- and 88-kDa bindins. (F–I) Extracts of whole-sperm from individual males (50 μg protein per lane). Arrows indicate oyster bindins determined by N-terminal peptide sequencing. (J–N) Western immunoblots using anti-bindin serum; preimmune serum gave no reaction. (O and P) Northern blots of 20 μg total RNA isolated from the spermatocytes of two males; O shows a male with only 48-kDa bindin; P shows a male with both 63- and 88-kDa bindins.
Bindin cDNA Variation.
Full-length bindin sequences were generated by 5′ and 3′ RACE of a spermatogenic cell cDNA template, using primers made from peptide sequences of the 48-kDa AR protein. Bindin has a signal sequence of 24 residues and an N terminus (Fig. 2, A25 to Q122) with no similarity to any protein. In the N terminus, five single amino acid substitutions occur in 4,315 aa from 39 sequences from 16 males [supporting information (SI) Fig. 7]. Two individuals had a tandem duplication of the 11-residue sequence of P84 to P94, and one individual lacked this sequence, suggesting unequal crossing-over (Fig. 2). The N terminus is followed by one to five tandem repeats of 134 residues with identity to a fucose-binding lectin whose crystal structure is solved and named the F-lectin fold (19). Oyster bindin F-lectin repeats are ≈45% similar to the consensus F-lectin fold (smart00607), and fucose-binding residues are conserved in each repeat (Fig. 2). Internal F-lectin repeats are separated by a linker (PGAK or PGAKGK); the stop codon follows the last repeat. Cloned PCRs showed that individual males have cDNAs with one to five F-lectin repeats (SI Fig. 8). Northern blots of total RNA, probed with a full-length F-lectin repeat, yield bands of the size expected for bindin mRNAs with one to five repeats with one or two bands hybridizing per individual (Fig. 1 O–P).
Fig. 2.
Deduced amino acid sequence of a five-repeat oyster bindin cDNA. Fucose-binding lectin repeats are in gray boxes; numbers inside each box indicate the repeat numbering scheme used. The fifth repeat lacks the linker sequence found between the other repeats, and the stop codon occurs directly after the last repeat (codon 809). Arrows indicate intron positions determined by sequencing genomic DNA. Black boxes mark the fucose-binding motif (H37, R64, R70) (19). A five-repeat oyster bindin cDNA is 808 aa, including the 24-residue signal-sequence (italics). The mature bindin starts at residue A25. The bracketed sequence (KKTVKPPRPPT) appears once but is absent, or repeated twice, in some molecules. Underlined sequences were obtained by peptide sequencing.
Bindin Is Single Copy with Variably Sized Introns.
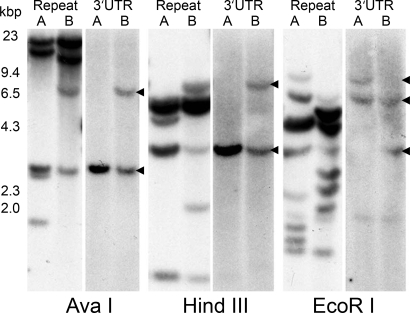
Genomic DNA from seven diploid males was digested with three restriction enzymes that do not cut in the full-length cDNA. When probed with a full-length repeat, Southern blots of all three restriction enzymes yielded multiple fragments from 1.5 to 22 kbp (Fig. 3). Of seven individuals, no two had identical hybridization patterns with the repeat probe. Genomic DNA sequences indicated an intron in the middle of each F-lectin repeat, between repeat positions 67 and 68 (Fig. 2, arrowheads). Assuming five F-lectin repeats in the genome, and a single intron in each repeat, there could be six restriction fragments (5′unique-R1|R1–R2|R2–R3|R3–R4|R4–R5|R5–3′UTR) for each gene. If every intron is a different size and contains at least one restriction site, a single 2N individual could yield 12 bands of hybridization. When probed with a full-length repeat, the AvaI and HindIII digests have four or five bands; EcoRI digests 8 to 10 bands. Band intensity varies; dark bands are presumably two or more same-size fragments. Southern blots probed with the 3′UTR sequence yield at most two bands per individual (Fig. 3, arrowheads), suggesting that C. gigas bindin is a single-copy gene. Confirmation of this conclusion awaits assembly of the complete genome sequence.
Fig. 3.
