Abstract
Borna disease virus (BDV) is a noncytolytic, neurotropic RNA virus that has a broad host range in warm-blooded animals, probably including humans. Recently, it was demonstrated that a 24-kDa phosphoprotein (P) of BDV directly binds to a multifunctional protein, amphoterin-HMGB1, and inhibits its function in cultured neural cells (W. Kamitani, Y. Shoya, T. Kobayashi, M. Watanabe, B. J. Lee, G. Zhang, K. Tomonaga, and K. Ikuta, J. Virol. 75:8742-8751, 2001). This observation suggested that expression of BDV P may cause deleterious effects in cellular functions by interference with HMGB1. In this study, we further investigated the significance of the binding between P and HMGB1. We demonstrated that P directly binds to the A-box domain on HMGB1, which is also responsible for interaction with a tumor suppression factor, p53. Recent works have demonstrated that binding between HMGB1 and p53 enhances p53-mediated transcriptional activity. Thus, we examined whether BDV P affects the transcriptional activity of p53 by interference with HMGB1. Mammalian two-hybrid analysis revealed that p53 and P competitively interfere with the binding of each protein to HMGB1 in a p53-deficient cell line, NCI-H1299. In addition, P was able to significantly decrease p53-mediated transcriptional activation of the cyclin G promoter. Furthermore, we showed that activation of p21waf1 expression was repressed in cyclosporine-treated BDV-infected cells, as well as p53-transduced NCI-H1299 cells. These results suggested that BDV P may be a unique inhibitor of p53 activity via binding to HMGB1.
Borna disease virus (BDV) belongs to the family Bornaviridae within the nonsegmented, negative-strand RNA viruses of the order Mononegavirales, which are characterized by low productivity, neurotropism, and nuclear localization of transcription and replication (29, 43). Although BDV was originally described as an agent of nonpurulent encephalomyelitis in horses in Germany, BDV infection has now been found in a wide range of vertebrate species (31, 37). BDV can cause neurological disturbances characterized by behavioral or neurodevelopmental abnormalities in various experimental animals, such as rats and mice, demonstrating that BDV infection may provide a useful model for analyzing neuropsychiatric or neurodegenerative disorders in humans (21). Although mounting evidence suggests that BDV infection also occurs in humans and that it may be related to psychiatric diseases (6, 25, 33, 44), the specific link between BDV infection and any of these human disorders has not yet been elucidated (38, 40).
BDV shows noncytolytic replication in cultured neuronal cells. In animal models of BDV infection, persistent infection is frequently found in specific areas in the brain (22). In rats infected with BDV as adults, a long-lasting persistence is established after the acute phase (22, 41). In contrast, infection of immunoincompetent rats, such as neonates, induces a persistent infection (22, 41). Numerous reports have shown that persistently BDV-infected rats show a variety of behavioral abnormalities and serious neurological disturbances, indicating that establishment of persistent infection may be critical for the induction of neuropathogenesis by this virus. Interestingly, unlike other central nervous system (CNS) viral infections, BDV has a unique manner of infection in which the expression levels of viral mRNAs, as well as the viral titer, seems not to shift from acute to persistent stages of infection (21). Furthermore, it has been demonstrated that the viral proteins are abundantly detected in the brain throughout the infection. These observations suggest that expression or accumulation of the viral products in infected cells may influence cellular functions at various levels, leading to neurological disorders characterized by BDV persistence, as well as maintenance of the persistent viral infection.
Molecular biological analysis has indicated that the BDV antigenome consists of at least six open reading frames, of which the p40/38 nucleoprotein (N), p10 protein (X), and p24 phosphoprotein (P) are abundantly expressed in persistently infected cells (43). In a recent study, it was revealed that BDV P binds directly to a multifunctional protein, HMGB1 (also called amphoterin or HMG-1), and inhibits its function in cultured neural cells (30). A line of evidence has suggested that HMGB1 plays important roles not only in the neurite outgrowth of neurons but also in cell survival through interaction with its cellular receptor, RAGE (receptor for advanced glycation end products) (36). Furthermore, it has been reported that HMGB1 has a function in the nucleus as a specific enhancer of p53 activity (26, 28). Previous works have demonstrated that HMGB1 binds directly to p53 and enhances the p53-mediated transcriptional activities of certain promoters that contain p53-specific binding sites. These observations raised the possibility that BDV P can influence the transcriptional activity of p53 through the interference of HMGB1 in the nuclei of infected cells.
