Abstract
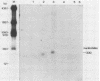
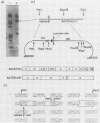
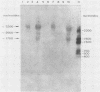
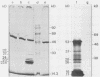
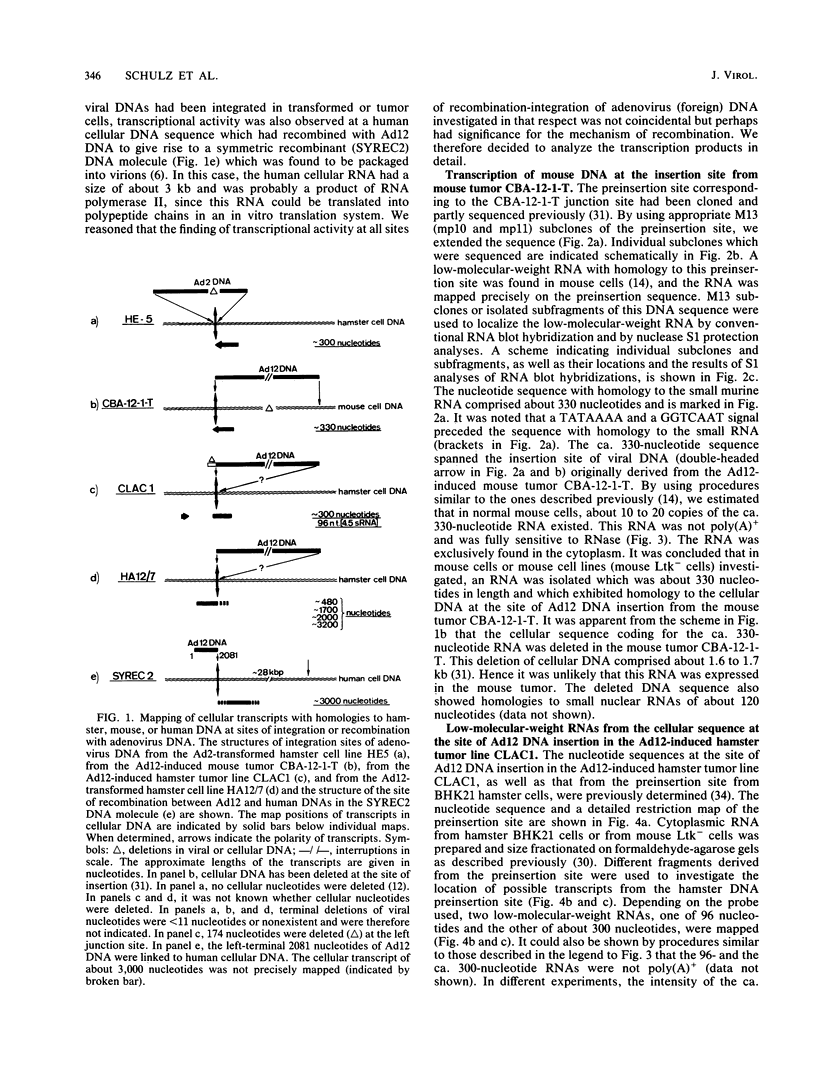
The nucleotide sequences of several sites of recombination between adenovirus DNA and hamster, mouse, or human cell DNAs were determined. These sites of recombination had been cloned from adenovirus type 2 (Ad2)- or type 12 (Ad12)-transformed cells, from Ad12-induced tumor cells, or from a symmetric recombinant between Ad12 DNA and human cell DNA. One important precondition for the generation of recombinants between host and foreign DNAs might be the establishment of a chromatin configuration that permits access of foreign DNA and of the recombination machinery to cellular DNA. Such favorable chromatin structures might arise during cellular DNA replication or transcription or both. As a first approach toward investigating these more complex problems of foreign DNA insertion, we determined transcriptional activities of cellular DNA sequences at viral junction sites. The sites of linkage investigated in this study with respect to their transcriptional activities were those previously cloned and sequenced (W. Doerfler, R. Gahlmann, S. Stabel, R. Deuring, U. Lichtenberg, M. Schulz, D. Eick, and R. Leisten, Curr. Top. Microbiol. Immunol. 109:193-228, 1983). In addition, a site from cell line HA12/7 which is described in this paper was also analyzed. The results presented demonstrate that the cellular DNA sequences involved in linkage to viral DNA at five completely different sites in DNA from three different species are transcribed into RNAs even in cells which have not been transformed or infected by adenovirus. Some of these RNAs were cytoplasmic and were not poly(A)+. Human cell DNA sequences at the junction to Ad12 DNA in SYREC2 DNA were transcribed into poly(A)+ cytoplasmic RNA which could be translated in vitro. These results are consistent with the notion that at least some of the cellular DNA sequences at sites of insertion of adenovirus (foreign) DNA are transcriptionally active and thus provide an opportunity for recombination.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BABLANIAN R., EGGERS H. J., TAMM I. STUDIES ON THE MECHANISM OF POLIOVIRUS-INDUCED CELL DAMAGE. I. THE RELATION BETWEEN POLIOVIRUS,-INDUCED METABOLIC AND MORPHOLOGICAL ALTERATIONS IN CULTURED CELLS. Virology. 