Southern blots of two diploid males (A and B) digested with three enzymes whose recognition sites are not in full-length bindin cDNA (AvaI, HindIII, and EcoRI). Shown is 10 μg of digested DNA per lane. Probes of the 3′ UTR show a maximum of two bands per individual, suggesting that oyster bindin is a single gene. Genomic DNA sequences indicate that each F-lectin repeat is bisected by an intron, and probes using a full-length repeat yield multiple bands. Of seven individuals analyzed, no two had identical patterns of hybridization with the repeat probe. HindIII-digested Lambda DNA size marker (kbp) is on the left.
Recombination.
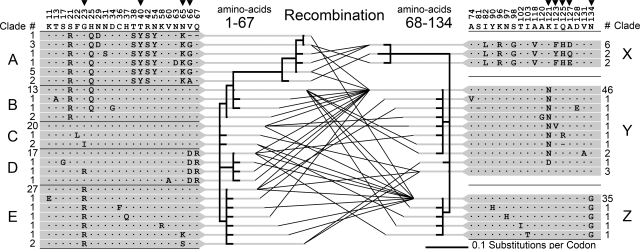
There are 41 unique F-lectin repeat amino acid sequences in the 107 repeats from 39 full-length oyster bindin cDNAs from 16 males from five global locations. A genetic algorithm finds evidence of a single recombination site between repeat amino acid positions 67 and 68, at the exact position of the F-lectin repeat intron (20). There are 24 unique sequences of repeat positions 1–67 (Fig. 4, clades A-E) and 18 unique sequences of repeat positions 68–134 (Fig. 4, clades X-Z). No internal recombination is evident in either half-repeat. Thus, recombination creates variability among F-lectin repeats by generating pairwise combinations of the 5′ and 3′ halves of each repeat (Fig. 4). These combinations are not random across clades, presumably because interclade combinations result from rare unequal crossover events (SI Table 1). Of the 39 full-length bindin cDNAs only five pairs are identical (two pairs of one repeat, two pairs of two repeats, and one pair of four repeats). All others are unique by sequence or length variation.
Fig. 4.
Recombination in the F-lectin repeat intron. Amino acid variability among 107 F-lectin repeats of 39 full-length bindin molecules is shown. Numbers above the consensus sequence indicate the amino acid position of variable sites within the repeat. Arrows indicate positively selected sites inferred by PAML. An algorithm that detects recombination finds evidence of a single recombination site between repeat positions 67 and 68. Phylogenetic relationships of the repeat halves on either side of this site are depicted (1–67, 24 unique sequences and 68–134, 18 unique sequences). The lines connecting sequence variants on either side of the site represent the full-length repeats sequenced in this study. Numbers adjacent to each variant indicate the number of times it was sampled. The 5′ and 3′ half-repeats are organized into clades (A–E and X–Z) each composed of a high-frequency allele and its variants. The pairwise combinations of 5′ and 3′ clades generated by recombination are summarized in SI Table 1.
Alternative Splicing.
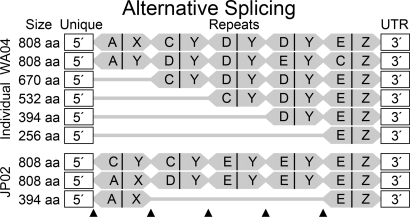
Bindin mRNAs differ by single F-lectin repeats of 402–414 bp. Mature bindin proteins differ by the approximate mass of one F-lectin repeat (≈12–15 kDa; Fig. 1). The 107 repeats (SI Fig. 9) occur in 39 full-length cDNAs as follows: 9 (one repeat), 7 (two repeats), 12 (three repeats), 7 (four repeats), and 4 (five repeats). Alternative splicing creates mRNA variants that are shortened by one or more full repeats relative to the genomic allele. N-terminal repeats or internal repeats can be excised by alternative splicing (Fig. 5). Splice combinations are not random. The number of repeats in the longest cDNAs approaches the number in the gene, limiting the splice variation available to long transcripts. The 5′ region is connected to clades A, B, and C more often, and clades D and E less often, than expected by chance (SI Table 2). Clade A connects only to the 5′ unique region, and clade Z connects only to the 3′ UTR. These are the first and last genomic repeats; the lettering of other clades represents their average position in the genomic repeat array.
Fig. 5.
Alternative splicing of oyster bindin. Six alternatively spliced full-length bindin cDNAs sequenced from a single individual (WA04) and three from another individual (JP02) are shown. Each repeat is shaded in gray with a vertical line indicating the boundary between repeat halves. 5′ and 3′ Clades are lettered according to Fig. 4. Arrows indicate the positions where alternative splicing occurs; splicing does not occur between the last repeat and the 3′ UTR. Splice patterns for the whole data set are summarized in SI Table 2.