This work was undertaken to further investigate the effects of P binding to HMGB1 and to elucidate its role in p53 activity in vivo. We demonstrate here that the A-box domain of HMGB1, which is responsible for binding to p53, also contributes to interaction with BDV P. Interestingly, we were able to show that p53 and P competitively interfere with the binding of each protein to HMGB1 in transiently transfected cells. Furthermore, P could significantly inhibit p53-mediated transcriptional activation of the cyclin G promoter in a p53-deficient cell line. We also showed that induction of p53 failed to up-regulate the expression of a cell cycle inhibitor, p21waf1, in cell lines expressing P and persistently BDV infected. These results indicated that BDV P could repress the transcriptional activity of p53 by binding to HMGB1, suggesting the possibility that expression of P may play a role in the modification of p53-mediated cellular responses in infected cells.
MATERIALS AND METHODS
Cell lines and virus.
C6 (rat glioma) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal calf serum (FCS). The OL cell line, derived from a human oligodendroglioma, was grown in DMEM-high glucose (4.5%) supplemented with 10% FCS. A BDV-infected cell line, C6BV, which is persistently infected with strain He/80 (8), was maintained under the same conditions as the parental cell line. NCI-H1299 cells (34), which are deficient in tumor suppression protein p53, were kindly provided by Y. Nakanishi (Kyushu University, Fukuoka, Japan) and A. F. Gazdar (Hamon Cancer Center, University of Texas Southwestern Medical Center, Dallas) and maintained in RPMI 1640 medium containing 10% FCS.
Plasmid construction.
The recombinant HMGB1 and BDV P used for far-Western blot analysis were expressed, using baculovirus and Escherichia coli expression systems, as fusion proteins with histidine (His) and glutathione S-transferase (GST), respectively. The construction of the baculovirus expression plasmid for rat HMGB1, pBac-HMGB1 (formerly referred to as pFastBac-AMP), and the E. coli expression plasmid for BDV P, pGEX-P, were described elsewhere (30). The generation of expression plasmids used for mammalian two-hybrid analysis, VP16-P and GAL4-HMGB1 (formally referred to as GAL4-AMP), was also described previously (30). A eukaryotic expression plasmid encoding full-length human p53 cDNA, pcD-p53, was provided by H. Nojima (Osaka University, Osaka, Japan). The construction of the expression vectors pcD-HMGB1 and pcD-P (also called pcD-AMP/FLAG and pcP-FLAG, respectively) was described in previous studies (30). The mutant forms of these expression plasmids were generated from a wild-type plasmid by using PCR and recloning techniques. The oligonucleotide primers used in PCR to create the mutant plasmids are available on request. The introduction of the correct sequences for each mutant was confirmed by DNA sequencing and Western blot analysis of protein production.
Expression of recombinant proteins.
Recombinant HMGB1-expressing baculoviruses were constructed using the BAC-TO-BAC Baculovirus Expression System (GIBCO/BRL) according to the manufacturer's recommendations. Purification of His-tagged HMGB1 produced in the baculovirus from Bac-HMGB1-infected High Five cells (Invitrogen, Carlsbad, Calif.) was performed using a His-Trap kit (Amersham Pharmacia Biotech, Uppsala, Sweden) according to the manufacturer's recommendations. Recombinant proteins expressed in E. coli were produced as fusion proteins with GST and purified using a glutathione-Sepharose 4B column. The purified recombinant proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue or transferred to a polyvinylidene difluoride (PVDF) membrane by electroblotting.
Far-Western assay.
The purified recombinant proteins were denatured by boiling them in sample buffer and subjected to SDS-PAGE analysis. After electrophoresis, the proteins were transferred onto PVDF membranes, which were then blocked with 5% skim milk in phosphate-buffered saline (PBS)-0.05% Tween-20 (PBS-T) at room temperature. As probes, purified GST- or His-tagged recombinant proteins were allowed to bind to the blotted proteins in PBS-T containing 5% skim milk overnight at 4°C with gentle agitation. The blots were washed three times with PBS-T for 15 min each and then incubated with antibody specific to the probe (anti-P or anti-HMGB1 rabbit polyclonal antibody) in PBS-T containing 5% skim milk for 1 h at room temperature. After being washed, the membranes were incubated with horseradish peroxidase-conjugated antibody. The specific reactions on the membranes were then detected by an ECL Western blotting kit (Amersham Pharmacia Biotech).
Mammalian two-hybrid analysis.
OL or NCI-H1299 cells were transfected with a luciferase reporter plasmid, pG5luc (Promega, Madison, Wis.), and test plasmids (0.5 to 1.0 μg) using TransFast transfection reagent (Promega) in six-well culture plates. Thirty-six to 48 h after transfection, the cells were lysed in 350 μl of lysis buffer for 15 min with shaking at room temperature. After centrifugation at 18,000 × g for 5 min at 4°C, the cell extracts were assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's recommendations.
