Abstract
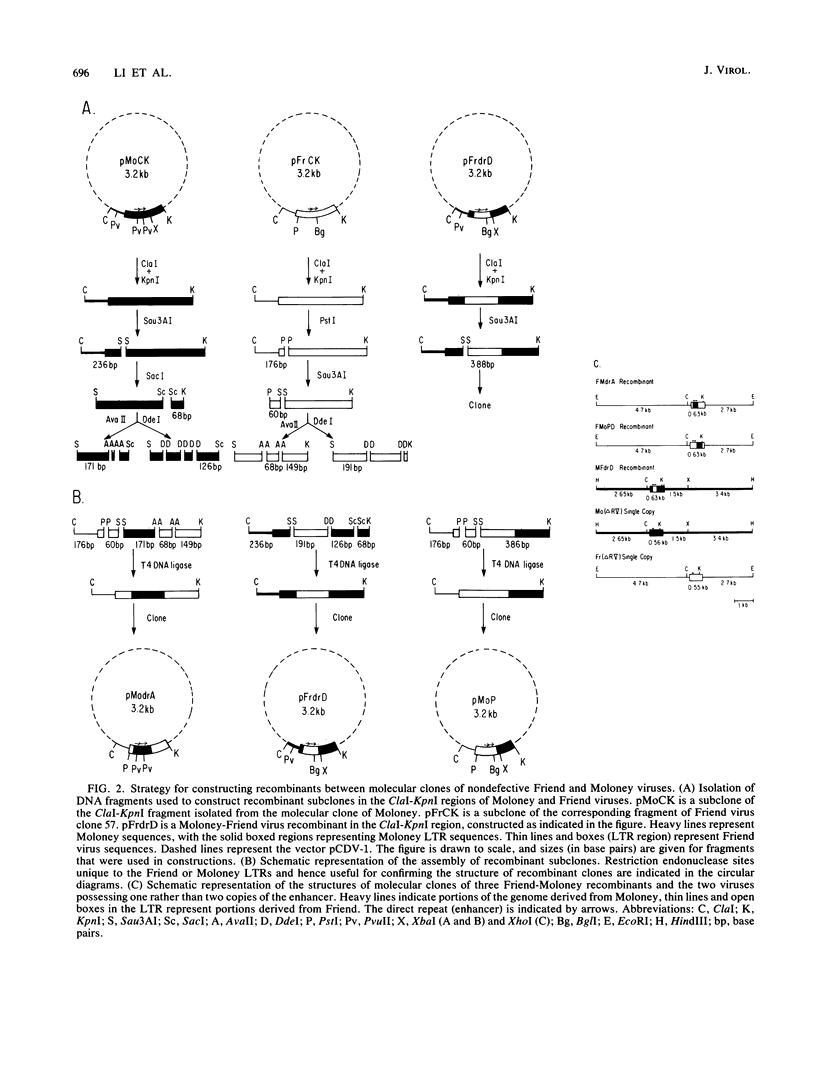
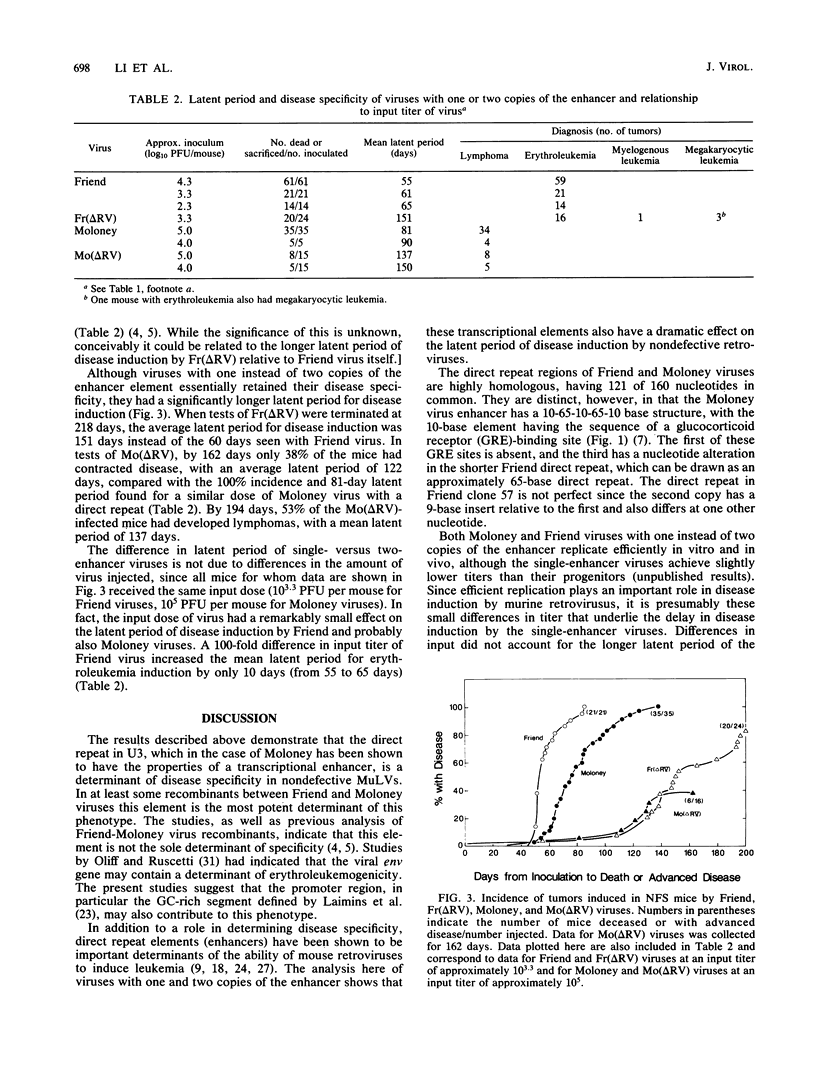
Moloney murine leukemia virus induces T cell lymphomas after injection into NFS mice, whereas the nondefective Friend virus induces erythroleukemias. Previous studies showed that sequences encompassing the viral transcriptional signals in U3 are the primary determinant of this phenotype in recombinants between these two viruses. To more precisely identify the sequences responsible, we constructed additional recombinants, within U3, between Friend and Moloney viruses and assayed these recombinants for for their disease specificity. We found that a fragment 191 bases long that included the direct repeat (enhancer) region plus 22 nucleotides to its 3' side from Friend virus was sufficient to convert Moloney virus to a virus that induced only erythroleukemias. A 171-base-long fragment of Moloney virus, including just the direct repeat, converted Friend virus to a virus that induced primarily lymphomas (about 85% of mice injected). We also constructed Moloney and Friend virus variants with one rather than two copies of the enhancer element. These viruses retained their disease specificity, although they exhibited a marked increase in the latent period of disease induction. Together the results suggest that 25 or fewer nucleotide differences, lying within and also just 3' of the direct repeat, are the primary determinant of the distinct disease specificities of nondefective Friend and Moloney viruses.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aaronson S. A., Todaro G. J. Development of 3T3-like lines from Balb-c mouse embryo cultures: transformation susceptibility to SV40. J Cell Physiol. 1968 Oct;72(2):141–148. doi: 10.1002/jcp.1040720208. [DOI] [PubMed] [Google Scholar]
- Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970 Jun 27;226(5252):1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Silver J. E., Frederickson T. N., Hopkins N., Hartley J. W. A 3' end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984 Oct;52(1):248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone R. D., Weber-Benarous A., Baorto D., Mulligan R. C. Regulated expression of a complete human beta-globin gene encoded by a transmissible retrovirus vector. Mol Cell Biol. 1987 Feb;7(2):887–897. doi: 10.1128/mcb.7.2.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Yamamoto K. R. Two different factors act separately or together to specify functionally distinct activities at a single transcriptional enhancer. Mol Cell Biol. 1986 Apr;6(4):993–1001. doi: 10.1128/mcb.6.