Positive Selection.
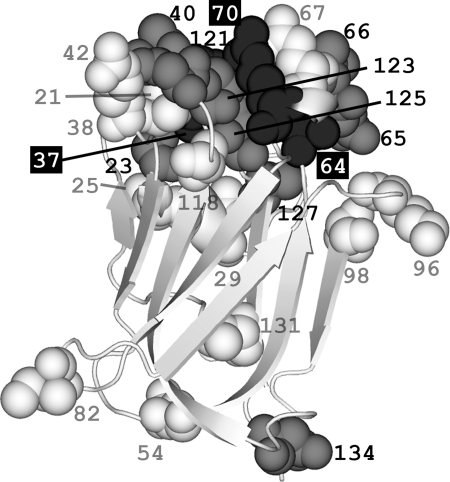
Divergence among F-lectin repeats is most apparent in the 5′ end of the repeat array (clades A and X); 18 of the 134 aa in these two clades (13.4%) differ from the consensus (Fig. 4). Maximum-likelihood methods identify nine positions subject to positive selection (SI Table 3). Four are in the 5′ half-repeat (positions 23, 40, 65, and 66; dN/dS = 6.023), and five are in the 3′ half-repeat (positions 121, 123, 125, 127, and 134; dN/dS = 5.045). Eight of these nine residues cluster on the fucose binding face of the F-lectin structure (Fig. 6 and SI Movie 1). The average pairwise distance between these eight sites is <99.99% (P = 0.009) of 106 randomly generated sets of eight substitutions on this structure. Polymorphic sites and singleton substitutions are not spatially clustered. Site 134 is distant from the fucose binding face (Fig. 6), but adjacent to the splice site and substituted (G|N) between the 3′ clades that are alternatively spliced and those that are not (Fig. 4).
Fig. 6.
Spatial locations of amino acid substitutions in oyster bindin repeats threaded onto the F-lectin crystal structure. Numbers refer to the amino acid position within the repeat. Eight of the nine positively selected sites (estimated dN/dS >1; gray spacefill) are clustered on the sugar binding face of the lectin and surround the 1H and 2R residues of the fucose-recognition motif (black spacefill; Fig. 2). The average distance between these eight positively selected sites is less than that expected by chance (n = 8, P < 0.009, 106 replicates). Polymorphic sites (white spacefill) have amino acid substitutions observed in two or more cDNA sequences. Singleton sites (not depicted) are substitutions observed in only one cDNA sequence. Polymorphic (n = 21, P < 0.783, 106 replicates) and singleton sites (n = 24, P < 0.082, 106 replicates) are not spatially clustered.
Discussion
Sperm bindin is extensively polymorphic in C. gigas. The F-lectin repeats have diversified by positive selection, as is typical of many GRPs (4). However, unlike any existing example, oyster bindin has evolved a repetitive architecture that creates diversity in protein size and sequence by two mechanisms: alternative splicing and recombination. Together, these two processes create intraspecific bindin polymorphism that exceeds that of any other known GRP.
The longest bindin cDNAs and proteins have five F-lectin repeats. Assuming that there are also five repeats in each allele and four (two recombinant and two parental) combinations of the 5′ and 3′ halves of each repeat, the number of unique bindin cDNAs that can be generated in one male by alternative splicing and recombination is 4,756 (SI Text). The details of oyster bindin expression are more complex than this simple estimate assumes. For example, it disregards the contribution of sequence variation to bindin polymorphism and incorrectly assumes that patterns of alternative splicing and recombination are independent and random. Regardless, it is clear from bindin sequence and size polymorphism that the bindin gene can produce a large number of variant proteins. Full-length bindin cDNA sequences show that individual males express mRNAs of all five sizes, but Northern and Western blots show that these same individuals typically translate only one or two of the possible protein size variants. We do not know whether these translated proteins are selected from the pool of alternatively spliced mRNA transcripts generated by each individual, or if they are the products of allele size variation resulting from unequal crossing-over. The latter seems more likely, because protein sizes detected by Western blotting vary by location, 48-kDa bindins are most common in New Zealand individuals; 75- and 88-kDa bindins are found only in Oregon samples.
