Abstract
The members of the MCM protein family are essential eukaryotic DNA replication factors that form a six-member protein complex. In this study, we use antibodies to four MCM proteins to investigate the structure of and requirements for the formation of fission yeast MCM complexes in vivo, with particular regard to Cdc19p (MCM2). Gel filtration analysis shows that the MCM protein complexes are unstable and can be broken down to subcomplexes. Using coimmunoprecipitation, we find that Mis5p (MCM6) and Cdc21p (MCM4) are tightly associated with one another in a core complex with which Cdc19p loosely associates. Assembly of Cdc19p with the core depends upon Cdc21p. Interestingly, there is no obvious change in Cdc19p-containing MCM complexes through the cell cycle. Using a panel of Cdc19p mutants, we find that multiple domains of Cdc19p are required for MCM binding. These studies indicate that MCM complexes in fission yeast have distinct substructures, which may be relevant for function.
INTRODUCTION
DNA replication in fission yeast, as in other eukaryotes, is precisely controlled to ensure the production of exactly one copy of the genome per cell division. Many elements contribute to this control, including cell cycle regulators, origin-associated factors, and the DNA replication machinery (reviewed in Forsburg, 1996; MacNeill and Nurse, 1997). An essential group of factors required for the regulation of DNA replication is the MCM protein family. The six members of this family are named for the original Saccharomyces cerevisiae mutants defective in minichromosome maintenance (reviewed in Tye, 1994; Kearsey et al., 1996). Homologs have been identified in many metazoans, including humans, Xenopus, mouse, and Drosophila (reviewed in Tye, 1994; Su et al., 1995; Botchan, 1996; Kearsey et al., 1996; Rowles and Blow, 1997). Each of the six MCM family members has a central core of homology that contains sequences related to DNA-dependent ATPases (Koonin, 1993).
In the fission yeast, Schizosaccharomyces pombe, the genes for four MCM proteins have been cloned and shown to be essential: cdc19+/nda1+ (MCM2, Miyake et al., 1993; Forsburg and Nurse, 1994), cdc21+ (MCM4, Coxon et al., 1992), nda4+ (MCM5, Miyake et al., 1993), and mis5+ (MCM6, Takahashi et al., 1994). As in other cell types, the S. pombe MCMs form a heteromeric complex (Okishio et al., 1996; Adachi et al., 1997). There are genetic interactions not only among the fission yeast MCM mutants (Forsburg and Nurse, 1994; Takahashi et al., 1994), but between these MCM mutants and other mutants affecting DNA replication (Grallert and Nurse, 1996; Forsburg et al., 1997). However, it is not clear why six closely related MCM proteins are essential for the formation of a functional MCM protein complex.
To further characterize the roles of individual MCM proteins in the regulation of DNA replication, we have been investigating the role of fission yeast Cdc19p in the formation of the MCM protein complex. In this report, we show that Cdc19p association with the MCM complex does not vary significantly during the cell cycle. The complex is unstable and we present evidence for a core structure containing at least two subunits. By characterizing the interactions between different MCMs, we demonstrate that Cdc19p weakly associates with a core subcomplex of tightly bound MCMs including Mis5p and Cdc21p. Cdc21p is essential for Cdc19p association with the core MCM proteins. Finally, using a panel of deletion and point mutations throughout Cdc19p, we find that multiple domains of this protein are required for association with the other MCM proteins. Thus, the fission yeast MCM complex is not a homogeneous structure, which may indicate distinct roles for individual MCM proteins.
MATERIALS AND METHODS
Yeast Strains and Plasmids
S. pombe strains were grown in Edinburgh minimal medium and supplemented with adenine, leucine, and uracil when required (Moreno et al., 1991). In this study, “wild type” refers to strain FY254 (h- ura4-D18 leu1–32 ade6-M210 can1–1, Forsburg and Nurse, 1994). cdc10 (FY562), cdc17 (FY322), cdc19 (FY243), cdc22 (FY583), and cdc25 (FY584) mutant strains were described by Forsburg et al. (1997). The genotype of the cdc21 mutant strain (FY786) is h- cdc21-M68 ura4-D18 leu1–32 ade6-M216. Unless noted, cultures were grown at 32°C. Cells were transformed using electroporation (Kelly et al., 1993). When required, the nmt1+ promoter was repressed with 5 μg/ml thiamine in the media (Maundrell, 1990). In cell cycle block experiments, strains were grown at 25°C to early exponential phase and then shifted to 36°C for 4 h.
For construction of the nda4-HA strain (FY803), a 1.3-kb fragment from BalI to SalI containing the C terminus of nda4+, tagged with a triple HA epitope, was isolated from plasmid pSLF204 (Forsburg et al., 1997), and cloned into SmaI–SalI cut plasmid pJK148 (Keeney and Boeke, 1994) to create plasmid pSLF244. The plasmid was linearized with EcoRI within the nda4+ sequence and integrated into a diploid strain, resulting in a tandem partial duplication at the nda4+ locus. After sporulation, leu1+ progeny were isolated. The presence of the nda4-HA allele and loss of the nda4+ allele were verified by Western blotting.
Plasmids carrying HA-tagged mis5+, cdc21+, and cdc19 variants were described by Forsburg et al. (1997). Strain FY863 contains the HA-mis5+ plasmid pSLF225 in a haploid strain with a Δmis5::his3+ disruption in the chromosome. Similarly, the strain FY862 contains the HA-cdc21+ plasmid pSLF221 in a strain with Δcdc21::his3+ in the chromosome (Liang, Hodson, and Forsburg, in preparation). The cdc19-M4 mutant was constructed and cloned into the HA-tagging nmt1+ expression vector pSLF172 (Forsburg and Sherman, 1997) as described previously (Forsburg et al., 1997). Plasmid pMF56, expressing N-terminally HA-tagged Cdc18p under control of the nmt1+ promoter was a kind gift of Marco Muzi-Falconi and Tom Kelly (Muzi-Falconi et al., 1996).
Two-Hybrid Screen
We constructed a “bait” protein corresponding to Cdc19-D7 fused to a GAL4 DNA-binding domain, for use in a two-hybrid screen (Fields and Song, 1989). The full-length Cdc19p bait protein was capable of transactivation in the absence of any “prey” molecule, possibly due to its acidic N terminus. All materials including plasmids, strains, and an S. pombe cDNA library were the generous gift of Steve Elledge (Baylor College of Medicine, Houston, TX). We screened approximately 1.5 million cDNA clones for β-galactosidase expression and isolated two clones that contained similarly truncated versions of cdc19+. We retested these clones and a reconstructed full-length cdc19+ fused to the GAL4 activation domain, against the original bait. All showed a positive interaction compared with control plasmids.
Antibodies
Antibodies to Mis5p, Nda4p, and Cdc21p were prepared as follows. MCM polypeptide fragments corresponding to the nonconserved N-terminal regions of the proteins were expressed in Escherichia coli as 6xHis-tagged fusion proteins in pRSET vectors (Invitrogen, San Diego, CA). pSGP11 has the 1230-bp SmaI–EcoRV fragment from pAC1 (Coxon et al., 1992) subcloned into PvuII-digested pRSETC and encodes a 47-kDa Cdc21p fragment (amino acid residues 125–534). The amino-terminal 1375 base pairs (bp) of nda4+ were subcloned as a PstI/SphI (end-filled) fragment from pTZ-nda4+ (Forsburg et al., 1997) into pRSETB digested with PstI and EcoRI (end-filled) to generate pSGP14. Subsequently, the 809-bp BglII/HindIII fragment from pSGP14 was subcloned into pRSETC digested with BglII and HindIII to generate pSGP24, which encodes a 31-kDa Nda4p fragment (amino acid residues 67–345). pSGP15 has the 895-bp XhoI/HindIII fragment from pTZ-mis5+ (Forsburg et al., 1997) subcloned into pRSETA digested with XhoI and HindIII and encodes a 34-kDa Mis5p fragment (amino acid residues 43–341).