1965 May;26:100–113. doi: 10.1016/0042-6822(65)90030-9. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Deuring R., Doerfler W. Proof of recombination between viral and cellular genomes in human KB cells productively infected by adenovirus type 12: structure of the junction site in a symmetric recombinant (SYREC). Gene. 1983 Dec;26(2-3):283–289. doi: 10.1016/0378-1119(83)90198-1. [DOI] [PubMed] [Google Scholar]
- Deuring R., Klotz G., Doerfler W. An unusual symmetric recombinant between adenovirus type 12 DNA and human cell DNA. Proc Natl Acad Sci U S A. 1981 May;78(5):3142–3146. doi: 10.1073/pnas.78.5.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R., Winterhoff U., Tamanoi F., Stabel S., Doerfler W. Site of linkage between adenovirus type 12 and cell DNAs in hamster tumour line CLAC3. Nature. 1981 Sep 3;293(5827):81–84. doi: 10.1038/293081a0. [DOI] [PubMed] [Google Scholar]
- Doerfler W., Gahlmann R., Stabel S., Deuring R., Lichtenberg U., Schulz M., Eick D., Leisten R. On the mechanism of recombination between adenoviral and cellular DNAs: the structure of junction sites. Curr Top Microbiol Immunol. 1984;109:193–228. doi: 10.1007/978-3-642-69460-8_9. [DOI] [PubMed] [Google Scholar]
- Doerfler W. Nonproductive infection of baby hamster kidney cells (BHK21) with adenovirus type 12. Virology. 1969 Aug;38(4):587–606. doi: 10.1016/0042-6822(69)90179-2. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Esche H., Lübbert H., Siegmann B., Doerfler W. The translational map of the Autographa californica nuclear polyhedrosis virus (AcNPV) genome. EMBO J. 1982;1(12):1629–1633. doi: 10.1002/j.1460-2075.1982.tb01365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
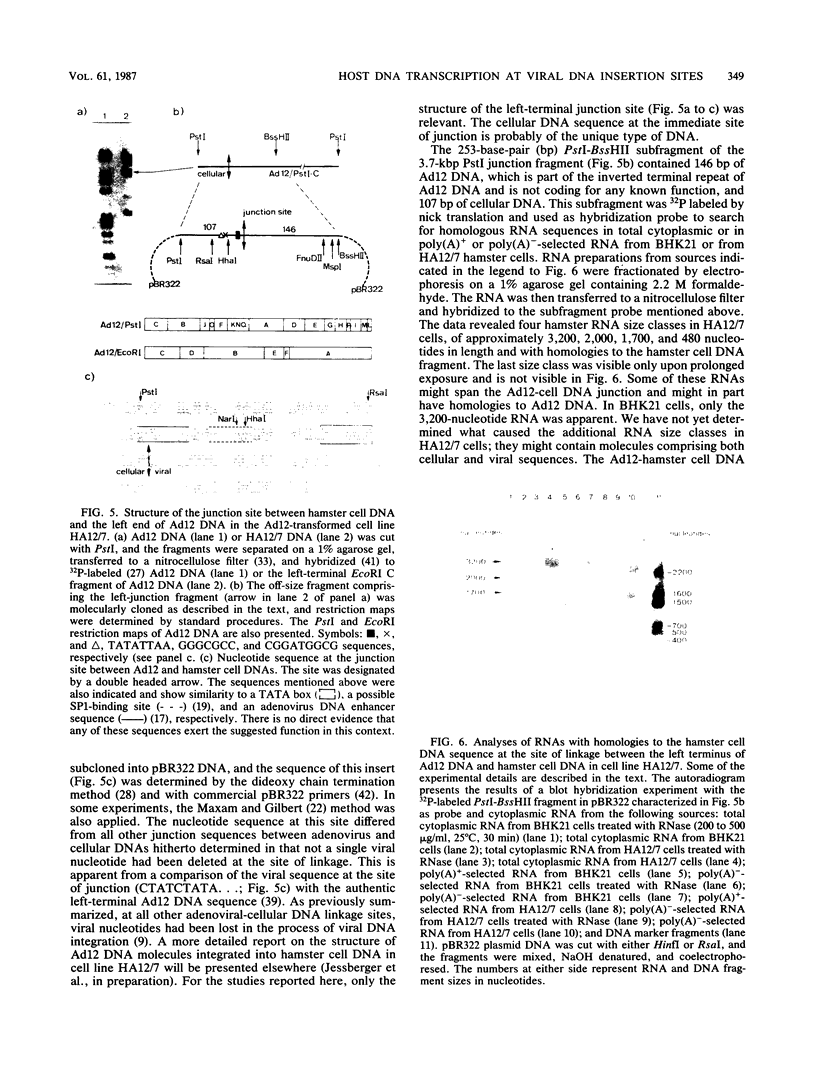
- Gahlmann R., Doerfler W. Integration of viral DNA into the genome of the adenovirus type 2-transformed hamster cell line HE5 without loss or alteration of cellular nucleotides. Nucleic Acids Res. 1983 Nov 11;11(21):7347–7361. doi: 10.1093/nar/11.21.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlmann R., Leisten R., Vardimon L., Doerfler W. Patch homologies and the integration of adenovirus DNA in mammalian cells. EMBO J. 1982;1(9):1101–1104. doi: 10.1002/j.1460-2075.1982.tb01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
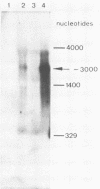