Interestingly, gene architectures capable of creating combinatorial diversity are common in other rapidly evolving recognition proteins, but have not been previously found among GRPs. The two known invertebrate egg receptors for sperm, abalone VERL (21) and sea urchin EBR-1 (22), have large and complex gene structures, as does sea urchin sperm bindin (9). However, these genes lack mechanisms of creating combinatorial diversity characteristic of, for example, immunoglobulins (3). In the snail Tegula, the major acrosomal protein is produced in only two alternatively spliced forms (23). Some GRPs have simple gene structures, but exist as members of diverse gene families. Abalone vitelline envelopes are composed of a suite of related proteins (24), as are the putative egg envelope sperm receptors of vertebrates (25). Abalone sperm lysin and sp18 protein are ancient paralogs that now perform separate roles in fertilization and perhaps bind egg proteins that are themselves the product of an ancient duplication (24). Mussel (Mytilus) sperm Lysin-M3, -M6, and -M7 are little more than single C-type lectin carbohydrate recognition domains, but exist as a gene family that may share genetic variation by concerted evolution (8).
Many GRPs have a history of divergent selection between species (4). In contrast, the maintenance and elimination of variation within species does not follow any simple pattern in the sperm GRP that have been studied (5). The spectrum of possible polymorphism syndromes exist: from coding monomorphism in abalone and some sea urchins, through moderate polymorphism in blue mussels and many sea urchin species, to the extreme polymorphism in oyster bindin reported here. These different modes of GRP evolution among species are interesting because they bear on the maintenance of adaptive phenotypic diversity and on the origin of reproductive isolation between incipient species.
Mechanisms of Polyspermy Avoidance and GRP Polymorphism.
Broadly defined, freely spawned eggs have two mechanisms for avoiding fusion with multiple sperm: global and local. Global polyspermy avoidance mechanisms act over the entire egg, eliminating excess sperm regardless of their compatibility with the egg. Examples of global blocks include: membrane depolarization, cortical granule exocytosis, mechanical barriers such as envelope lifting, and target size modifications such as localized fertilization sites and egg size (14). Elimination of excess sperm can also result from local recognition and rejection. Local polyspermy avoidance mechanisms act on individual sperm, favoring the subset with compatible molecular recognition systems. Examples of local blocks include: sperm activation, chemoattraction, acrosome reaction inhibition, egg envelope binding and lysis, plasma membrane binding and fusion, and mechanisms of pronucleus choice in cases of physiological polyspermy (14).
Eggs use both types of polyspermy avoidance mechanism, but different taxa use different global and local blocks, and the relative contribution of each to the total polyspermy block varies among species. When sperm are not limiting, the absolute intensity of both sperm competition and sexual conflict increases with increasing sperm density. But, the relative intensity of these two conflicts depends on the mechanism of polyspermy avoidance. A species whose polyspermy block relies exclusively on global mechanisms (a strong electrical block) would experience intense sperm competition at high density because, in the absence of local blocks, all of the sperm competing for an egg are capable of fertilizing it. Global blocks reduce the opportunity for multiple fusions, but not the number of sperm competing for a given egg. In contrast, a species that relies exclusively on local blocks (molecular recognition at sperm–egg binding) would experience intense sexual conflict at high density. Local blocks reduce the effective sperm density by eliminating incompatible sperm, but they do not reduce the potential for multiple fertilizations by the subset of sperm that can overcome the recognition block.
Diploid models of sexual conflict suggest that female diversification occurs over a broad range of parameter values and that male polymorphism can arise as a secondary response when sexual conflict and female preference are strong (13). These results depend on the assumption that the phenotypic effects of recognition alleles are nonadditive, something we know little about. Sea urchin bindin alleles show fitter-allele dominance. In a given cross heterozygotes have phenotypes that resemble the more-compatible parental allele (26). Systems with such a heterozygote advantage are also more apt to maintain polymorphism (5). Models of sexual conflict suggest that species experiencing strong sexual conflict and female preference at high sperm density, those with an effective local and weak global block to polyspermy, should have a greater chance of maintaining male polymorphism than species with effective global and weak local blocks, whose fertilization dynamics are dominated by sperm competition at high density.
Polyspermy Avoidance in Oysters and Other Taxa.
The relative contributions of global and local polyspermy avoidance mechanisms to the total block have not been quantified in any group. But, data on the existence of various polyspermy blocks in abalone, sea urchins, mussels, and oysters are available. Abalone eggs have an effective global electrical block and resist polyspermy at sperm–egg ratios 100 times that required for complete fertilization (14). Recognition blocks act between abalone species, but intraspecific patterns of sperm–egg recognition have not yet been examined (6). Mussels and sea urchins have evolved multiple global blocks to polyspermy, suggesting that no single mechanism is perfectly effective (14). Experimental fertilization in laboratory and field conditions establish the existence of a local block in sea urchins, possibly mediated by interaction between sperm bindin and its egg receptor EBR-1 (26, 27). A tight correspondence between ease of fertilization and polyspermy susceptibility implies that this recognition block is an important polyspermy avoidance mechanism, consistent with the moderate bindin polymorphism observed in many sea urchin species (28).