Polypeptide fragments of the MCM proteins were purified from 500-ml cultures of E. coli BL21(DE3)pLysS cells harboring pSGP11, pSGP24, or pSGP15 to obtain His-tagged fragments of Cdc21p, Nda4p, or Mis5p, respectively. Uninduced cultures were grown at 37°C to 0.5 OD595 and then induced for expression with 0.4 mM isopropyl-β-d-thiogalactopyranoside for 3 h. Cells were harvested in 50-ml aliquots, and pellets were stored at −70°C. For purification of the recombinant proteins, two bacterial pellets were resuspended in a total of 7.5 ml Buffer B (8 M urea, 0.1 M Na2HPO4, and 10 mM Tris, pH 8.0), and the His-tagged proteins were purified with a Ni-NTA agarose column (QIAGEN, Chatsworth, CA) with urea-based buffers, as per the manufacturer’s recommendations. During dialysis against PBS, the purified protein precipitated and was solubilized in 0.1% SDS. Rabbits were injected subcutaneously with the purified proteins and injected with four subsequent boosts. Antibodies were precipitated from crude sera by ammonium sulfate precipitation followed by dialysis against PBS, and affinity purified from Western blots using bacterially produced polypeptide fragments.
Anti-Cdc19p affinity-purified polyclonal serum 5616 was described previously (Forsburg et al., 1997). Monoclonal anti-HA 12CA5 antibody was a kind gift of Jill Meissenholder and Tony Hunter. Monoclonal anti-α-tubulin antibody (T5168) was purchased from Sigma Chemical (St. Louis, MO).
Protein Extracts and Immunoblotting
Cell lysates were prepared by glass bead lysis (Moreno et al., 1991) in lysis buffer (50 mM HEPES, pH 7.0, 50 mM potassium acetate, 5 mM magnesium acetate, and 100 mM sorbitol) with the addition of 1 mM ATP, 1 mM DTT, and protease inhibitors. Lysates were cleared by spinning at 16,000 × g for 20 min. When noted, total protein concentrations were determined by BCA protein assay (Pierce, Rockford, IL). For Western blotting, samples were boiled in SDS sample buffer (100 mM Tris, pH 6.8, 20% glycerol, 4% SDS, 200 mM dithiothreitol, 0.02% bromophenol blue), fractionated by SDS-PAGE (Protogel, National Diagnostics, Atlanta, GA) and transferred to Immobilon-P (Millipore, Bedford, MA). Samples were run on 7% SDS-PAGE gels, except where noted. Detection was performed using HRP-conjugated anti-rabbit or anti-mouse secondary antibodies and ECL (Amersham, Arlington, IL). Films were digitally scanned into Canvas 5.0.2 for the Macintosh, and composite images were printed on a Fujix (Tokyo, Japan) printer.
Gel Filtration
Wild-type cell lysate was cleared at 100,000 × g for 20 min, and 5 mg total protein were loaded on a Superose 6 gel filtration column (Pharmacia, Piscataway, NY). Elution buffer was as follows: 50 mM HEPES, pH 7.0, 50 mM potassium acetate, 5 mM magnesium acetate, 100 mM sorbitol. Glycerol (10%) was substituted for the sorbitol when required; 0.75-ml fractions were collected, and 10 μl of each fraction were diluted with an equal volume of SDS sample buffer and boiled, and 15 μl were loaded on SDS-polyacrylamide gels for analysis. Markers used were gel filtration standards (Bio-Rad, Richmond, CA).
Immunoprecipitations
Approximately 1–2 μg immunoglobulin of appropriate antibody were added to cleared cell lysates (usually 250–750 μg total protein). Lysis buffer was added to bring the total volume to ∼300–400 μl. Samples were incubated overnight at 4°C, with gentle agitation. After the addition of 50 μl of Protein A-Sepharose CL-4B (Sigma, 1:1 in lysis buffer), incubation was continued for 1.5 h. Samples were spun briefly to pellet the Sepharose beads. The pellets were washed four times with 750 μl of lysis buffer or, where noted, with modified RIPA buffer (50 mM Tris, pH 7.5, 150 mM sodium chloride, 1% Nonidet P-40, 0.5% sodium deoxycholate). Immunoprecipitates were boiled in 70 μl SDS sample buffer, and 10 μl were loaded on SDS-polyacrylamide gels for analysis.
Flow Cytometry
Cells were fixed in ice-cold 70% ethanol and stained for flow cytometry, as described previously (Sazer and Sherwood, 1990). Flow cytometry was performed on a FACScan (Becton Dickinson, Nutley, NJ), and data analysis was carried out using Cell Quest software for the Macintosh.
RESULTS
Antibodies to S. pombe MCMs
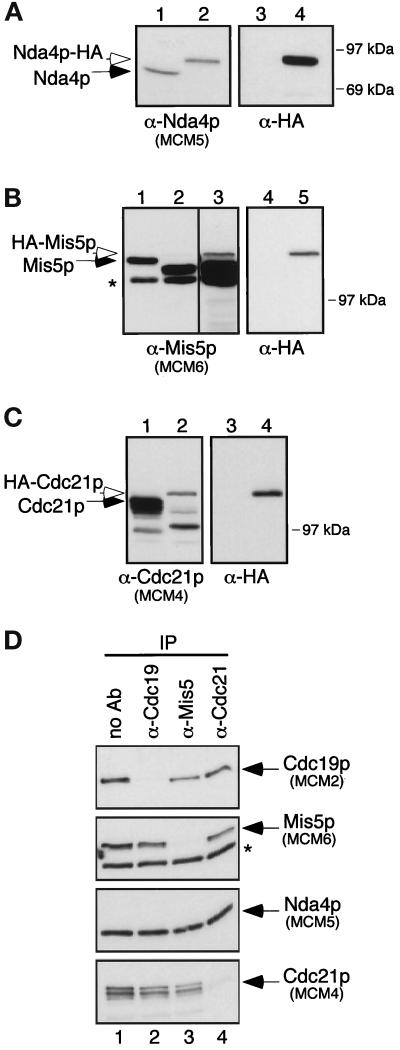
To characterize MCM complexes, we raised polyclonal rabbit antibodies to the four fission yeast MCM proteins. Characterization of the anti-Cdc19p antibody was previously described (Forsburg et al., 1997). Antibodies to Nda4p, Mis5p, and Cdc21p were raised against bacterially produced protein fragments corresponding to the nonconserved N-terminal halves of each protein (see MATERIALS AND METHODS). Affinity-purified antibodies to Nda4p, Mis5p, and Cdc21p recognized bands of 80 kDa, 120 kDa, and 110 kDa, respectively. To verify the specificity of these antibodies on Western blots, we compared lysates from wild-type cells to lysates from cells in which the cognate MCM protein was HA-tagged. As expected, the HA-tagged version of each MCM migrated more slowly than the endogenous protein, and the wild-type protein was not detected in strains expressing only the tagged version (Figure 1, A–C).
Figure 1.