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Jolicoeur P. Thymotropism of murine leukemia virus is conferred by its long terminal repeat. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4203–4207. doi: 10.1073/pnas.80.14.4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Villemur R., Jolicoeur P. The high leukemogenic potential of Gross passage A murine leukemia virus maps in the region of the genome corresponding to the long terminal repeat and to the 3' end of env. J Virol. 1983 Jul;47(1):24–32. doi: 10.1128/jvi.47.1.24-32.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller D. V., Hopkins N. RNase T1-resistant oligonucleotides of B-tropic murine leukemia virus from BALB/c and five of its NB-tropic derivatives. J Virol. 1977 Jul;23(1):188–195. doi: 10.1128/jvi.23.1.188-195.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautsch J. W., Wilson M. C. Delayed de novo methylation in teratocarcinoma suggests additional tissue-specific mechanisms for controlling gene expression. Nature. 1983 Jan 6;301(5895):32–37. doi: 10.1038/301032a0. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Goff S., Shields A., Yoshimura F., Mitra S., Baltimore D. In vitro synthesis of a 9 kbp terminally redundant DNA carrying the infectivity of Moloney murine leukemia virus. Cell. 1979 Apr;16(4):863–874. doi: 10.1016/0092-8674(79)90101-6. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Rigby P. W., Lane D. P. Negative regulation of viral enhancers in undifferentiated embryonic stem cells. Cell. 1985 Sep;42(2):519–526. doi: 10.1016/0092-8674(85)90109-6. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. At least four viral genes contribute to the leukemogenicity of murine retrovirus MCF 247 in AKR mice. J Virol. 1985 Jan;53(1):158–165. doi: 10.1128/jvi.53.1.158-165.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto A., Adachi A., Sakai K., Matsuyama M. Long terminal repeat of Friend-MCF virus contains the sequence responsible for erythroid leukemia. Virology. 1985 Feb;141(1):30–42. doi: 10.1016/0042-6822(85)90180-1. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. Retroviruses and embryogenesis: microinjection of Moloney leukemia virus into midgestation mouse embryos. Cell. 1980 Jan;19(1):181–188. doi: 10.1016/0092-8674(80)90399-2. [DOI] [PubMed] [Google Scholar]
- Jainchill J. L., Aaronson S. A., Todaro G. J. Murine sarcoma and leukemia viruses: assay using clonal lines of contact-inhibited mouse cells. J Virol. 1969 Nov;4(5):549–553. doi: 10.1128/jvi.4.5.549-553.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Zimmermann W., Oliff A., Friedrich R. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J Virol. 1984 Mar;49(3):828–840. doi: 10.1128/jvi.49.3.828-840.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Levinson B., Khoury G., Vande Woude G., Gruss P. Activation of SV40 genome by 72-base pair tandem repeats of Moloney sarcoma virus. Nature. 1982 Feb 18;295(5850):568–572. doi: 10.1038/295568a0. [DOI] [PubMed] [Google Scholar]