A primary prediction, that oysters have poor global blocks, has been confirmed by three independent studies (15–17). A Na+- dependent electrical block exists, but fertilized oyster eggs can fuse with fresh sperm up to 15 min after fusion with the first sperm (17). No direct evidence of a recognition block exists in oysters, but such weak global blocks imply a local mechanism; oysters live and spawn at high density and sperm–egg ratio is probably high at fertilization. Interestingly, when exposed to a fixed number of sperm per egg, a fixed number of additional sperm gain entry, ≈6 per 10,000 after each exposure during the first 5 min after the initial insemination (15). It is difficult to see how a simple global mechanism could cause such a pattern; slow blocks should persist, fast blocks should be exhausted after the first insemination. Perhaps only a fraction of oyster sperm are compatible with a given female's eggs. A plasma membrane fusion block exists (17) and could be mediated by bindin and its receptor. We predict this egg receptor will also be extraordinarily diverse.
Materials and Methods
Oysters.
Live C. gigas came from aquaculture facilities in Newport, Oregon, Willapa Bay, Washington, and Nelson, New Zealand. Samples of spermatogenic cells fixed in 50–70% ethanol were from oyster farms in Aomori and Hokkaido, Japan and Santa Catarina, Brazil (SI Table 4). Oysters from Oregon and from New Zealand are known diploids.
AR Isolation.
ARs were isolated as described (12). Individual males were opened, and spermatogenic tissue was scraped into buffered seawater (BSW; 10 mM Mes in filtered seawater, pH 6.0). Tissue was stirred and filtered through cheesecloth to release sperm. The sperm were washed twice by centrifugation and resuspension in 100 vol of BSW (1,000 × g, 15 min, 4°C) and twice by centrifugation and resuspension in 100 vol of isolation medium (IM; 10 mM Mes, 10 mM sodium acetate, 10 mM benzamidine, and 0.2 mg/ml soybean trypsin inhibitor in filtered sea water, pH 6.0). Washed, resuspended sperm were vigorously stirred, and a 10% vol/vol solution of sodium sarkosyl was added to a final concentration of 0.5%. The sperm suspension gelled when the DNA was released. The viscosity was decreased by homogenization in a 40-ml glass ball tissue grinder. The homogenized AR suspension was centrifuged (26,000 × g, 30 min, 4°C), and the resulting AR pellet was resuspended in fresh IM plus 0.1% sarkosyl and homogenized with a 10-ml glass ball homogenizer. The homogenate was centrifuged again, and the AR wash was repeated without sarkosyl until phase-contrast microscopy (×1,200 magnification) showed the ARs were free of other structures. Fragments of nuclei containing the 34-kDa sperm nuclear basic protein (18) could not be seen by phase-contrast, but this protein could be extracted with 0.6 M KCl (Fig. 1 B and C).
Protein Electrophoresis and Sequencing.
Whole sperm or isolated ARs were dissolved in 10% SDS, total protein concentration was determined (Lowry), and the extracted proteins were electrophoresed on 12.5% SDS/PAGE and stained with Coomassie blue. For N-terminal sequencing, SDS/PAGE-separated proteins were electro-transferred to PVDF membrane and Coomassie-stained. Protein bands were excised and sequenced at the University of California at San Diego Protein Sequencing Laboratory. For internal sequences, bands were excised from Coomassie-stained SDS/PAGE gels and lyophilized. Trypsinization, peptide isolation, and sequencing were done by the Protein and Nucleic Acid Facility at Stanford University.
Western Blots.
Antibody was raised by standard protocol to 63-kDa bindin bands excised from SDS/PAGE gels that were negatively stained with CuCl2. Whole preimmune and immune sera were used at 1:200 to 1:500 dilutions. Gel lanes of 25–50 μg of whole sperm protein (Fig. 1 F–I), electrophoresed on 12.5% SDS/PAGE gels, were electro-transferred to nitrocellulose membrane (Protran; Whatman) and blocked in 5% nonfat dry milk containing 10 mg/ml BSA. Blots were incubated in primary sera for 2 h, washed five times in excess TBST [150 mM NaCl/10 mM Tris/0.05% (vol/vol) Tween 20, pH 7.5] and incubated for 1 h in a 1:50,000 dilution of HRP-conjugated goat anti-rabbit IgG (CalBiochem). After five washes in TBST, blots were developed in SuperSignal West Dura Extended Duration Substrate (Pierce) following the manufacturer's protocol.