Characterization of antibodies to Nda4p, Mis5p, and Cdc21p. For panels A–C, cell lysates were prepared from appropriate strains and 10 μg of total protein were separated by SDS-PAGE. Duplicate filters were Western blotted with antibodies to the appropriate MCM protein or to the HA-tag, as indicated. (A) Characterization of anti-Nda4p. Lanes 1 and 3: lysate from wild-type cells; lanes 2 and 4: lysate from strain FY803. (B) Characterization of anti-Mis5p. Lanes 1 and 4: wild-type; lanes 2, 3, and 5: FY863, a haploid strain carrying a disruption of mis5+ and containing HA-mis5+ on a plasmid, grown in the presence of thiamine. Lane 3 is a longer film exposure of lane 2 to allow visualization of HA-Mis5p. A degradation product (lacking the HA epitope) of HA-Mis5p is visible in lanes 2 and 3. (C) Characterization of anti-Cdc21p. Lanes 1 and 3: wild type; lanes 2 and 4: FY862, a haploid strain carrying a disruption of cdc21+ and containing HA-cdc21+ on a plasmid, grown in the presence of thiamine. This antibody recognizes two predominant bands, similar to results of Maiorano et al. (1996). (D) Depleted supernatants from anti-MCM immunoprecipitations. Wild-type cell lysate (300 μg) was immunoprecipitated with each anti-MCM antibody. Equal amounts of the resulting depleted supernatants (∼5 μg total protein) were separated by SDS-PAGE, and duplicate filters were immunoblotted with antibodies to each MCM, as indicated. Lane 1: no antibody; lane 2: anti-Cdc19p; lane 3: anti-Mis5p; lane 4: anti-Cdc21p. Asterisks (*) in panels B and D show a protein that cross-reacts with the anti-Mis5p antibody.
To test the effectiveness of these antibodies in immunoprecipitating each MCM protein, we immunoprecipitated cell lysates with each antibody and blotted depleted supernatants with antibodies to the MCMs. Antibodies to Cdc19p, Mis5p, and Cdc21p were able to immunodeplete almost all of the respective protein (Figure 1D), but the anti-Nda4p antibody immunoprecipitated very little of the available Nda4p (our unpublished results). The anti-Mis5p antibody immunoprecipitated Mis5p, but not the cross-reacting protein recognized by the same antibody (Figure 1D, lane 3, asterisk). Interestingly, when each MCM protein was immunodepleted from the lysate, some, but not all, of the other MCMs disappeared from the lysate. This suggests that while MCM proteins associate with one another, a substantial fraction is either disassociated or in a complex that lacks at least one member of the family.
S. pombe MCM Complexes Are Unstable
The six fission yeast MCM proteins are known to associate with one another in a heteromeric complex similar to that in other organisms (Okishio et al., 1996; Adachi et al., 1997). Using sucrose gradients (our unpublished results) and gel filtration analysis, we investigated the structure of this heteromer. We found evidence for a large complex with a predicted molecular mass of approximately 500 kDa in agreement with other reports (our unpublished results and Adachi et al., 1997). However, we also detected the Nda4 protein in significantly lower molecular mass ranges, which suggested it was less stably associated with the other members.
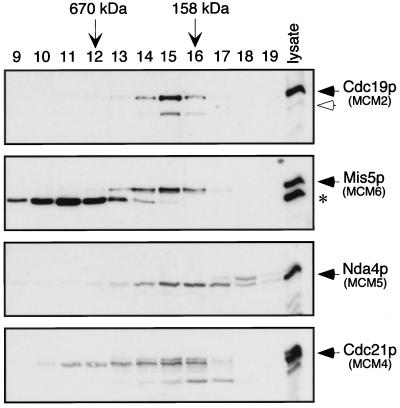
We repeated the experiment using a more stringent lysis buffer that omitted glycerol. This shifted the profile of MCM proteins to lower molecular mass ranges. The four MCMs we monitored eluted from the column in overlapping fractions, with mobilities corresponding to globular proteins ranging in size from 100–700 kDa (Figure 2). Cdc19p was detected in fractions 13–16, corresponding to molecular masses of 500–150 kDa, with the peak protein level in fraction 15 (∼250 kDa). Similarly, most of the Mis5 and Cdc21 proteins were present in fractions 13–16, with a small amount of Cdc21p detected at even higher molecular masses, in fractions 11 and 12. The levels of these two proteins also peaked in fraction 15, well below the predicted size of the full complex.
Figure 2.
Fission yeast MCM protein complexes break down during gel filtration in the absence of glycerol. Wild-type cell lysate was fractionated over a Superose 6 gel filtration column, using lysis buffer as the elution buffer. Samples from each fraction were separated by SDS-PAGE, and duplicate filters were immunoblotted with antibodies to fission yeast MCM proteins, as indicated. The asterisk (*) shows a protein that cross-reacts with the Mis5p antibody (see Figure 1B). Gel filtration markers, bovine thyroglobulin (670 kDa) and bovine γ-globulin (158 kDa), are indicated by the vertical arrows.
Unlike the others, Nda4p was spread out toward the lower molecular mass range, detected mainly in fractions 14–18 (350–50 kDa) and peaking in fractions 15 and 16. (The slower migrating band seen in fraction 18 of the α-Nda4 blot [Figure 2] is a cross-reacting protein.) MCM5 is easily disassociated from other MCMs in other systems (Burkhart et al., 1995; Lei et al., 1996; Thömmes et al., 1997); thus, the weak affinity of Nda4p (MCM5) for the MCM complex is a conserved feature.
The behavior of the MCM proteins in these experiments is consistent with their presence in multimeric protein complexes, ranging from dimer to tetramer or greater. However, our data also suggest that the single heteromeric MCM complex readily breaks down into component parts that may contain a subset of MCM proteins.
The MCM Complex Contains a Tightly Associated Core
We investigated the strength of association between different MCMs using reciprocal coimmunoprecipitation and washing immunoprecipitated material with gentle or harsh buffers. Because the anti-Nda4p antibody fails to immunoprecipitate Nda4p, we used strain FY803, in which the endogenous nda4+ gene was replaced with an HA-tagged nda4+ gene. Antibodies to the HA epitope were used to immunoprecipitate the functional Nda4-HA protein. This experiment was also repeated with a wild-type strain with the same results (our unpublished results).
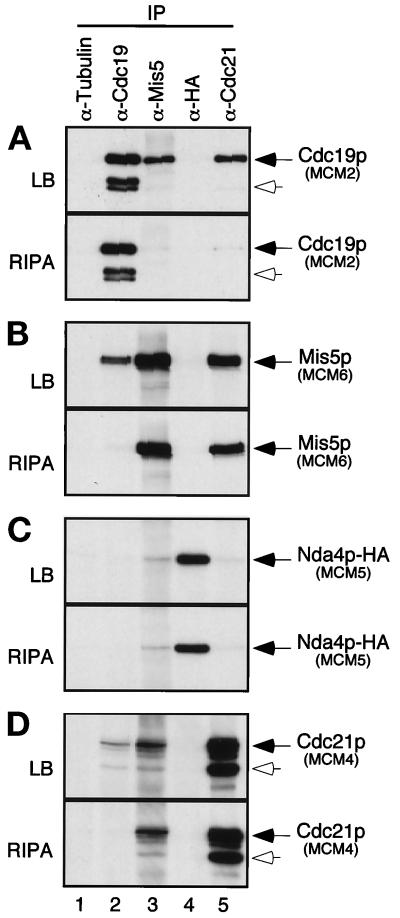
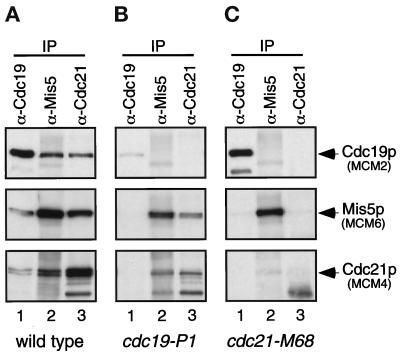
Antibodies to α-tubulin, Cdc19p, Mis5p, Cdc21p, and the HA epitope were used to immunoprecipitate the respective proteins from identical amounts of FY803 cell lysate. Immunoprecipitates were washed in lysis buffer (low salt, low detergent, see MATERIALS AND METHODS), and proteins were eluted by boiling in SDS sample buffer. The presence of MCMs in each immunoprecipitate was determined by Western blot analysis (Figure 3,A–D, top panels). The MCM proteins were not detected in immunoprecipitates with an irrelevant antibody (anti-α-tubulin); however, multiple MCM proteins were detected in immunoprecipitates with anti-MCM antibodies. Cdc19p was precipitated with antibodies to itself, to Mis5p, and to Cdc21p (Figure 3A, top). Similarly, Mis5p and Cdc21p were each precipitated using anti-Cdc19p, anti-Mis5p, and anti-Cdc21p antibodies (Figure 3, B and D, top panels). Thus, these reciprocal immunoprecipitation experiments show that Cdc19p, Mis5p, and Cdc21p associate as expected.