- Gahlmann R., Schulz M., Doefler W. Low molecular weight RNAs with homologies to cellular DNA at sites of adenovirus DNA insertion in hamster or mouse cells. EMBO J. 1984 Dec 20;3(13):3263–3269. doi: 10.1002/j.1460-2075.1984.tb02288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada F., Kato N. Nucleotide sequences of 4.5S RNAs associated with poly(A)-containing RNAs of mouse and hamster cells. Nucleic Acids Res. 1980 Mar 25;8(6):1273–1285. doi: 10.1093/nar/8.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada F., Takeuchi Y., Kato N. Molecular cloning and in vitro transcription of rat 4.5S RNAH genes. Nucleic Acids Res. 1986 Feb 25;14(4):1629–1642. doi: 10.1093/nar/14.4.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Hohn B. In vitro packaging of lambda and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Tjian R. Sp1 binds to promoter sequences and activates herpes simplex virus 'immediate-early' gene transcription in vitro. Nature. 1985 Sep 12;317(6033):179–182. doi: 10.1038/317179a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lübbert H., Doerfler W. Transcription of overlapping sets of RNAs from the genome of Autographa californica nuclear polyhedrosis virus: a novel method for mapping RNAs. J Virol. 1984 Oct;52(1):255–265. doi: 10.1128/jvi.52.1.255-265.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Morrison D. A. Transformation and preservation of competent bacterial cells by freezing. Methods Enzymol. 1979;68:326–331. doi: 10.1016/0076-6879(79)68023-0. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Ricciardi R. P., Miller J. S., Roberts B. E. Purification and mapping of specific mRNAs by hybridization-selection and cell-free translation. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4927–4931. doi: 10.1073/pnas.76.10.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellner J., Stüber K., Doerfler W. Computer analyses on the structure of junction sites between adenovirus DNA and cellular DNA. Biochim Biophys Acta. 1986 Jun 20;867(3):114–123. doi: 10.1016/0167-4781(86)90071-0. [DOI] [PubMed] [Google Scholar]
- Schirm S., Doerfler W. Expression of viral DNA in adenovirus type 12-transformed cells, in tumor cells, and in revertants. J Virol. 1981 Sep;39(3):694–702. doi: 10.1128/jvi.39.3.694-702.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M., Doerfler W. Deletion of cellular DNA at site of viral DNA insertion in the adenovirus type 12-induced mouse tumor CBA-12-1-T. Nucleic Acids Res. 1984 Jun 25;12(12):4959–4976. doi: 10.1093/nar/12.12.