Abstract
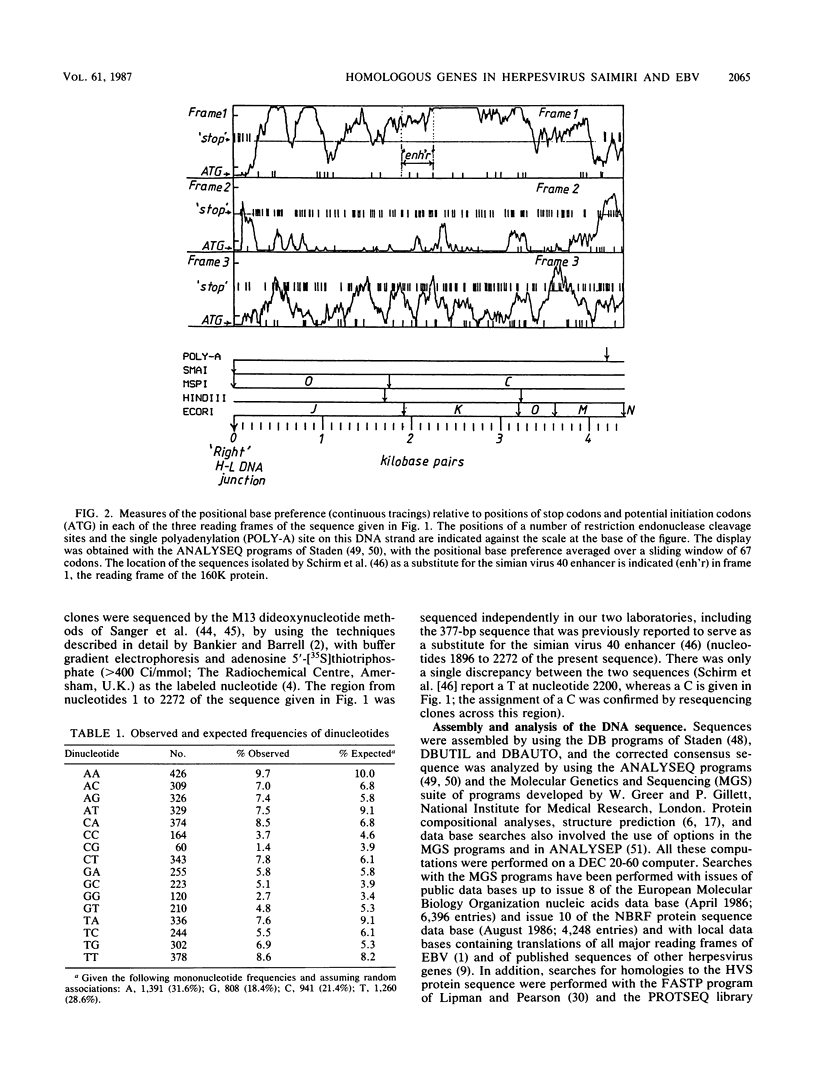
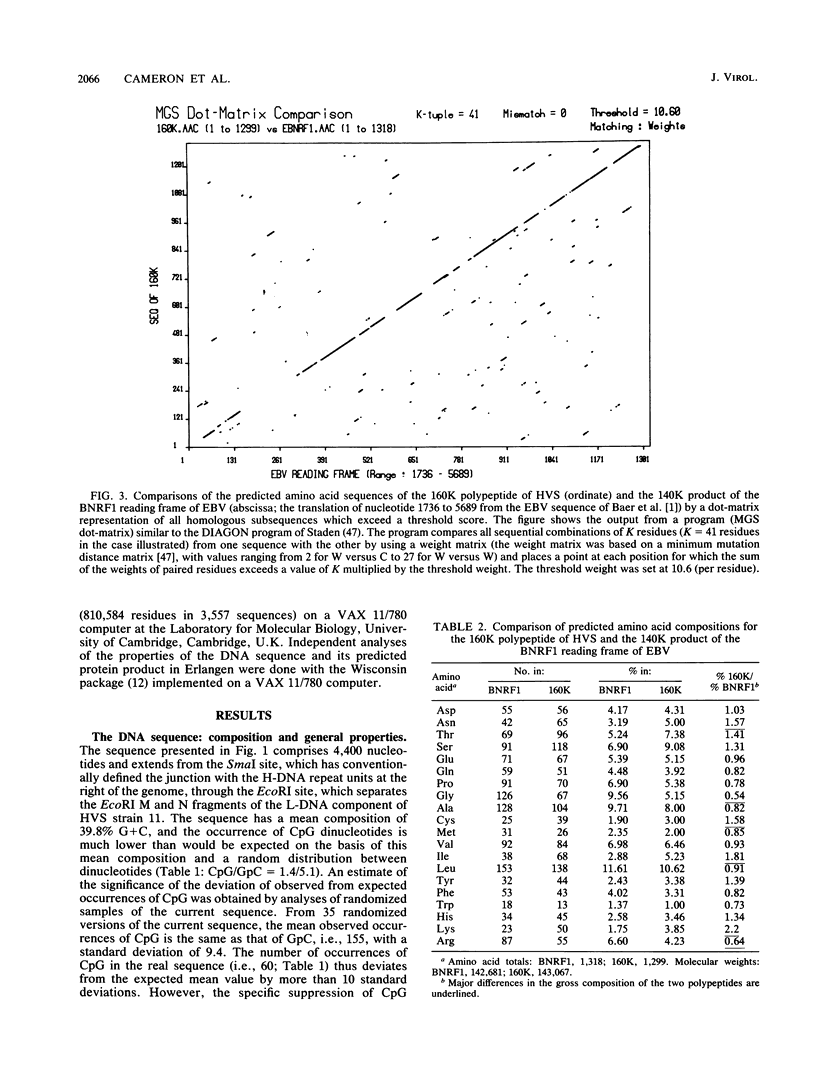
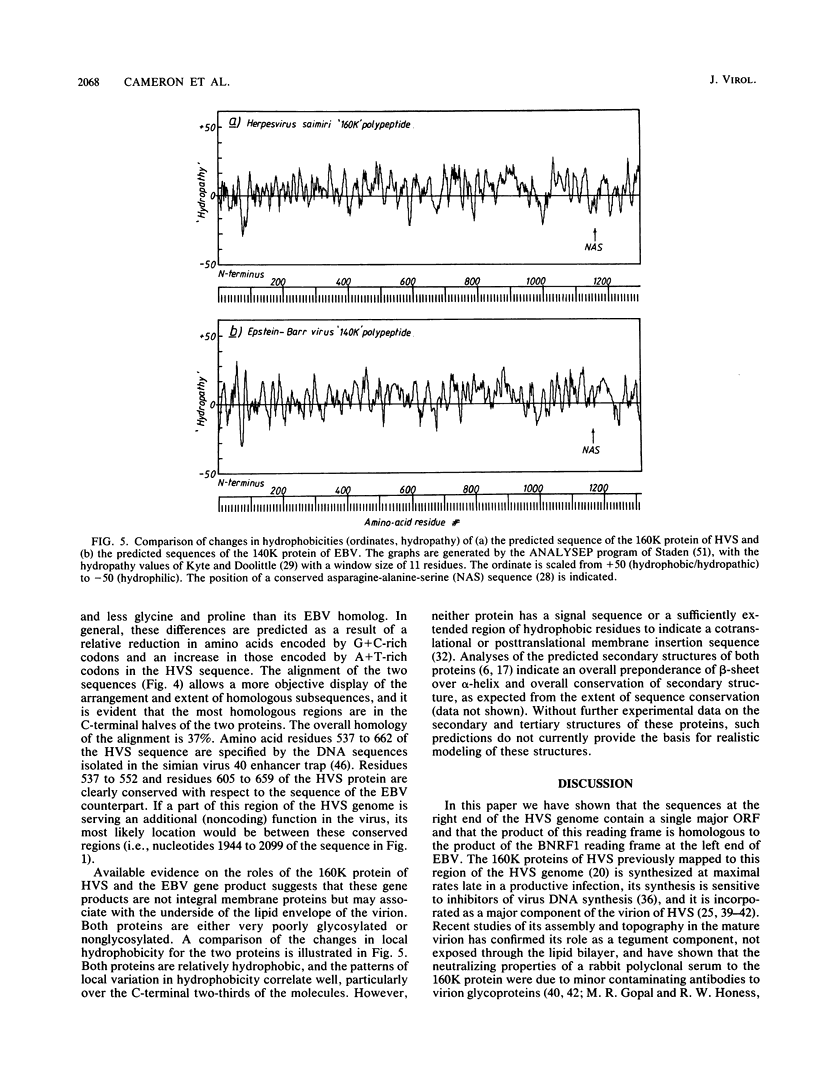
The sequence of 4.4 kilobase pairs (kbp) from the conventional right terminus of the A + T-rich light-DNA (L-DNA) sequences of the herpesvirus saimiri (HVS) genome contains a leftward-directed open reading frame (ORF) for a 1,299-residue protein. The molecular weight predicted for the protein (143,000) is in good agreement with the estimates of 150,000 to 160,000 for the major nonglycosylated polypeptide of the virion tegument (the 160K polypeptide), previously shown to be encoded by this region of the genome. The first initiation codon of the ORF is only 250 nucleotides from the junction of the L-DNA component with the G + C-rich terminal reiterations (i.e., heavy or H-DNA) of the genome. An unusually A + T-rich sequence (43 of 45 nucleotides are A or T, relative to a mean composition of 40% G + C for the ORF) occurs some 75 bp 5' to this initiation codon, and the first adenylation signal (AATAAA) on this DNA strand occurs 18 bp 3' to the termination codon. The amino acid sequence predicted for the 160K protein of HVS is homologous over most of its length to the 1,318-residue protein encoded by the leftmost major ORF of the G + C-rich genome of Epstein-Barr virus (BNRF1, the 140K nonglycosylated membrane antigen). No homology to either of these proteins is evident among the products predicted from the complete sequence of the alpha herpesvirus varicella-zoster virus. Thus gamma herpesviruses with coding sequences which differ in mean nucleotide composition by some 20% G + C have homologous proteins encoded at similar positions with respect to genome termini, with the right end of HVS being homologous to the left end of Epstein-Barr virus.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Bankier A. T., Dietrich W., Baer R., Barrell B. G., Colbère-Garapin F., Fleckenstein B., Bodemer W. Terminal repetitive sequences in herpesvirus saimiri virion DNA. J Virol. 1985 Jul;55(1):133–139. doi: 10.1128/jvi.55.1.133-139.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodemer W., Knust E., Angermüller S., Fleckenstein B. Immediate-early transcription of Herpesvirus saimiri. J Virol. 1984 Aug;51(2):452–457. doi: 10.1128/jvi.51.2.452-457.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Costa R. H., Draper K. G., Kelly T. J., Wagner E. K. An unusual spliced herpes simplex virus type 1 transcript with sequence homology to Epstein-Barr virus DNA. J Virol. 