Figure 3.
Coimmunoprecipitation of fission yeast MCMs demonstrates different strengths of association. Identical amounts of FY803 lysate were immunoprecipitated (∼1.5 × 108 cell equivalents; ∼750 μg total protein per immunoprecipitate) with the antibodies shown. Immunoprecipitates were washed nonstringently with lysis buffer (LB) (A, B, C, and D, top panels) or stringently with modified RIPA buffer (RIPA) (A, B, C, and D, bottom panels). Samples were separated by SDS-PAGE. Lane 1: immunoprecipitation with irrelevant antibody (α-tubulin), lane 2: α-Cdc19p, lane 3: α-Mis5p, lane 4: α-HA, lane 5: α-Cdc21p. Identical blots were probed with antibodies to Cdc19p (A), Mis5p (B), HA epitope tag (C), and Cdc21p (D). Open arrows show degradation products of Cdc19p (A) and Cdc21p (D). FY803 is a nda4-HA integrant, and anti-HA antibody was used to immunoprecipitate/immunoblot Nda4p-HA. Similar results were observed for a wild-type strain (our unpublished results).
Cdc19p, Mis5p, and Cdc21p were not detected in the anti-HA immunoprecipitate, which precipitated the epitope-tagged Nda4p-HA. A small amount of Nda4p-HA was detectable in the anti-Mis5p immunoprecipitate and barely detectable in the anti-Cdc21p immunoprecipitate, but not in the anti-Cdc19p immunoprecipitate (Figure 3C, top). Nda4p does associate with Cdc19p (e.g., Figure 5B), but the amount of protein is below the level of detection in this experiment. It is possible that the reason most other MCMs were not seen in the anti-Nda4p-HA immunoprecipitate is that Nda4p may be particularly loosely associated with the other MCMs, and the epitope tag may further weaken its association under our assay conditions (although we note that Nda4p-HA is clearly functional, since it is the only Nda4p in the cell, Figure 1). In addition, the epitope tag may not be accessible to the antibody when the protein is in the complex.
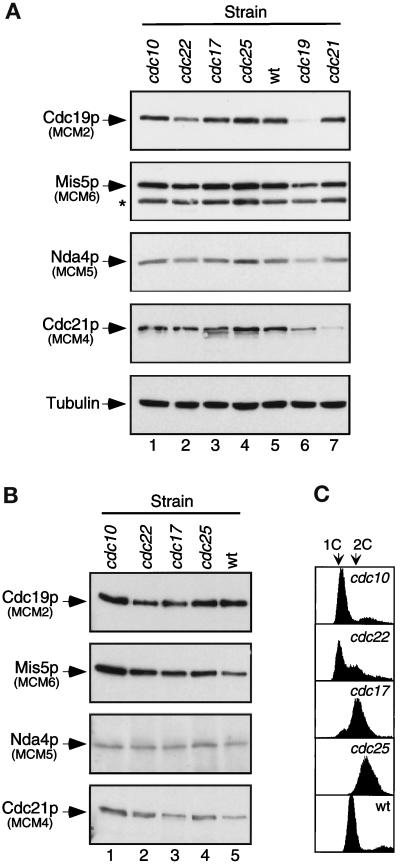
Figure 5.
Cdc19p-containing MCM complexes do not change during the cell cycle. Each strain was shifted to 36°C for 4 h before harvest and lysis. Lanes 1: cdc10-V50 (G1 phase arrest, FY562); lanes 2: cdc22-M45 (early S phase, FY583); lanes 3: cdc17-K42 (late S phase, FY322); lanes 4: cdc25–22 (G2/M phase, FY584); lanes 5: wild-type asynchronous culture (FY254); lane 6: cdc19-P1 (S phase, FY243); lane 7: cdc21-M68 (S phase, FY786). (A) Equal amounts of cell lysate (10 μg total protein) were immunoblotted with antibodies to fission yeast MCMs (7% SDS-polyacrylamide gels) or α-tubulin (12% SDS-polyacrylamide gel). (B) Identical amounts of lysate (250 μg total protein) were immunoprecipitated with anti-Cdc19p antibody. Immunoprecipitates were washed with lysis buffer and immunoblotted for fission yeast MCM proteins, as indicated. (C) Flow cytometry measuring DNA content (X-axis) versus cell number (Y-axis) was used to verify cell cycle arrest.
We compared identical immunoprecipitates washed with a modified RIPA buffer (moderate salt, high detergent; Figure 3, bottom panels) to those washed with lysis buffer (low salt, low detergent; Figure 3, top panels). The harsh buffer removed Cdc19p from anti-Mis5p and anti-Cdc21p immunoprecipitates (Figure 3A, bottom) and removed Mis5p and Cdc21p from an anti-Cdc19p immunoprecipitate (Figure 3, B and D, lane 2, bottom panels). Thus, the more stringent buffer disrupts the interaction between Cdc19p and Mis5p or Cdc21p. In contrast, there was little change in the interaction between Mis5p and Cdc21p under gentle or harsh conditions (Figure 3, B and D, lanes 3 and 5, compare top and bottom panels). This suggests that the association between Mis5p and Cdc21p is much stronger than their association with Cdc19p.
Thus, while the S. pombe MCM proteins interact with each other, the strengths of their affinities vary: Cdc19p forms a loose (RIPA-sensitive) association with Mis5p and Cdc21p, and the latter two proteins form a tight (RIPA-resistant) association. This suggests that a subset of MCM proteins interacts tightly in a core complex with which other MCMs associate more peripherally. This agrees with the gel filtration analysis that showed the complexes break down into dimer- or tetramer-sized structures.
Cdc21p Is Required for Cdc19p Association with the Core Complex
Next, we investigated whether these protein interactions are disrupted by temperature-sensitive mutations in cdc19 (Nasmyth and Nurse, 1981; Forsburg and Nurse, 1994) or cdc21 (Nasmyth and Nurse, 1981; Coxon et al., 1992). Lysates were prepared from asynchronous wild-type cells, and from cdc19-P1 and cdc21-M68 mutants incubated at the restrictive temperature for 4 h. Equal amounts of each lysate were immunoprecipitated with antibodies to Cdc19p, Mis5p, or Cdc21p. The immunoprecipitates were washed in lysis buffer and assayed for the presence of S. pombe MCM proteins. Because the levels of associated proteins in the immunoprecipitates were below the limit of detection of the anti-Nda4p antibody, it was excluded from this analysis.
Both cdc19 and cdc21 mutants have reduced levels of the respective mutant protein at restrictive temperatures (Figure 5A, lanes 6 and 7). In the cdc19-P1 mutant at the restrictive temperature, a small amount of Cdc19tsp was precipitated with antibodies to Cdc19p; however no Cdc19tsp was detected in anti-Mis5p and anti-Cdc21p immunoprecipitates (Figure 4B, top panel), and Mis5p and Cdc21p were not detected in the anti-Cdc19p immunoprecipitate (Figure 4B, lane 1). Thus, in the cdc19ts mutant at the restrictive temperature, Cdc19tsp is unable to bind to Mis5p and Cdc21p. However, these latter two proteins still associate with one another (Figure 4B, middle and bottom panels, lanes 2 and 3).
Figure 4.