RNA, cDNA, and Sequencing.
Total RNA was isolated from live or fixed spermatogenic cells with the RNeasy kit (Qiagen). cDNA synthesis using SuperScript II reverse transcriptase (Invitrogen) and 3′ and 5′ RACE using the FirstChoice RLM-RACE Kit (Ambion) were done following the manufacturer's directions. Full-length bindin cDNA sequences were amplified with primers that bracket the ORF: forward (5′-GAAGTTTACCAGAATGCTCG3-′, start codon underlined), and reverse (5′-GTAAGCCTCTTTCTCTCTA-3′, in the 3′UTR). PCR products were either sequenced directly or cloned into pCR4-TOPO (Invitrogen) using Big Dye version 3 (Applied Biosystems). Sequences were edited with Sequencher (Gene Codes). GenBank accession numbers for C. gigas bindin cDNAs with one to five F-lectin repeats are: EF219425 to EF219429. Shell morphology does not identify oyster species, so mitochondrial cytochrome oxidase subunit I (COI) primers were used to amplify 323 bp of the COI gene of every individual in this study: forward (5′-GCTATGTTTTTAGACCCCGTG-3′) and reverse (5′-CCAGCAAGGTGAAGGCTTAG-3′). Species-diagnostic COI sequences of five Crassostrea species are available in GenBank, and all individuals in this study had COI sequences identical to C. gigas.
Southern Blots.
Twenty micrograms of purified sperm genomic DNA was digested for 18 h at 37°C in 40-μl reactions with 20 units of each restriction enzyme (AvaI, HindIII, EcoRI; New England Biolabs). Digests were ethanol-precipitated, and 10 μg of DNA per lane was electrophoresed in 0.7% agarose. Capillary transfer of DNA fragments to Hybond-N membrane (Amersham) and processing of blots for hybridization with α-32P-dCTP random primed labeled probes (DECAprime II DNA Labeling Kit; Ambion) were done with standard procedures. The full-length repeat probe was 414 bp. The 3′UTR probe was 229 bp made by amplifying cDNA with the forward primer (5′-ATTAGACTCTGTTAACCAA-3′) and the reverse primer (5′-AGCTTACCGATTTGAAAC-3′). Overnight hybridization and stringency washing of blots were done by standard protocols.
Northern Blots.
Total RNA was isolated by dissolving spermatogenic tissue in 4 M guanidine thiocyanate and ultracentrifugation through CsCl gradient (29). RNA pellets were dissolved in 2% SDS and quantified by spectrophotometry. Twenty micrograms of each sample was separated on 1.5% agarose gels. Gels were capillary transferred to Hybond-N membrane (Amersham Bioscience) following the manufacturer's directions. Blots were probed with a 32P-labeled F-lectin repeat sequence. Hybridization and washing were done following the Hybond-N directions.
Statistical Analysis.
The 107 F-lectin repeats from 39 cDNAs were manually aligned, and linkers were trimmed from internal repeats to a final length of 134 aa. Recombination was tested by using a genetic algorithm for recombination detection (GARD) on the Datamonkey server (20). Alignments were split into two parts (amino acids 1–67 and 68–134), reflecting evidence of a single recombination site located at the intron between amino acid positions 67–68 of each repeat. Estimates of phylogeny and of selection were done on each half-repeat separately. Phylogenetic relationships were inferred by using PAUP 4.0b10 (neighbor-joining uncorrected p distance, 100,000 bootstrap replicates, 50% majority-rule consensus) (30). Site-specific dN/dS substitution rate ratios were estimated for each codon by maximum likelihood with nested models implemented in PAML 3.14b (31). EsyPred 3D was used to model the 3D structure of the F-lectin repeat consensus sequence based on alignments with the fucose-binding lectin module of a Streptococcus pneumoniae virulence factor (Protein Data Bank ID code 2J22; 28.7% amino acid identity) (32, 33). Three classes of amino acid polymorphism (selected, polymorphic, and singleton) were mapped onto the inferred 3D structure. Selected sites had an estimated dN/dS ratio >1. Polymorphic sites were amino acid substitutions (from consensus) observed in more than one sequence. Singleton sites were amino acid substitutions observed in only one sequence. Spatial clustering of selected, polymorphic, and singleton sites on the inferred structure was assessed by comparing the average distance between the α-carbons of observed amino acid substitutions with the distribution of 106 random data sets simulated with the observed number of substitutions (Perl code by SAS).