Neurite outgrowth assay.
Laminin-coated eight-well chamber slides (Lab-Tek Nunc Inc., Naperville, Ill.) were covered with purified GST or GST-P fusion proteins at 4°C overnight as previously described (30). The chambers were washed twice with cold PBS and blocked with 1% bovine serum albumin. After the slides were washed with serum-free DMEM, C6 cells were added to the chambers and cultured in DMEM without FCS at 37°C for 3 to 4 h to induce cell process outgrowth of the cells. The cells were then fixed with 4% paraformaldehyde and stained with hematoxylin.
Luciferase reporter gene assay.
NCI-H1299 cells were cotransfected with the pGL-cyclin G-Luc reporter plasmid (27), which was a gift from M. Oren (Institute of Science, Rehovot, Israel), and each combination of tested plasmids: pcD-p53, pcD-HMGB1, pcD-P, and pcD-PΔM1. For luciferase assays, cells were seeded at 1.5 × 105 per well and grown overnight. To keep the total amount of transfected DNA constant in each sample, the pcDNA3 plasmid was incubated. To estimate the transfection efficiencies, the expression of recombinant proteins in the transfected cells was analyzed by immunoblotting. Luciferase activity was detected as described above.
Detection of p21waf1 protein in p53-induced cells.
NCI-H1299 cells were cotransfected with each combination of tested plasmids: pcD-p53, pcD-P, pcD-PΔM1, and pcDNA3. C6 rat glioma cells and BDV-infected C6BV cells were treated with 60 μM cyclosporine (CsA) to induce p53 expression (35). Twenty-four hours after transfection or CsA treatment, the cells were collected and cell lysis was achieved by the addition of 2× sampling buffer followed by boiling the cells for 5 min. The protein samples were then subjected to SDS-PAGE analysis and transferred onto PVDF membranes. The membranes were subsequently incubated with the corresponding primary antibodies as follows: mouse anti-p53 (BD Biosciences, San Diego, Calif.) and anti-p21waf1 (Santa Cruz Biotechnology, Santa Cruz, Calif.) or anti-tubulin monoclonal antibody (Sigma-Aldrich). Antibody reactions were detected as described above. The images on X-ray films were captured electronically, and the intensity of each reactive band was quantified using NIH Image software.
RESULTS
A short amino acid stretch on BDV P is necessary for binding to HMGB1.
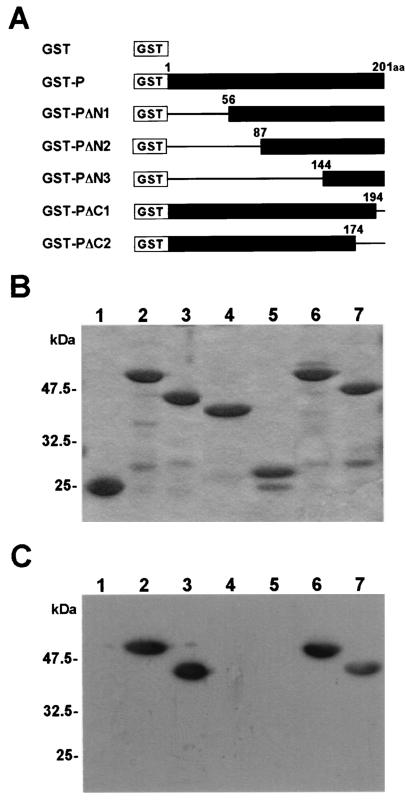
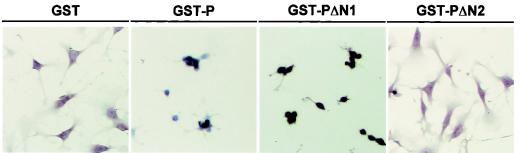
In a previous study, we demonstrated that BDV P binds specifically to HMGB1 and inhibits the functions of HMGB1 in cultured neural cells. To understand the significance of the binding between P and HMGB1 in more detail, we first examined the amino acid regions of P and HMGB1 required for the interaction. To map the binding domain to HMGB1 on P, we constructed a series of GST-P fusions containing a deletion in either the N or C terminus (Fig. 1A). Purified GST fusion proteins were analyzed on SDS-PAGE and stained with Coomassie brilliant blue (Fig. 1B). We performed far-Western blotting with mutant P using baculovirus-expressed HMGB1 as a probe. As shown in Fig. 1C, HMGB1 could clearly detect truncated proteins lacking 55 (GST-PΔN1) and 7 (GST-PΔC1) or 27 (GST-PΔC2) amino acids in the N and C termini of P, respectively, while mutants that lacked the N-terminal 86 (GST-PΔN2) or 143 (GST-PΔN3) amino acids were unable to bind to the probe. This observation suggested that the region encompassing the amino acid sequence between positions 56 and 86 of P is essential for the interaction with HMGB1. To determine whether this region is also important for interference with the HMGB1 function, we examined the cell process outgrowth of neural cells in the presence of mutant P. C6 glioma cells were cultured on laminin-coated plates with the truncated mutants. As a negative control, the cells were cultured with purified GST protein (Fig. 2). As described in a previous report (30), wild-type P efficiently inhibited the cell process outgrowth of the cells, while the deletion mutant lacking the N-terminal 86 amino acids did not show any effects on the cell processes of the cells, indicating that the region between amino acids 56 and 86 of P is necessary for interference with the HMGB1 function, as well as for binding to the protein.