MCM complex formation is defective in MCM mutants. (A) Wild-type (FY254), (B) cdc19-P1 mutant (FY243), or (C) cdc21-M68 mutant (FY786) fission yeast strains were incubated at 36°C for 4 h before harvest and lysis. Equal amounts of lysate (300 μg total protein) were immunoprecipitated with antibodies to Cdc19p (lanes 1), Mis5p (lanes 2), or Cdc21p (lanes 3). Immunoprecipitates were washed with lysis buffer, and samples were immunoblotted and probed for the presence of MCM proteins, as indicated.
In the cdc21-M68 mutant at the restrictive temperature, the Cdc21ts protein likewise shows reduced association with Cdc19p and Mis5p (Figure 4C, lanes 2 and 3). Strikingly however, the interaction between Cdc19p and Mis5p is greatly reduced in the cdc21 mutant cells, even though these two proteins are present at normal levels (Figure 4C, lanes 1 and 2; see below). This suggests that the presence of functional Cdc21p is required for the interaction between Cdc19p and Mis5p. Cdc19p (MCM2) may bind to the core structure that contains Cdc21p (MCM4). Alternatively, Cdc21p (MCM4) may bridge the interaction between Cdc19p (MCM2) and Mis5p (MCM6).
The Cdc19p–MCM Complex Does Not Vary during the Cell Cycle
We further investigated the role of Cdc19p by looking for significant changes in S. pombe MCM interactions with Cdc19p at different stages of the cell cycle. First, we measured the level of MCMs in cell lysates prepared from asynchronous wild-type cells, and from the following mutants, which arrest at the indicated cell cycle stages: cdc10-V50 (G1/START, Nurse et al., 1976), cdc22-M45 (early S, Nasmyth and Nurse, 1981), cdc17-K42 (late S, Nasmyth, 1977), cdc25–22 (G2/M, Fantes, 1979), cdc19-P1 (S phase, Nasmyth and Nurse, 1981; Forsburg and Nurse, 1994), cdc21-M68 (S phase, Nasmyth and Nurse, 1981; Coxon et al., 1992). Flow cytometric analysis confirms that the cells were blocked as expected (Figure 5C; cdc19 and cdc21, our unpublished results). Figure 5A shows that the levels of different MCM proteins do not vary in the cell cycle-arrested mutants, as seen previously for Cdc19p (Forsburg et al., 1997). MCM levels are also unaffected in mcm mutant strains. We note that isoforms of Cdc21p with increased mobility are more apparent in cells arrested in late S or G2, suggesting that Cdc21p may be modified or degraded. These isoforms are also visible in asynchronous culture. No mobility changes are apparent for the other three MCMs we examined.
Next, we asked whether the profile of interactions with Cdc19p changes during the cell cycle. We immunoprecipitated Cdc19p from equal amounts of the wild-type, cdc10, cdc22, cdc17, and cdc25 lysates. Immunoprecipitates were washed with lysis buffer and probed for the presence of MCM proteins via Western blot analysis. Figure 5B shows that Mis5p, Nda4p, and Cdc21p coimmunoprecipitate with Cdc19p at all stages of the cell cycle. There is no change in the profile of proteins coprecipitated in any of the samples. This suggests that the bulk of Cdc19p is associated with the same panel of MCM proteins throughout the cell cycle.
Cdc18p Is Not Detected in MCM Complexes
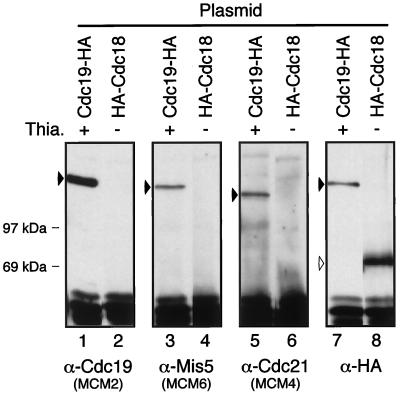
The Cdc18p protein has been reported to interact physically and genetically with the ORC proteins Orp1p and Orp2p (Grallert and Nurse, 1996; Leatherwood et al., 1996). In turn, Orp1p has been shown to interact with Cdc21p (Grallert and Nurse, 1996). Thus, Orp1p may link the MCMs to Cdc18p or vice versa. We examined whether Cdc18p could associate with MCM proteins under conditions in which MCMs associate with each other. We transformed wild-type cells with a plasmid expressing a functional, epitope-tagged Cdc18p (3 copies of the HA epitope tag on the N terminus) under control of the thiamine-repressible nmt1+ promoter (Maundrell, 1990; Muzi-Falconi et al., 1996). Because the protein is very unstable, no HA-Cdc18p is detected when the promoter is repressed by thiamine in the media, but it is detected when expression is induced by growth without thiamine (Muzi-Falconi et al., 1996). We also transformed wild-type cells with a plasmid expressing Cdc19p-HA under control of nmt1+. We can detect expression of Cdc19p-HA in the presence of thiamine, suggesting that this protein is more stable (Forsburg et al., 1997). The HA-tagged proteins were immunoprecipitated with antibodies to the HA epitope and blotted with antibodies to Cdc19p, Mis5p, and Cdc21p (Figure 6, lanes 1–6).
Figure 6.
MCMs do not associate with HA-Cdc18p under conditions in which an MCM complex forms. Wild-type fission yeast cells (FY254) were transformed with a plasmid expressing Cdc19p-HA, grown with thiamine (promoter repressed, lanes 1, 3, 5, and 7), or a plasmid expressing HA-Cdc18p, grown without thiamine (promoter induced, lanes 2, 4, 6, and 8). Lysates from each strain were immunoprecipitated with anti-HA antibody. Immunoprecipitates were washed with lysis buffer and immunoblotted with antibodies to Cdc19p (lanes 1 and 2), Mis5p (lanes 3 and 4), Cdc21p (lanes 5 and 6), or HA epitope (lanes 7 and 8), as indicated. Approximately 1.5 × 108 cell equivalents (∼750 μg total protein) of cell lysate were used for each immunoprecipitation.
Cdc19-HA lanes show that even in the presence of endogenous wild-type Cdc19p, MCM proteins coimmunoprecipitate with the Cdc19-HA protein (Figure 6, lanes 3 and 5). Thus, the HA tag does not affect Cdc19p binding to the MCMs tested, and we have shown previously that Cdc19p-HA is fully functional (Forsburg et al., 1997). Interestingly, immunoprecipitates of Cdc19p-HA do not coprecipitate wild-type Cdc19p, which has a faster mobility on SDS-polyacrylamide gels (Forsburg et al., 1997). This suggests that each complex contains only one molecule of Cdc19p, in agreement with purification data (Adachi et al., 1997).
When HA-Cdc18p was immunoprecipitated under the same conditions, no MCM proteins were detected (Figure 6, lanes 2, 4, and 6). Thus, even under conditions in which the MCM proteins interact strongly with one another, no association can be detected between Cdc18p and any of the MCM proteins we tested.
Multiple Domains of Cdc19p Are Required for MCM Complex Formation
To investigate the regions of Cdc19p that are important in MCM binding, we used a panel of Cdc19p mutants and determined the effect of these mutations on the association between Cdc19p and other MCM proteins. Characteristics of the Cdc19 deletion mutants and point mutants were previously described (Forsburg et al., 1997), except for Cdc19-M4, which changes C360 to S in the putative zinc finger. As is the case for the other mutants affecting this region, cdc19-M4 is unable to complement the cdc19-P1 temperature-sensitive strain (our unpublished results). The mutant proteins were all tagged with three copies of the HA epitope on the C terminus and expressed in wild-type S. pombe under control of the nmt1+ promoter (Forsburg et al., 1997). The cells were grown at 32°C in the presence of thiamine, such that the tagged proteins were expressed at levels significantly below that of the endogenous wild-type Cdc19p (Forsburg et al., 1997). As observed previously (Figure 6), the HA tag on the endogenously expressed Cdc19p does not affect its ability to associate with other MCMs, and in vivo analysis has shown that Cdc19p-HA functions like wild type (Forsburg et al., 1997).
