Abstract
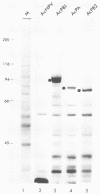
Each of the influenza virus polymerase (P) genes PB1, PB2, and PA was inserted into a baculovirus vector under the control of the polyhedrin promoter. In insect (Spodoptera frugiperda) cells infected by each baculovirus recombinant containing a P gene insert, a large amount of the encoded P protein was synthesized. Gel electrophoretic analysis of the total proteins in infected cells revealed the presence of a new protein band corresponding to the encoded P protein that was abundant enough to be stained with Coomassie blue. In cells infected simultaneously with both the PB1 and PB2 baculovirus recombinants, a PB1-PB2 complex was formed that was immunoprecipitated with an antiserum specific for either PB1 or PB2. In cells infected simultaneously with all three P baculovirus recombinants, a PB1-PB2 complex lacking the PA protein was formed. Formation of this PB1-PB2 complex partially mimics events that occur in influenza virus-infected cells, where all three P proteins form a complex with each other (B. M. Detjen, C. St. Angelo, M. G. Katze, and R. M. Krug, J. Virol. 61:16-22, 1987). These results indicate that the ability of PB1 and PB2 to form a complex is an intrinsic property of these two proteins that does not require the participation of other influenza viral gene products. Possible reasons for the absence of the PA protein from the immunoprecipitable P protein complex in insect cells infected by the three P baculovirus recombinants are discussed.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blaas D., Patzelt E., Kuechler E. Identification of the cap binding protein of influenza virus. Nucleic Acids Res. 1982 Aug 11;10(15):4803–4812. doi: 10.1093/nar/10.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam-Markson J., Jaudon C., Krug R. M. Expression of a functional influenza viral cap-recognizing protein by using a bovine papilloma virus vector. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4326–4330. doi: 10.1073/pnas.82.13.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J., Ulmanen I., Krug R. M. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell. 1983 Sep;34(2):609–618. doi: 10.1016/0092-8674(83)90393-8. [DOI] [PubMed] [Google Scholar]
- Detjen B. M., St Angelo C., Katze M. G., Krug R. M. The three influenza virus polymerase (P) proteins not associated with viral nucleocapsids in the infected cell are in the form of a complex. J Virol. 1987 Jan;61(1):16–22. doi: 10.1128/jvi.61.1.16-22.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRACE T. D. Establishment of four strains of cells from insect tissues grown in vitro. Nature. 1962 Aug 25;195:788–789. doi: 10.1038/195788a0. [DOI] [PubMed] [Google Scholar]
- Hink W. F. Established insect cell line from the cabbage looper, Trichoplusia ni. Nature. 1970 May 2;226(5244):466–467. doi: 10.1038/226466b0. [DOI] [PubMed] [Google Scholar]
- Inglis S. C., Carroll A. R., Lamb R. A., Mahy B. W. Polypeptides specified by the influenza virus genome I. Evidence for eight distinct gene products specified by fowl plague virus. Virology. 1976 Oct 15;74(2):489–503. doi: 10.1016/0042-6822(76)90355-x. [DOI] [PubMed] [Google Scholar]
- Krystal M., Li R., Lyles D., Pavlakis G., Palese P. Expression of the three influenza virus polymerase proteins in a single cell allows growth complementation of viral mutants. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2709–2713. doi: 10.1073/pnas.83.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Plotch S. J., Bouloy M., Ulmanen I., Krug R. M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981 Mar;23(3):847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- Smith G. E., Summers M. D., Fraser M. J. Production of human beta interferon in insect cells infected with a baculovirus expression vector. Mol Cell Biol. 1983 Dec;3(12):2156–2165. doi: 10.1128/mcb.3.12.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. E., Vlak J. M., Summers M. D. Physical Analysis of Autographa californica Nuclear Polyhedrosis Virus Transcripts for Polyhedrin and 10,000-Molecular-Weight Protein. J Virol. 1983 Jan;45(1):215–225. doi: 10.1128/jvi.45.1.215-225.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I., Broni B. A., Krug R. M. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7355–7359. doi: 10.1073/pnas.78.12.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I., Broni B., Krug R. M. Influenza virus temperature-sensitive cap (m7GpppNm)-dependent endonuclease. J Virol. 1983 Jan;45(1):27–35. doi: 10.1128/jvi.45.1.27-35.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]