Supplementary Material
ACKNOWLEDGMENTS.
We thank Mark Camara and Christopher Langdon (Oregon State University, Hatfield Marine Science Center, Newport), Hideki Katow and Makoto Osada (Tohoku University, Sendai, Japan), Chia Wei Li and Chia Wei Yei (Taiwan National University, Taipei, Taiwan), Norio Suzuki and Kiyoshi Sano (Hokkaido University, Sapporo, Japan), Noritaka Hirohashi (Ochanomizu University, Tokyo), Motonori Hoshi (University of the Air, Chiba City, Japan), Rafael Aquino and Paulo Mourão (Federal University of Rio de Janiero, Rio de Janiero, Brazil), Youn-Ho Lee (Korea Polar Research Institute, Incheon, Korea), and Michael McCartney (University of North Carolina, Wilmington), for animals. Mark Hildebrand and Masashi Kinukawa for help with experiments, and Dennis Hedgecock and Ronald Burton for discussion. G.W.M. and V.D.V. were supported by National Institutes of Health Grant HD12986. S.A.S. and W.J.S. were supported by National Institutes of Health Grant HD42563 and National Science Foundation Grant DEB-0716761. S.A.S. is a recipient of an Natural Sciences and Engineering Research Council Postgraduate Scholarships-Doctoral Fellowship. S.L.A. was supported by the Royal Society of New Zealand International Science and Technology Fund.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF219425–EF219429).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711862105/DC1.
References
- 1.Wang X, Hughes AL, Tsukamoto T, Ando T, Kao T. Evidence that intragenic recombination contributes to allelic diversity of the S-RNase gene at the self-incompatibility (S) locus in Petunia inflata. Plant Physiol. 2001;125:1012–1022. doi: 10.1104/pp.125.2.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hattori D, et al. Dscam diversity is essential for neuronal wiring and self-recognition. Nature. 2007;449:223–227. doi: 10.1038/nature06099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Litman GW, Dishaw LJ, Cannon JP, Haire RN, Rast JP. Alternative mechanisms of immune receptor diversity. Curr Opin Immunol. 2007;19:526–534. doi: 10.1016/j.coi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 5.Haygood R. Sexual conflict and protein polymorphism. Evol Int J Org Evol. 2004;58:1414–1423. doi: 10.1111/j.0014-3820.2004.tb01723.x. [DOI] [PubMed] [Google Scholar]
- 6.Kresge N, Vacquier VD, Stout CD. Abalone lysin: The dissolving and evolving sperm protein. BioEssays. 2001;23:95–103. doi: 10.1002/1521-1878(200101)23:1<95::AID-BIES1012>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 7.Riginos C, McDonald JH. Positive selection on an acrosomal sperm protein, M7 lysin, in three species of the mussel genus Mytilus. Mol Biol Evol. 2003;20:200–207. doi: 10.1093/molbev/msg021. [DOI] [PubMed] [Google Scholar]
- 8.Springer SA, Crespi BJ. Adaptive gamete-recognition divergence in a hybridizing Mytilus population. Evol Int J Org Evol. 2007;61:772–783. doi: 10.1111/j.1558-5646.2007.00073.x. [DOI] [PubMed] [Google Scholar]
- 9.Metz EC, Palumbi SR. Positive selection and sequence rearrangements generate extensive polymorphism in the gamete recognition protein bindin. Mol Biol Evol. 1996;13:397–406. doi: 10.1093/oxfordjournals.molbev.a025598. [DOI] [PubMed] [Google Scholar]
- 10.Geyer LB, Palumbi SR. Reproductive character displacement and the genetics of gamete recognition in tropical sea urchins. Evol Int J Org Evol. 2003;57:1049–1060. doi: 10.1111/j.0014-3820.2003.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 11.Hylander BL, Summers RG. An ultrastructural analysis of the gametes and early fertilization in two bivalve molluscs, Chama macerophylla and Spisula solidissima, with special reference to gamete binding. Cell Tissue Res. 1977;182:469–489. doi: 10.1007/BF00219830. [DOI] [PubMed] [Google Scholar]
- 12.Brandriff B, Moy GW, Vacquier VD. Isolation of sperm bindin from the oyster (Crassostrea gigas). Gamete Res. 1978;1:89–99. [Google Scholar]