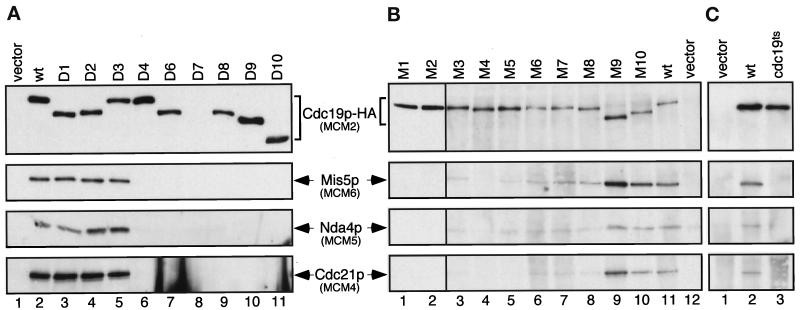
To assay the MCM complex-forming abilities of these mutant proteins, the HA-tagged proteins were immunoprecipitated from cell lysates with an antibody to the HA epitope. Western blot analysis was used to detect MCM proteins that coimmunoprecipitated with the mutant Cdc19-HA proteins. We note that in each case, wild-type Cdc19p was also present in the cell, so the mutant proteins must compete with wild type for association. The Cdc19-D7 mutant protein is missing the epitope recognized by the anti-Cdc19p antibody and is therefore not detected in the immunoblot with this antibody (Figure 7A, lane 8). The presence of this protein at comparable levels was verified by reprobing the blot with anti-HA antibody (our unpublished results).
Figure 7.
Mutants of Cdc19p reduce or abolish MCM complex formation. (A) Deletion mutants, (B) point mutants, or (C) the temperature-sensitive allele (cdc19-P1) of Cdc19p, all with a C-terminal triple HA epitope tag, were expressed on plasmids in wild-type fission yeast at 32°C (A and B) or 25°C (C). The mutant form of Cdc19p was selectively immunoprecipitated with antibodies to the HA epitope and compared with immunoprecipitates from cells with vector only (pSLF173), or cells expressing wild-type Cdc19p-HA from an identical plasmid (pSLF176). Immunoprecipitates were washed with lysis buffer, and MCM proteins were detected by immunoblotting with antibodies to Cdc19p, Mis5p, Nda4p, or Cdc21p, as indicated. All proteins were expressed under control of the nmt1+ promoter under repressed conditions (plus thiamine). Approximately 1.5 × 108 cell equivalents (∼750 μg total protein) of cell lysate were used for each immunoprecipitation.
Of the Cdc19 deletion mutants, MCM proteins coprecipitated with D1, D2, and D3 (Figure 7A; summarized in Table 1). These three are deletions in N-terminal regions of Cdc19p and are able to complement cdc19ts and Δcdc19 mutations (Table 1 and Forsburg et al., 1997). The other deletions affect various regions of Cdc19p. Each of these fails to complement the cdc19-P1 temperature-sensitive allele, and all are defective in MCM interactions (Table 1). These results suggest that multiple regions of Cdc19p are required for interactions with the other MCM proteins, and association with each MCM is equally affected. Again, we note that there is no evidence for the association of wild-type Cdc19p with any HA-tagged derivative, arguing that there is only one molecule of Cdc19p per complex.
Table 1.
Summary of Cdc19 mutantsa
| Mutant | Mutation | Region affected | Comp.b | MCM binding |
|---|---|---|---|---|
| wt | + | ++ | ||
| D1 | Δ50-79 | Acidic region | + | ++ |
| D2 | Δ80-104 | Acidic region | + | ++ |
| D3 | Δ137-143 | Acidic region | + | ++ |
| D4 | Δ45-126 | Acidic region | − | − |
| D6 | Δ736-830 | C terminus | − | − |
| D7 | Δ1-204 | N terminus | − | − |
| D8 | Δ528-613 | MCM homology core | − | − |
| D9 | Δ615-724 | MCM homology core | − | − |
| D10 | Δ528-724 | MCM homology core | − | − |
| M1 | C334S | Putative zinc finger | − | − |
| M2 | C337S | Putative zinc finger | − | − |
| M3 | C357S | Putative zinc finger | − | − |
| M4 | C360S | Putative zinc finger | −c | − |
| M5 | G534R | Putative NTP binding site | − | − |
| M6 | K540A | Putative NTP binding site | − | + |
| M7 | K540R | Putative NTP binding site | + | + |
| M8 | D598A | Putative NTP binding site | − | + |
| M9 | R7E R9D | Putative NLS1d | − | ++ |
| M10 | A113T R116E R117E | Putative NLS2d | − | ++ |
| P1 (ts) | P257L T272I | − | − |
Mutants were described by Forsburg et al. (1997).
Complementation of the cdc19-P1 mutant at 36°C. Data from Forsburg et al. (1997).
Our unpublished data.
NLS, nuclear localization sequence.
The Cdc19 point mutants presented a more complicated picture. Unlike the deletion mutants, there was a wide variation in the amounts of MCMs bound to the Cdc19 point mutants. Normal levels of MCMs coprecipitated with M9 and M10, while greatly reduced levels of MCMs were bound to M6, M7, and M8. Coprecipitating MCM proteins were essentially undetectable in M1, M2, M3, M4, and M5 immunoprecipitates (Figure 7B; summarized in Table 1). Point mutations in several different regions of the protein reduced or abolished binding; there is no simple correlation between a single domain and binding that might define a single MCM association region.
Interestingly, none of the mutations in Cdc19p cause a loss in the binding of only one or two of the other MCMs. Either all or none of the MCMs could bind, suggesting that Cdc19p is tied to the other MCMs via a single partner (e.g., Cdc21p) or via interactions with the core structure.
In addition to the Cdc19 point mutants listed, we tested the MCM binding of the HA-tagged Cdc19ts protein in a similar assay. The cdc19-P1 allele was cloned into the same HA expression vector and expressed in wild-type cells at the permissive temperature (25°C). Under these conditions in which wild-type protein is present to compete for binding, MCM association was not detected (Figure 7C, lane 3). Nor was binding apparent upon incubation at the restrictive temperature (our unpublished results), consistent with the experiment with the cdc19-P1 strain (Figure 4B). Interestingly, if MCM binding is required for function, the Cdc19ts protein must be able to bind MCMs when it is the only protein in the cell. We presume that Cdc19tsp has a reduced affinity for other MCMs and, in the presence of wild-type Cdc19p, competition prevents detectable association.
Does Cdc19p Self-Associate?
During the course of our experiments, we carried out a two-hybrid screen using a truncated derivative of Cdc19p lacking the acidic N terminus as bait (the full-length protein bait transactivated by itself; our unpublished results). Interestingly, the screen isolated a similarly truncated cdc19 clone from a cDNA library. Both this truncated prey and a reconstructed full-length protein interacted with the bait (our unpublished results; see MATERIALS AND METHODS). We therefore examined whether or not Cdc19p self-associates in vivo. First, we expressed the Cdc19-HA protein in wild-type cells. The HA tag fused at either the N-terminal or C-terminal end of the protein confers a mobility shift in SDS-PAGE (Forsburg and Sherman, 1997; Forsburg et al., 1997). When we immunoprecipitated with anti-HA and Western blotted for Cdc19p, the wild-type Cdc19p protein was not detected whether we used the N-terminal or C-terminal HA-tagged derivative (Figure 6 and our unpublished results). Thus, Cdc19p-HA and Cdc19p do not coimmunoprecipitate. Second, we constructed a mutant, cdc19-D7, that corresponds to the original two-hybrid bait. This mutant is not able to complement a cdc19ts mutant and lacks the epitope recognized by our anti-Cdc19p antibody (Forsburg et al., 1997). Cdc19p-D7 is completely defective in binding other MCM proteins, and it does not coimmunoprecipitate with wild-type Cdc19p (Figure 7, lane 8, and our unpublished results). Thus, although a truncated derivative of Cdc19p is able to self-associate in a two-hybrid screen, we find no evidence that this interaction occurs in vivo.