FIG. 1.
Identification of the portion of BDV P necessary for interaction with HMGB1. (A) Schematic representation of GST-P fusion proteins. The numbers indicate amino acid (aa) positions in BDV P. (B) Expression of GST-P fusion proteins. The purified fusion proteins were analyzed by SDS-PAGE and stained with Coomassie brilliant blue. (C) Far-Western blot analysis of GST-P fusion proteins. The purified GST-P proteins were transferred onto PVDF membranes. His-tagged HMGB1 protein was used as a probe. The specific binding was detected by an antibody to HMGB1. (B and C) Lanes 1, GST alone; lanes 2, GST-P; lanes 3, GST-PΔN1; lanes 4, GST-PΔN2; lanes 5, GST-PΔN3; lanes 6, GST-PΔC1; lanes 7, GST-PΔC2.
FIG. 2.
BDV P lacking 86 amino acids in the N terminus does not affect cell process outgrowth of rat glioma cells. A neurite outgrowth assay was performed in laminin-coated chamber plates covered with purified GST or GST-P fusion proteins. C6 rat glioma cells were cultured in DMEM without FCS at 37°C for 3 to 4 h to induce cell process outgrowth of the cells. The cells were then fixed and stained with hematoxylin.
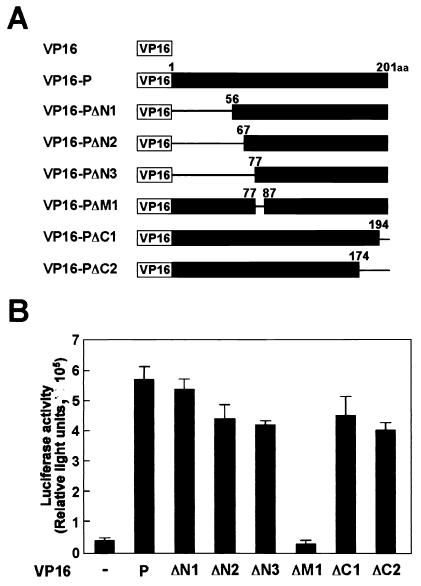
To determine the region on P binding to HMGB1 in vivo in more detail, we next used a GAL4-VP16-based mammalian two-hybrid system in human OL cells (Fig. 3). The cDNAs corresponding to HMGB1 and mutant P were fused to the GAL4 DNA binding domain and the VP16 transactivating domain, respectively. Forty-eight hours after transfection, the luciferase activity was measured in cell extracts as an index of the protein-protein interactions (Fig. 3B). As previously reported, the interaction between P and HMGB1 was clearly demonstrated in cells transfected with VP16-P and GAL4-HMGB1. Transfection of mutant forms of P containing deletions in the N-terminal 55, 66, or 76 amino acids also showed significant luciferase activities compared with the negative control plasmids (Fig. 3B). In contrast, no interaction was detected in the cells transfected with mutant P lacking the region from amino acids 77 to 86 of the protein. We repeated this experiment at least six times and obtained similar results in each experiment. This result indicated that a short amino acid stretch on P, K77LVTELAENS86, is important for interaction with HMGB1 in vivo.
FIG. 3.
Mammalian two-hybrid analysis determines the domain of BDV P essential for interaction with HMGB1. (A) Schematic representation of VP16-P fusion proteins. The numbers indicate amino acid (aa) positions in BDV P. VP16-PΔM1 lacks amino acids between positions 78 and 86 of BDV P. (B) Mammalian two-hybrid analysis of VP16-fused BDV P. OL cells were transfected with the luciferase reporter plasmid pG5luc, GAL4-HMGB1, and the indicated VP16-P fusion plasmids. −, VP16 plasmid (negative control). Forty-eight hours after transfection, the cell extracts were assayed for luciferase activity. The values are expressed as means plus standard errors of the mean.