- Linney E., Davis B., Overhauser J., Chao E., Fan H. Non-function of a Moloney murine leukaemia virus regulatory sequence in F9 embryonal carcinoma cells. 1984 Mar 29-Apr 4Nature. 308(5958):470–472. doi: 10.1038/308470a0. [DOI] [PubMed] [Google Scholar]
- Lung M. L., Hering C., Hartley J. W., Rowe W. P., Hopkins N. Analysis of the genomes of mink cell focus-inducing murine type-C viruses: a progress report. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 2):1269–1274. doi: 10.1101/sqb.1980.044.01.138. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Okayama H., Berg P. A cDNA cloning vector that permits expression of cDNA inserts in mammalian cells. Mol Cell Biol. 1983 Feb;3(2):280–289. doi: 10.1128/mcb.3.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A. I., Hager G. L., Chang E. H., Scolnick E. M., Chan H. W., Lowy D. R. Transfection of molecularly cloned Friend murine leukemia virus DNA yields a highly leukemogenic helper-independent type C virus. J Virol. 1980 Jan;33(1):475–486. doi: 10.1128/jvi.33.1.475-486.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff A., Ruscetti S. A 2.4-kilobase-pair fragment of the Friend murine leukemia virus genome contains the sequences responsible for friend murine leukemia virus-induced erythroleukemia. J Virol. 1983 Jun;46(3):718–725. doi: 10.1128/jvi.46.3.718-725.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe W. P., Pugh W. E., Hartley J. W. Plaque assay techniques for murine leukemia viruses. Virology. 1970 Dec;42(4):1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Vogt M., Haggblom C., Swift S., Haas M. Envelope gene and long terminal repeat determine the different biological properties of Rauscher, Friend, and Moloney mink cell focus-inducing viruses. J Virol. 1985 Jul;55(1):184–192. doi: 10.1128/jvi.55.1.184-192.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F. K., Davison B., Chaffin K. Murine leukemia virus long terminal repeat sequences can enhance gene activity in a cell-type-specific manner. Mol Cell Biol. 1985 Oct;5(10):2832–2835. doi: 10.1128/mcb.5.10.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wachter R., Fiers W. Preparative two-dimensional polyacrylamide gel electrophoresis of 32 P-labeled RNA. Anal Biochem. 1972 Sep;49(1):184–197. doi: 10.1016/0003-2697(72)90257-6. [DOI] [PubMed] [Google Scholar]


