Abstract
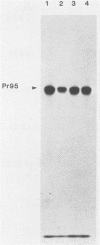
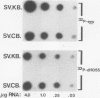
Expression of the Rous sarcoma virus envelope gene (env) from a simian virus 40 (SV40) late-region replacement vector is dependent on the position of env within the SV40 late-region sequences. The difference in expression levels appeared to be due to differences in the efficiency with which the env-specific transcripts were translated, because transcription levels from different constructions were similar. Deletion of the nucleotides encoding the agnoprotein initiator codon, located upstream of the env sequences in the poorly expressed construct, resulted in high levels of env expression. The agnoprotein initiator codon and overlapping open reading frame thus act as strong barriers to further ribosome scanning and prevent initiation at the env AUG codon. We conclude that AUG codons present in the late region of SV40 can reduce expression of inserted genes positioned downstream. Nevertheless, intrinsic properties of the gene may determine its ultimate level of expression.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bos T. J., Davis A. R., Nayak D. P. NH2-terminal hydrophobic region of influenza virus neuraminidase provides the signal function in translocation. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2327–2331. doi: 10.1073/pnas.81.8.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady J., Radonovich M., Vodkin M., Natarajan V., Thoren M., Das G., Janik J., Salzman N. P. Site-specific base substitution and deletion mutations that enhance or suppress transcription of the SV40 major late RNA. Cell. 1982 Dec;31(3 Pt 2):625–633. doi: 10.1016/0092-8674(82)90318-x. [DOI] [PubMed] [Google Scholar]
- Ganem D., Nussbaum A. L., Davoli D., Fareed G. C. Propagation of a segment of bacteriophage lamda-DNA in monkey cells after covalent linkage to a defective simian virus 40 genome. Cell. 1976 Mar;7(3):349–359. doi: 10.1016/0092-8674(76)90164-1. [DOI] [PubMed] [Google Scholar]
- Gething M. J., Sambrook J. Cell-surface expression of influenza haemagglutinin from a cloned DNA copy of the RNA gene. Nature. 1981 Oct 22;293(5834):620–625. doi: 10.1038/293620a0. [DOI] [PubMed] [Google Scholar]
- Goff S. P., Berg P. Construction of hybrid viruses containing SV40 and lambda phage DNA segments and their propagation in cultured monkey cells. Cell. 1976 Dec;9(4 Pt 2):695–705. doi: 10.1016/0092-8674(76)90133-1. [DOI] [PubMed] [Google Scholar]
- Gruss P., Khoury G. Expression of simian virus 40-rat preproinsulin recombinants in monkey kidney cells: use of preproinsulin RNA processing signals. Proc Natl Acad Sci U S A. 1981 Jan;78(1):133–137. doi: 10.1073/pnas.78.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer D. H., Leder P. Expression of the chromosomal mouse Beta maj-globin gene cloned in SV40. Nature. 1979 Sep 6;281(5726):35–40. doi: 10.1038/281035a0. [DOI] [PubMed] [Google Scholar]
- Hardwick J. M., Shaw K. E., Wills J. W., Hunter E. Amino-terminal deletion mutants of the Rous sarcoma virus glycoprotein do not block signal peptide cleavage but can block intracellular transport. J Cell Biol. 1986 Sep;103(3):829–838. doi: 10.1083/jcb.103.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter E., Hill E., Hardwick M., Bhown A., Schwartz D. E., Tizard R. Complete sequence of the Rous sarcoma virus env gene: identification of structural and functional regions of its product. J Virol. 1983 Jun;46(3):920–936. doi: 10.1128/jvi.46.3.920-936.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. F., Baltimore D. Polypeptide cleavages in the formation of poliovirus proteins. Proc Natl Acad Sci U S A. 1968 Sep;61(1):77–84. doi: 10.1073/pnas.61.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay G., Nomura S., Anderson C. W., Khoury G. Identification of the SV40 agnogene product: a DNA binding protein. Nature. 1981 May 28;291(5813):346–349. doi: 10.1038/291346a0. [DOI] [PubMed] [Google Scholar]
- Keller J. M., Alwine J. C. Activation of the SV40 late promoter: direct effects of T antigen in the absence of viral DNA replication. Cell. 1984 Feb;36(2):381–389. doi: 10.1016/0092-8674(84)90231-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Mechanism of mRNA recognition by eukaryotic ribosomes during initiation of protein synthesis. Curr Top Microbiol Immunol. 1981;93:81–123. doi: 10.1007/978-3-642-68123-3_5. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 1984 Mar 15;308(5956):241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. Regulation of protein synthesis in virus-infected animal cells. Adv Virus Res. 1986;31:229–292. doi: 10.1016/S0065-3527(08)60265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Selection of initiation sites by eucaryotic ribosomes: effect of inserting AUG triplets upstream from the coding sequence for preproinsulin. Nucleic Acids Res. 1984 May 11;12(9):3873–3893. doi: 10.1093/nar/12.9.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Petersen R. B., Hensel C. H., Hackett P. B. Identification of a ribosome-binding site for a leader peptide encoded by Rous sarcoma virus RNA. J Virol. 1984 Sep;51(3):722–729. doi: 10.1128/jvi.51.3.722-729.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipas J. M., Peden K. W., Nathans D. Mutational analysis of simian virus 40 T antigen: isolation and characterization of mutants with deletions in the T-antigen gene. Mol Cell Biol. 1983 Feb;3(2):203–213. doi: 10.1128/mcb.3.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J., Condra J. H., Arnheiter H., Lazzarini R. A. Expression of a recombinant DNA gene coding for the vesicular stomatitis virus nucleocapsid protein. J Virol. 1983 Feb;45(2):773–781. doi: 10.1128/jvi.45.2.773-781.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveda M. M., Lai C. J. Functional expression in primate cells of cloned DNA coding for the hemagglutinin surface glycoprotein of influenza virus. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5488–5492. doi: 10.1073/pnas.78.9.5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills J. W., Hardwick J. M., Shaw K., Hunter E. Alterations in the transport and processing of Rous sarcoma virus envelope glycoproteins mutated in the signal and anchor regions. J Cell Biochem. 1983;23(1-4):81–94. doi: 10.1002/jcb.240230109. [DOI] [PubMed] [Google Scholar]
- Wills J. W., Srinivas R. V., Hunter E. Mutations of the Rous sarcoma virus env gene that affect the transport and subcellular location of the glycoprotein products. J Cell Biol. 1984 Dec;99(6):2011–2023. doi: 10.1083/jcb.99.6.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]