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smithies O., Gregg R. G., Boggs S. S., Koralewski M. A., Kucherlapati R. S. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985 Sep 19;317(6034):230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stabel S., Doerfler W., Friis R. R. Integration sites of adenovirus type 12 DNA in transformed hamster cells and hamster tumor cells. J Virol. 1980 Oct;36(1):22–40. doi: 10.1128/jvi.36.1.22-40.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
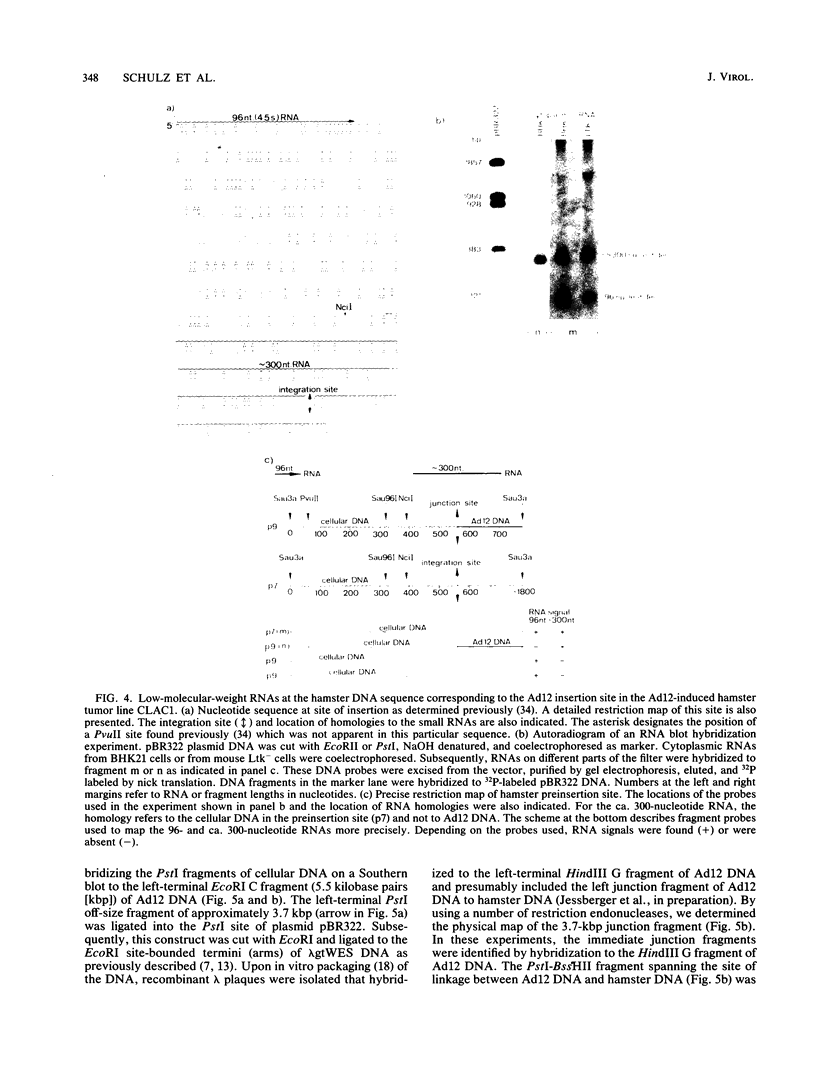
- Stabel S., Doerfler W. Nucleotide sequence at the site of junction between adenovirus type 12 DNA and repetitive hamster cell DNA in transformed cell line CLAC1. Nucleic Acids Res. 1982 Dec 20;10(24):8007–8023. doi: 10.1093/nar/10.24.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohl W. A., Rouse H., Teets K., Schlesinger R. W. The response of BHK21 cells to infection with type 12 adenovirus. 3. Transformation and restricted replication of superinfecting type 2 adenovirus. Arch Gesamte Virusforsch. 1970;31(1):93–112. doi: 10.1007/BF01241669. [DOI] [PubMed] [Google Scholar]
- Subramani S., Berg P. Homologous and nonhomologous recombination in monkey cells. Mol Cell Biol. 1983 Jun;3(6):1040–1052. doi: 10.1128/mcb.3.6.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugisaki H., Sugimoto K., Takanami M., Shiroki K., Saito I., Shimojo H., Sawada Y., Uemizu Y., Uesugi S., Fujinaga K. Structure and gene organization in the transformed Hind III-G fragment of Ad12. Cell. 1980 Jul;20(3):777–786. doi: 10.1016/0092-8674(80)90324-4. [DOI] [PubMed] [Google Scholar]
- Sutter D., Westphal M., Doerfler W. Patterns of integration of viral DNA sequences in the genomes of adenovirus type 12-transformed hamster cells. Cell. 1978 Jul;14(3):569–585. doi: 10.1016/0092-8674(78)90243-x. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Suggs S. V., Miyoshi K., Bhatt R., Itakura K. A set of synthetic oligodeoxyribonucleotide primers for DNA sequencing in the plasmid vector pBR322. Gene. 1981 Dec;16(1-3):21–26. doi: 10.1016/0378-1119(81)90057-3. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winocour E., Keshet I. Indiscriminate recombination in simian virus 40-infected monkey cells. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4861–4865. doi: 10.1073/pnas.77.8.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]