Abstract
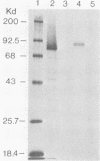
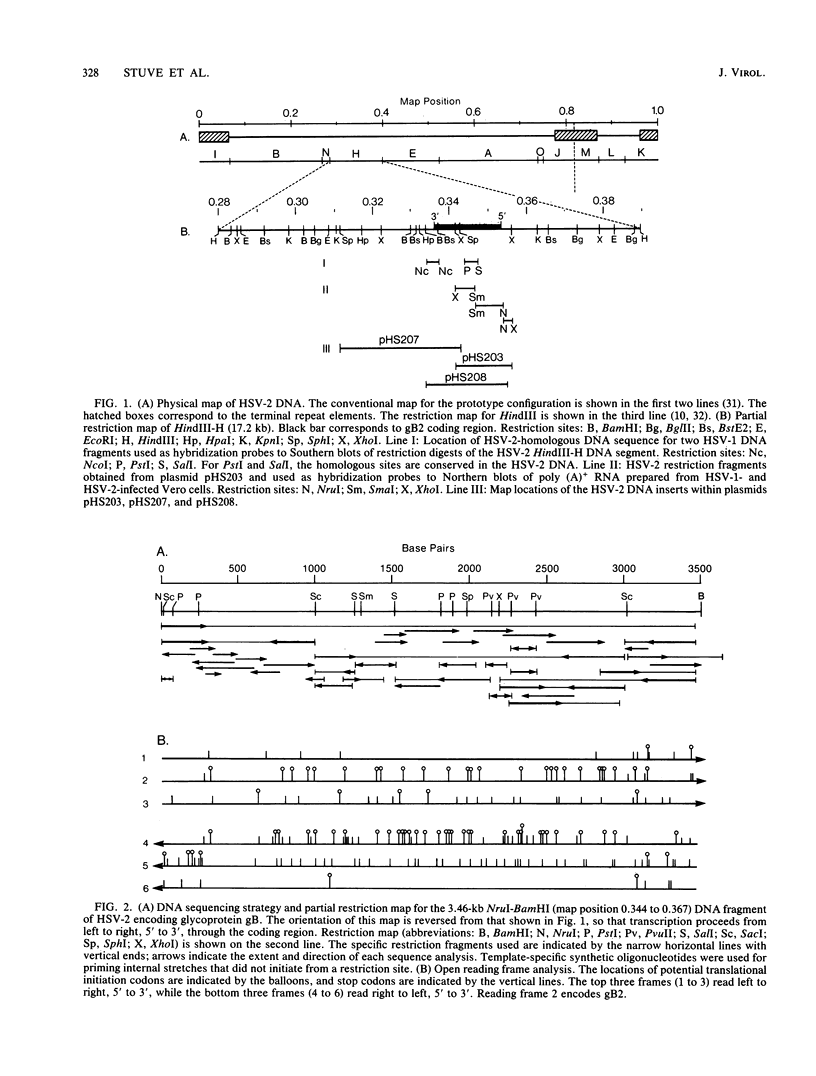
The gene for glycoprotein gB2 of herpes simplex virus type 2 strain 333 was cloned, sequenced, and expressed in mammalian cells. The gB2 protein had an overall nucleotide and amino acid sequence homology of 86% with the cognate gB1 protein. However, of the 125 amino acid substitutions or deletions, only 12.5% were conservative replacements. These differences were clustered within an NH2-terminal region, a central region, and a COOH-terminal region, resulting in domains of near identity broken by small regions of marked divergence. Regions of greatest homology included a 90-amino-acid stretch starting at residue 484 and 39 amino acids spanning residues 835 to 873, which cover a rate-of-entry locus mapped to Ala-552 and a syn locus mapped to Arg-857, respectively, in gB1 by Bzik et al. (D. J. Bzik, B. A. Fox, N. A. DeLuca, and S. Person, Virology 133:301-314, 1984). Pellett et al. (P. E. Pellett, K. G. Kousoulas, L. Pereira, and B. Roizman, J. Virol. 53:243-253, 1985) mapped the mutations in three monoclonal antibody-resistant gB1 mutants between amino acids 273 and 443. These epitopes are included in a region of 98 residues identical between gB1 and gB2. The identity of this protein was verified by placing a truncated gene lacking the 303 carboxyl-terminal amino acids of gB2 into mammalian COS and CHO cells. Expression was demonstrated by immunofluorescence and radioimmunoprecipitation. This protein will be purified from the stable CHO cell lines and compared with gB1 for immunogenicity and protective efficacy in animal challenge models.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Kellems R. E., Bertino J. R., Schimke R. T. Selective multiplication of dihydrofolate reductase genes in methotrexate-resistant variants of cultured murine cells. J Biol Chem. 1978 Mar 10;253(5):1357–1370. [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology. 1984 Aug;137(1):185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- Bzik D. J., Fox B. A., DeLuca N. A., Person S. Nucleotide sequence specifying the glycoprotein gene, gB, of herpes simplex virus type 1. Virology. 1984 Mar;133(2):301–314. doi: 10.1016/0042-6822(84)90397-0. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Corey L., Spear P. G. Infections with herpes simplex viruses (1). N Engl J Med. 1986 Mar 13;314(11):686–691. doi: 10.1056/NEJM198603133141105. [DOI] [PubMed] [Google Scholar]
- Cortini R., Wilkie N. M. Physical maps for HSV type 2 DNA with five restriction endonucleases. J Gen Virol. 1978 May;39(2):259–280. doi: 10.1099/0022-1317-39-2-259. [DOI] [PubMed] [Google Scholar]
- Eberle R., Courtney R. J. Assay of type-specific and type-common antibodies to herpes simplex virus types 1 and 2 in human sera. Infect Immun. 1981 Mar;31(3):1062–1070. doi: 10.1128/iai.31.3.1062-1070.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981 Feb;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Holland L. E., Sandri-Goldin R. M., Goldin A. L., Glorioso J. C., Levine M. Transcriptional and genetic analyses of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J Virol. 1984 Mar;49(3):947–959. doi: 10.1128/jvi.49.3.947-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp T. P., Woods K. R. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S. C., Ivatt R. J. Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem. 1981;50:555–583. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- Kudler L., Jones T. R., Russell R. J., Hyman R. W. Heteroduplex analysis of cloned fragments of herpes simplex virus DNAs. Virology. 1983 Jan 15;124(1):86–99. doi: 10.1016/0042-6822(83)90292-1. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunberg J. H., Kaufman R. J., Chang A. C., Cohen S. N., Schimke R. T. Structure and genomic organization of the mouse dihydrofolate reductase gene. Cell. 1980 Feb;19(2):355–364. doi: 10.1016/0092-8674(80)90510-3. [DOI] [PubMed] [Google Scholar]