1985 May;54(2):317–328. doi: 10.1128/jvi.54.2.317-328.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., McGeoch D. J. Evolutionary comparisons of the S segments in the genomes of herpes simplex virus type 1 and varicella-zoster virus. J Gen Virol. 1986 Apr;67(Pt 4):597–611. doi: 10.1099/0022-1317-67-4-597. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. DNA sequence of the major capsid protein gene of herpes simplex virus type 1. J Gen Virol. 1986 Oct;67(Pt 10):2279–2286. doi: 10.1099/0022-1317-67-10-2279. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Location and orientation of homologous sequences in the genomes of five herpesviruses. J Gen Virol. 1983 Sep;64(Pt 9):1927–1942. doi: 10.1099/0022-1317-64-9-1927. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolyniuk M., Pritchett R., Kieff E. Proteins of Epstein-Barr virus. I. Analysis of the polypeptides of purified enveloped Epstein-Barr virus. J Virol. 1976 Mar;17(3):935–949. doi: 10.1128/jvi.17.3.935-949.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolyniuk M., Wolff E., Kieff E. Proteins of Epstein-Barr Virus. II. Electrophoretic analysis of the polypeptides of the nucleocapsid and the glucosamine- and polysaccharide-containing components of enveloped virus. J Virol. 1976 Apr;18(1):289–297. doi: 10.1128/jvi.18.1.289-297.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk L. A., Wolfe L. G., Deinhardt F. Isolation of Herpesvirus saimiri from blood of squirrel monkeys (Saimiri sciureus). J Natl Cancer Inst. 1972 May;48(5):1499–1505. [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gibson T., Stockwell P., Ginsburg M., Barrell B. Homology between two EBV early genes and HSV ribonucleotide reductase and 38K genes. Nucleic Acids Res. 1984 Jun 25;12(12):5087–5099. doi: 10.1093/nar/12.12.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompels U., Minson A. The properties and sequence of glycoprotein H of herpes simplex virus type 1. Virology. 1986 Sep;153(2):230–247. doi: 10.1016/0042-6822(86)90026-7. [DOI] [PubMed] [Google Scholar]
- Hell W., Modrow S., Wolf H. Mapping of herpesvirus saimiri proteins on the viral genome: proteins dependent and not dependent on viral DNA synthesis. J Virol. 1985 Nov;56(2):414–418. doi: 10.1128/jvi.56.2.414-418.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Bodemer W., Cameron K. R., Niller H. H., Fleckenstein B., Randall R. E. The A+T-rich genome of Herpesvirus saimiri contains a highly conserved gene for thymidylate synthase. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3604–3608. doi: 10.1073/pnas.83.11.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W. Herpes simplex and 'the herpes complex': diverse observations and a unifying hypothesis. The eighth Fleming lecture. J Gen Virol. 1984 Dec;65(Pt 12):2077–2107. doi: 10.1099/0022-1317-65-12-2077. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Watson D. H. Unity and diversity in the herpesviruses. J Gen Virol. 1977 Oct;37(1):15–37. doi: 10.1099/0022-1317-37-1-15. [DOI] [PubMed] [Google Scholar]
- Hudson G. S., Bankier A. T., Satchwell S. C., Barrell B. G. The short unique region of the B95-8 Epstein-Barr virus genome. Virology. 1985 Nov;147(1):81–98. doi: 10.1016/0042-6822(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Keil G., Fleckenstein B., Bodemer W. Structural proteins of Herpesvirus saimiri. J Virol. 1983 Sep;47(3):463–470. doi: 10.1128/jvi.47.3.463-470.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust E., Schirm S., Dietrich W., Bodemer W., Kolb E., Fleckenstein B. Cloning of Herpesvirus saimiri DNA fragments representing the entire L-region of the genome. Gene. 1983 Nov;25(2-3):281–289. doi: 10.1016/0378-1119(83)90232-9. [DOI] [PubMed] [Google Scholar]
- Kornfeld R., Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Littler E., Zeuthen J., McBride A. A., Trøst Sørensen E., Powell K. L., Walsh-Arrand J. E., Arrand J. R. Identification of an Epstein-Barr virus-coded thymidine kinase. EMBO J. 1986 Aug;5(8):1959–1966. doi: 10.1002/j.1460-2075.1986.tb04450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Davison A. J. DNA sequence of the herpes simplex virus type 1 gene encoding glycoprotein gH, and identification of homologues in the genomes of varicella-zoster virus and Epstein-Barr virus. Nucleic Acids Res. 1986 May 27;14(10):4281–4292. doi: 10.1093/nar/14.10.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Dolan A., Frame M. C. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the exonuclease gene and neighbouring genes. Nucleic Acids Res. 1986 Apr 25;14(8):3435–3448. doi: 10.1093/nar/14.8.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J. On the predictive recognition of signal peptide sequences. Virus Res. 1985 Oct;3(3):271–286. doi: 10.1016/0168-1702(85)90051-6. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- O'Hare P., Honess R. W. Identification of a subset of herpesvirus saimiri polypeptides synthesized in the absence of virus DNA replication. J Virol. 