- 13.Hayashi TI, Vose M, Gavrilets S. Genetic differentiation by sexual conflict. Evol Int J Org Evol. 2007;61:516–529. doi: 10.1111/j.1558-5646.2007.00059.x. [DOI] [PubMed] [Google Scholar]
- 14.Gould MC, Stephano JL. Polyspermy prevention in marine invertebrates. Microsc Res Technol. 2003;61:379–388. doi: 10.1002/jemt.10351. [DOI] [PubMed] [Google Scholar]
- 15.Alliegro MC, Wright DA. Polyspermy inhibition in the oyster, Crassostrea virginica. J Exp Zool. 1983;227:127–137. doi: 10.1002/jez.1402270117. [DOI] [PubMed] [Google Scholar]
- 16.Stephano JL, Gould M. Avoiding polyspermy in oyster (Crassostrea gigas). Aquaculture. 1988;73:295–307. [Google Scholar]
- 17.Togo T, Morisawa M. Mechanisms for blocking polyspermy in oocytes of the oyster Crassostrea gigas. J Exp Zool. 1999;283:307–314. [Google Scholar]
- 18.Eirin-Lopez JM, Lewis JD, Howe LA, Ausio J. Common phylogenetic origin of protamine-like (PL) proteins and histone H1: Evidence from bivalve PL genes. Mol Biol Evol. 2006;23:1304–1317. doi: 10.1093/molbev/msk021. [DOI] [PubMed] [Google Scholar]
- 19.Bianchet MA, Odom EW, Vasta GR, Amzel LM. A novel fucose recognition fold involved in innate immunity. Nat Struct Biol. 2002;9:628–634. doi: 10.1038/nsb817. [DOI] [PubMed] [Google Scholar]
- 20.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 21.Swanson WJ, Vacquier VD. The abalone egg vitelline envelope receptor for sperm lysin is a giant multivalent molecule. Proc Natl Acad Sci USA. 1997;94:6724–6729. doi: 10.1073/pnas.94.13.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamei N, Glabe CG. The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev. 2003;17:2502–2507. doi: 10.1101/gad.1133003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellberg ME, Moy GW, Vacquier VD. Positive selection and propeptide repeats promote rapid interspecific divergence of a gastropod sperm protein. Mol Biol Evol. 2000;17:458–466. doi: 10.1093/oxfordjournals.molbev.a026325. [DOI] [PubMed] [Google Scholar]
- 24.Aagaard JE, Yi X, MacCoss MJ, Swanson WJ. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc Natl Acad Sci USA. 2006;103:17302–17307. doi: 10.1073/pnas.0603125103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Litscher ES, Wassarman PM. Egg extracellular coat proteins: From fish to mammals. Histol Histopathol. 2007;22:337–347. doi: 10.14670/HH-22.337. [DOI] [PubMed] [Google Scholar]
- 26.Palumbi SR. All males are not created equal: Fertility differences depend on gamete recognition polymorphisms in sea urchins. Proc Natl Acad Sci USA. 1999;96:12632–12637. doi: 10.1073/pnas.96.22.12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitan DR, Ferrell DL. Selection on gamete recognition proteins depends on sex, density, and genotype frequency. Science. 2006;312:267–269. doi: 10.1126/science.1122183. [DOI] [PubMed] [Google Scholar]
- 28.Levitan DR, Terhorst CP, Fogarty ND. The risk of polyspermy in three congeneric sea urchins and its implications for gametic incompatibility and reproductive isolation. Evol Int J Org Evol. 2007;61:2007–2014. doi: 10.1111/j.1558-5646.2007.00150.x. [DOI] [PubMed] [Google Scholar]
- 29.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 30.Swofford DL. PAUP*: Phylogenetic analysis Using Parsimony and Other Methods. Sunderland, MA: Sinauer; 2001. version 4. [Google Scholar]
- 31.Yang ZH. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 32.Lambert C, Leonard N, De Bolle X, Depiereux E. ESyPred3D: Prediction of proteins 3D structures. Bioinformatics. 2002;18:1250–1256. doi: 10.1093/bioinformatics/18.9.1250. [DOI] [PubMed] [Google Scholar]
- 33.Boraston AB, Wang D, Burke RD. Blood group antigen recognition by a Streptococcus pneumoniae virulence factor. J Biol Chem. 2006;281:35263–35271. doi: 10.1074/jbc.M607620200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.