- Pachl C., Burke R. L., Stuve L. L., Sanchez-Pescador L., Van Nest G., Masiarz F., Dina D. Expression of cell-associated and secreted forms of herpes simplex virus type 1 glycoprotein gB in mammalian cells. J Virol. 1987 Feb;61(2):315–325. doi: 10.1128/jvi.61.2.315-325.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachl C., Schubach W., Eisenman R., Linial M. Expression of c-myc RNA in bursal lymphoma cell lines: identification of c-myc-encoded proteins by hybrid-selected translation. Cell. 1983 Jun;33(2):335–344. doi: 10.1016/0092-8674(83)90415-4. [DOI] [PubMed] [Google Scholar]
- Pellett P. E., Kousoulas K. G., Pereira L., Roizman B. Anatomy of the herpes simplex virus 1 strain F glycoprotein B gene: primary sequence and predicted protein structure of the wild type and of monoclonal antibody-resistant mutants. J Virol. 1985 Jan;53(1):243–253. doi: 10.1128/jvi.53.1.243-253.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L., Dondero D. V., Gallo D., Devlin V., Woodie J. D. Serological analysis of herpes simplex virus types 1 and 2 with monoclonal antibodies. Infect Immun. 1982 Jan;35(1):363–367. doi: 10.1128/iai.35.1.363-367.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Rafield L. F., Knipe D. M. Characterization of the major mRNAs transcribed from the genes for glycoprotein B and DNA-binding protein ICP8 of herpes simplex virus type 1. J Virol. 1984 Mar;49(3):960–969. doi: 10.1128/jvi.49.3.960-969.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector J. T., Lausch R. N., Oakes J. E. Use of monoclonal antibodies for analysis of antibody-dependent immunity to ocular herpes simplex virus type 1 infection. Infect Immun. 1982 Oct;38(1):168–174. doi: 10.1128/iai.38.1.168-174.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979 Mar;16(3):481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- Ruyechan W. T., Morse L. S., Knipe D. M., Roizman B. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol. 1979 Feb;29(2):677–697. doi: 10.1128/jvi.29.2.677-697.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seela F., Tran-Thi Q. H., Franzen D. Poly(7-deazaguanylic acid), the homopolynucleotide of the parent nucleoside of queuosine. Biochemistry. 1982 Aug 31;21(18):4338–4343. doi: 10.1021/bi00261a024. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Chasin L. A. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4216–4220. doi: 10.1073/pnas.77.7.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahlne A., Svennerholm B., Lycke E. Evidence for herpes simplex virus type-selective receptors on cellular plasma membranes. J Gen Virol. 1979 Jul;44(1):217–225. doi: 10.1099/0022-1317-44-1-217. [DOI] [PubMed] [Google Scholar]
- Vahlne A., Svennerholm B., Sandberg M., Hamberger A., Lycke E. Differences in attachment between herpes simplex type 1 and type 2 viruses to neurons and glial cells. Infect Immun. 1980 Jun;28(3):675–680. doi: 10.1128/iai.28.3.675-680.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]
- van der Eb A. J., Graham F. L. Assay of transforming activity of tumor virus DNA. Methods Enzymol. 1980;65(1):826–839. doi: 10.1016/s0076-6879(80)65077-0. [DOI] [PubMed] [Google Scholar]
- von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984 Feb 25;173(2):243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]