1983 Apr;46(1):279–283. doi: 10.1128/jvi.46.1.279-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett P. E., Biggin M. D., Barrell B., Roizman B. Epstein-Barr virus genome may encode a protein showing significant amino acid and predicted secondary structure homology with glycoprotein B of herpes simplex virus 1. J Virol. 1985 Dec;56(3):807–813. doi: 10.1128/jvi.56.3.807-813.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J. P., McGeoch D. J. DNA sequence of the region in the genome of herpes simplex virus type 1 containing the genes for DNA polymerase and the major DNA binding protein. Nucleic Acids Res. 1985 Nov 25;13(22):8143–8163. doi: 10.1093/nar/13.22.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R. E., Honess R. W., O'Hare P. Proteins specified by herpesvirus saimiri: identification and properties of virus-specific polypeptides in productively infected cells. J Gen Virol. 1983 Jan;64(Pt 1):19–35. doi: 10.1099/0022-1317-64-1-19. [DOI] [PubMed] [Google Scholar]
- Randall R. E., Honess R. W. Proteins of herpesvirus saimiri: identification of two virus polypeptides released into the culture medium of productively infected cells. J Gen Virol. 1980 Dec;51(Pt 2):445–449. doi: 10.1099/0022-1317-51-2-445. [DOI] [PubMed] [Google Scholar]
- Randall R. E., Honess R. W. Proteins specified by Herpesvirus saimiri: purification and properties of a single polypeptide which elicits virus-neutralizing antibody. J Gen Virol. 1982 Jan;58(Pt 1):149–161. doi: 10.1099/0022-1317-58-1-149. [DOI] [PubMed] [Google Scholar]
- Randall R. E., Newman C., Honess R. W. Isolation and characterization of monoclonal antibodies to structural and nonstructural herpesvirus saimiri proteins. J Virol. 1984 Dec;52(3):872–883. doi: 10.1128/jvi.52.3.872-883.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Weber F., Schaffner W., Fleckenstein B. A transcription enhancer in the Herpesvirus saimiri genome. EMBO J. 1985 Oct;4(10):2669–2674. doi: 10.1002/j.1460-2075.1985.tb03986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Graphic methods to determine the function of nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Measurements of the effects that coding for a protein has on a DNA sequence and their use for finding genes. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):551–567. doi: 10.1093/nar/12.1part2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. The current status and portability of our sequence handling software. Nucleic Acids Res. 1986 Jan 10;14(1):217–231. doi: 10.1093/nar/14.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]