DISCUSSION
The MCM family is very well conserved throughout evolution, with organisms as diverse as yeasts and vertebrates each having six MCM family members that associate in large complexes. The fact that these six proteins are each essential (reviewed in Tye, 1994; Chong et al., 1996; Kearsey et al., 1996) suggests that these proteins are either dependent on each other for a common function or else contribute different essential functions to a multifunctional complex. In this study, we examined the domains of one MCM protein, Cdc19p (MCM2), that are required for complex formation and investigated the architecture of MCM complexes formed by four of the fission yeast MCM proteins.
In characterizing the behavior of MCM proteins, we found that the complexes containing Cdc19p do not change their membership during the cell cycle. Similar observations have been reported in S. cerevisiae (Dalton and Hopwood, 1997). Thus, MCM function is unlikely to be regulated by MCM complex assembly but may require modification or association with other molecules. One candidate for association is Cdc18p/CDC6, a protein essential for initiation of DNA replication that is required for the loading of MCM proteins onto chromatin (Donovan et al., 1997; Rowles and Blow, 1997). However, we found no evidence for HA-Cdc18p-MCM interaction under conditions in which MCMs associate with one another. Any association between Cdc18p and MCM proteins may be too transient to observe under our conditions in asynchronous cells.
To investigate the domains of Cdc19p required for MCM complex formation, we employed a panel of Cdc19 deletion and point mutations that we characterized previously for complementation and overexpression phenotypes (Forsburg et al., 1997). We used a competition assay to investigate association of other MCM proteins with the mutant Cdc19p derivatives. Most nonfunctional Cdc19p mutants that contain an intact MCM core homology domain (Cdc19-M1, M2, M3, M9, M10, D4, D7) were toxic when overexpressed in the cdc19ts strain, but not in wild-type (Forsburg et al., 1997). A simple model for such “synthetic dosage lethality” of these mutants is that they sequester other MCMs into nonfunctional complexes, by virtue of out-competing Cdc19tsp. This is consistent with our observation that temperature-sensitive Cdc19p has reduced affinity for other MCMs. However, while competitive binding to other MCM proteins may explain the overproduction phenotype of mutants M9 and M10, which bind MCMs normally, it is not likely for M1, M2, M3, D4, and D7, which do not bind other MCMs (Figure 7). The synthetic dosage lethality of these five mutants could be caused by titrating away an as-yet-unidentified protein that binds to the C-terminal half of Cdc19p.
Not surprisingly, we have found no functional Cdc19 mutant that is incapable of binding MCM proteins. Our data show that ability to bind other MCMs is required for Cdc19p function, but that MCM association is not sufficient for complementation. However, reduced Cdc19p binding to MCMs does not necessarily abrogate function, as several mutants with reduced binding are still able to complement. We found that the profile of binding was the same for all the MCMs: individual Cdc19p mutants either bound all of the MCMs tested, or bound none of them. Therefore, no domain is specific for interaction with a single other MCM. Further, most Cdc19p mutants either abolished or severely reduced MCM binding, suggesting that a single “MCM interaction domain” cannot be defined.
Our failure to identify a minimal MCM interaction domain in Cdc19p suggests two possible models. First, multiple domains of Cdc19p may be required to contact other MCMs, with physical interactions occurring throughout the protein. Alternatively, distant regions of the protein may be required for a more localized “interaction domain” to assume the correct structure. By this model, some of the mutants we constructed may disrupt protein folding, although in all cases the proteins are expressed and appear stable. Our experiments cannot distinguish between these possibilities.
In characterizing the role of Cdc19p within the MCM complex, we find that the fission yeast MCM complex is not homogeneous. First, we find that although the fission yeast MCM proteins are found in large complexes (Okishio et al., 1996; Adachi et al., 1997), these complexes are prone to break down. Even when one MCM protein is immunodepleted, there are still substantial amounts of the other MCMs remaining in the lysates, and in gel filtration, the complex is unstable. Similar results have been observed in S. cerevisiae (Lei et al., 1996). This could reflect the breakdown of the MCM complex during lysis or that some fraction of MCM proteins in the cell is present in MCM subcomplexes.
Our analysis suggests that MCM subcomplexes are not random, because similar subcomplexes are found in different organisms. The model in Figure 8 summarizes our results. This model of MCM association is consistent with reports of MCM complexes in other organisms, including mouse, human, Xenopus, and S. cerevisiae, which suggests the presence of several MCM subcomplexes (Burkhart et al., 1995; Musahl et al., 1995; Kimura et al., 1996; Lei et al., 1996; Dalton and Hopwood, 1997; Fujita et al., 1997; Ishimi, 1997; Kubota et al., 1997; Thömmes et al., 1997). We found that all four MCMs we tested coimmunoprecipitate as expected, but some associations are more easily disrupted than others. For example, only a small fraction of the available Nda4p seems to immunoprecipitate with the other proteins. This agrees with our observation that Nda4p (MCM5) elutes in lower molecular weight fractions during gel filtration analysis and is consistent with observations in other systems that MCM5 is quite easily separated from other MCMs (Burkhart et al., 1995; Kimura et al., 1996; Lei et al., 1996; Thömmes et al., 1997). Thus, we describe Nda4p (MCM5) as a peripheral MCM protein, and this feature is conserved across different species and assay methods.
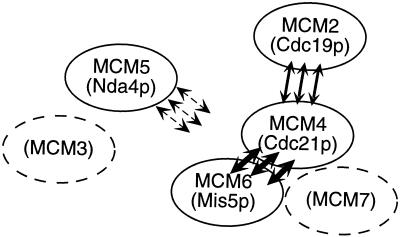
Figure 8.
A model for MCM protein interaction. MCM4 (Cdc21p) and MCM6 (Mis5p) associate tightly as part of a core complex (thick arrows). MCM2 (Cdc19p) binds to this core complex through its interactions with Cdc21p (thin arrows). MCM5 (Nda4p) binds loosely (dashed arrows), although our experiments do not address whether this interaction occurs through MCM4 (Cdc21p), MCM2 (Cdc19p), or others. MCM3 and MCM7, not yet cloned in fission yeast, are indicated by dashed circles; their places in the model are inferred based on the work of others (Burkhart et al., 1995; Musahl et al., 1995; Kimura et al., 1996; Lei et al., 1996; Ishimi, 1997; Thömmes et al., 1997).
On the other hand, Mis5p (MCM6) and Cdc21p (MCM4) associate very strongly and their coimmunoprecipitation is not disrupted by stringent washing. We suggest that these MCMs form part of a tightly associated core MCM complex. This also agrees with recent work from metazoan systems suggesting that MCM4, MCM6, and MCM7 proteins form a stable trimer (Musahl et al., 1995; Ishimi et al., 1996; Kimura et al., 1996; Ishimi, 1997). The interaction between Cdc19p and the core proteins is weaker and is disrupted by washing immunoprecipitates with a stringent buffer. Our data also indicate that Cdc21p is essential for complex formation by Cdc19p, because association between Cdc19p (MCM2) and Mis5p (MCM6) is greatly reduced in cdc21 mutants. Given this observation, we propose that Cdc19p binds the core complex via its interaction with Cdc21p or a core structure containing Cdc21p. We suggest that fission yeast MCM complexes include a tightly bound core of at least Mis5p (MCM6) and Cdc21p (MCM4), loosely associated MCMs, such as Cdc19p (MCM2), and very loosely associated MCMs, such as Nda4p (MCM5). Again, our observations are consistent with those from others that suggest the stable MCM4–6-7 trimer is bound by MCM2 (Musahl et al., 1995; Kimura et al., 1996; Ishimi, 1997; Thömmes et al., 1997). This shows that the behavior of MCM subcomplexes is conserved across eukaryotic systems and suggests that this substructure has functional relevance. A recent report suggests the intriguing possibility that the core MCM trimer of MCM4 (Cdc21p), MCM6 (Mis5p), and MCM7 functions as a helicase (Ishimi, 1997). Peripheral MCM molecules may regulate a core activity, associate with other replication factors, or direct the complex to the chromatin. More careful study of individual members of the family and the architecture of the complex will be important in elucidating the exact structure of the MCM complex and its role in the regulation of DNA replication.
ACKNOWLEDGMENTS
We thank Marco Muzi-Falconi and Tom Kelly for the pMF56 plasmid; Jill Meissenholder and Tony Hunter for the 12CA5 antibody; Steve Elledge for the fission yeast two-hybrid materials; Jeff Hodson for expert technical assistance and providing strains FY862 and FY863; Martin Latterich for helpful discussions and fast performance liquid chromatography training; and Debbie Liang and Mike McKeown for critical reading of the manuscript. This work was supported by American Cancer Society grant RPG-95–012-04-CCG (S.L.F.) and by National Institutes of Health training grants HD-07495 (D.A.S.) and CA-09370 (S.G.P.). D.A.S is a Salk Institute Association fellow. S.L.F. is a Scholar of the Leukemia Society of America.
REFERENCES
- Adachi Y, Usukura J, Yanagida M. A globular complex formation by Nda1 and the other five members of the MCM protein family in fission yeast. Genes Cells. 1997;2:467–479. doi: 10.1046/j.1365-2443.1997.1350333.x. [DOI] [PubMed] [Google Scholar]
- Botchan M. Coordinating DNA replication with cell division — current status of the licensing concept. Proc Natl Acad Sci USA. 1996;93:9997–10000. doi: 10.1073/pnas.93.19.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhart R, Schulte D, Hu D, Musahl C, Gohring F, Knippers R. Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur J Biochem. 1995;228:431–438. [PubMed] [Google Scholar]
- Chong JP, Thömmes P, Blow JJ. The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- Coxon A, Maundrell K, Kearsey SE. Fission yeast cdc21+ belongs to a family of proteins involved in an early step of chromosome replication. Nucleic Acids Res. 1992;20:5571–5577. doi: 10.1093/nar/20.21.5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S, Hopwood B. Characterization of Cdc47p-minichromosome maintenance complexes in Saccharomyces cerevisiae: identification of Cdc45p as a subunit. Mol Cell Biol. 1997;17:5867–5875. doi: 10.1128/mcb.17.10.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes P. Epistatic gene interactions in the control of division in fission yeast. Nature. 1979;279:428–430. doi: 10.1038/279428a0. [DOI] [PubMed] [Google Scholar]
- Fields S, Song OK. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Forsburg SL. Regulation of S phase in the fission yeast Schizosaccharomyces pombe. In: Blow JJ, editor. Eukaryotic DNA Replication. Oxford, United Kingdom: Oxford University Press; 1996. pp. 197–228. [Google Scholar]
- Forsburg SL, Nurse P. The fission yeast cdc19+ gene encodes a member of the MCM family of replication proteins. J Cell Sci. 1994;107:2779–2788. doi: 10.1242/jcs.107.10.2779. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Sherman DA. General purpose tagging vectors for fission yeast. Gene. 1997;191:191–195. doi: 10.1016/s0378-1119(97)00058-9. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Sherman DA, Ottilie S, Yasuda JR, Hodson JA. Mutational analysis of Cdc19p, an MCM replication protein in fission yeast. Genetics. 1997;147:1025–1041. doi: 10.1093/genetics/147.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Kiyono T, Hayashi Y, Ishibashi M. In vivo interaction of human MCM heterohexameric complexes with chromatin. Possible involvement of ATP. J Biol Chem. 1997;272:10928–10935. doi: 10.1074/jbc.272.16.10928. [DOI] [PubMed] [Google Scholar]
- Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- Ishimi Y, Ichinose S, Omori A, Sato K, Kimura H. Binding of human minichromosome maintenance proteins with histone H3. J Biol Chem. 1996;271:24115–24122. doi: 10.1074/jbc.271.39.24115. [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Maiorano D, Holmes EC, Todorov IT. The role of MCM proteins in the cell cycle control of genome duplication. Bioessays. 1996;18:183–190. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kimura H, Ohtomo T, Yamaguchi M, Ishii A, Sugimoto K. Mouse MCM proteins: complex formation and transportation to the nucleus. Genes Cells. 1996;1:977–993. doi: 10.1046/j.1365-2443.1996.840284.x. [DOI] [PubMed] [Google Scholar]
- Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherwood J, Lopez-Girona A, Russell P. Interaction of Cdc2 and Cdc18 with a fission yeast ORC2-like protein. Nature. 1996;379:360–363. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Tye BK. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill SA, Nurse P. Cell cycle control in fission yeast. In: Pringle J, Broach J, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 697–763. [Google Scholar]
- Maiorano D, Van Assendelft GB, Kearsey SE. Fission yeast cdc21, a member of the MCM protein family, is required for onset of S phase and is located in the nucleus throughout the cell cycle. EMBO J. 1996;15:861–872. [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast: a highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Miyake S, Okishio N, Samejima I, Hiraoka Y, Toda T, Saitoh I, Yanagida M. Fission yeast genes nda1+ and nda4+, mutations of which lead to S-phase block, chromatin alteration and Ca2+ suppression, are members of the CDC46/MCM2 family. Mol Biol Cell. 1993;4:1003–1015. doi: 10.1091/mbc.4.10.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Musahl C, Schulte D, Burkhart R, Knippers R. A human homologue of the yeast replication protein Cdc21. Interactions with other Mcm proteins. Eur J Biochem. 1995;230:1096–1101. doi: 10.1111/j.1432-1033.1995.tb20660.x. [DOI] [PubMed] [Google Scholar]
- Muzi-Falconi M, Brown GW, Kelly TJ. cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Temperature-sensitive lethal mutants in the structural gene for DNA ligase in the yeast Schizosaccharomyces pombe. Cell. 1977;12:1109–1120. doi: 10.1016/0092-8674(77)90173-8. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Okishio N, Adachi Y, Yanagida M. Fission yeast Nda1 and Nda4, MCM homologs required for DNA replication, are constitutive nuclear proteins. J Cell Sci. 1996;109:319–326. doi: 10.1242/jcs.109.2.319. [DOI] [PubMed] [Google Scholar]
- Rowles A, Blow JJ. Chromatin proteins involved in the initiation of DNA replication. Curr Opin Genet Dev. 1997;7:152–157. doi: 10.1016/s0959-437x(97)80123-2. [DOI] [PubMed] [Google Scholar]
- Sazer S, Sherwood SW. Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J Cell Sci. 1990;97:509–516. doi: 10.1242/jcs.97.3.509. [DOI] [PubMed] [Google Scholar]
- Su TT, Follette PJ, O’Farrell PH. Qualifying for the license to replicate. Cell. 1995;81:825–828. doi: 10.1016/0092-8674(95)90000-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamada H, Yanagida M. Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol Biol Cell. 1994;5:1145–1158. doi: 10.1091/mbc.5.10.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thömmes P, Kubota Y, Takisawa H, Blow JJ. The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye B-K. The MCM2–3-5 proteins: are they replication licensing factors? Trends Cell Biol. 1994;4:160–166. doi: 10.1016/0962-8924(94)90200-3. [DOI] [PubMed] [Google Scholar]