BDV P interacts with the A-box domain of HMGB1.
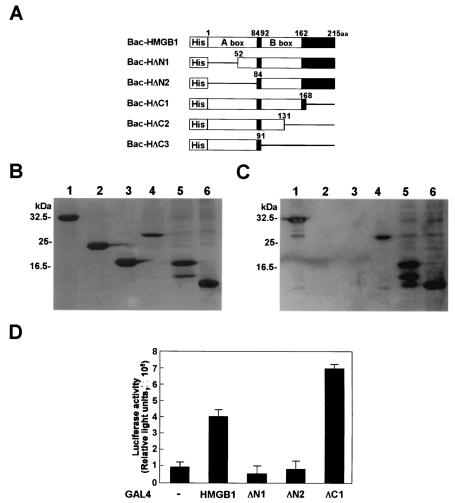
We next determined the HMGB1 domain responsible for interaction with BDV P. Because HMGB1 consists of three domains, two basic domains (A and B boxes) and a highly acidic C-terminal domain, we introduced deletions corresponding to each domain of the protein (Fig. 4A). The truncated HMGB1 was expressed in a baculovirus system and subjected to far-Western blot analysis using wild-type P as a probe. As shown in Fig. 4C, the mutants which contain a deletion in the A-box domain (Bac-HΔN1 and -HΔN2) could not bind to P, whereas HMGB1 lacking the B-box and/or the C-terminal domain (Bac-HΔC1 and -HΔC2) were clearly detected by the P probe. Furthermore, the P probe clearly detected a mutant that contains only the A-box domain (Bac-HΔC3), indicating that the A box of HMGB1 is sufficient for interaction with BDV P.
FIG. 4.
Identification of the portion of HMGB1 necessary for binding to BDV P. (A) Schematic representation of His-tagged HMGB1 fusion proteins. The numbers indicate amino acid positions in HMGB1. The solid box between amino acids (aa) 162 and 215 represents the acidic C-terminal domain. (B) Expression of His-HMGB1 fusion proteins. The purified fusion proteins were analyzed on SDS-PAGE and stained with Coomassie brilliant blue. (C) Far-Western blot analysis of His-HMGB1 fusion proteins. The purified HMGB1 mutants were transferred onto PVDF membranes. GST-fused BDV P was used as a probe. The specific binding was detected by a BDV P monoclonal antibody. (B and C) Lanes 1, Bac-HMG1; lanes 2, Bac-HΔN1; lanes 3, Bac-HΔN2; lanes 4, Bac-HΔC1; lanes 5, Bac-HΔC2; lanes 6, Bac-HΔC3. (D) Mammalian two-hybrid analysis of HMGB1 mutant proteins. OL cells were transfected with the luciferase reporter plasmid pG5luc, VP16-P, and the indicated GAL4-HMGB1 mutant plasmids. −, GAL4 plasmid (negative control). Forty-eight hours after transfection, the cell extracts were assayed for luciferase activity. The luciferase activities represent means plus standard errors of the mean.
To verify the role of the A-box domain in the interaction between HMGB1 and BDV P in vivo, we also performed mammalian two-hybrid analysis using the deletion mutants of HMGB1 fused to the GAL4 DNA binding domain. Transfection of the tested plasmids into OL cells confirmed that luciferase activity was not detected in the cells transfected with the GAL4-HΔN1 and -HΔN2 constructs, indicating that the A-box domain of HMGB1 is the only region responsible for binding to P in vivo (Fig. 4D).
Competitive binding to HMGB1 between BDV P and p53.
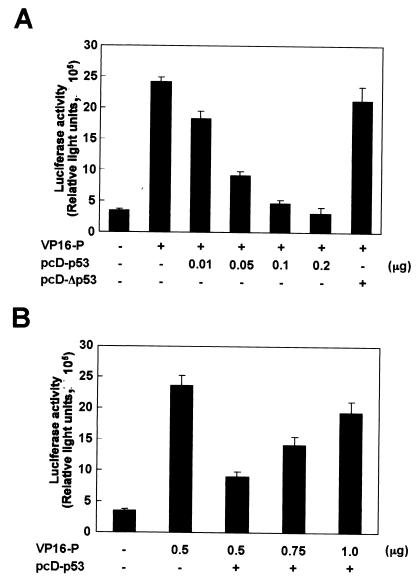
Previous studies have demonstrated that the A-box region of HMGB1 is important for interaction with several cellular transcription factors, including p53, Hox, steroid hormone receptors, and POU domain-containing factors, resulting in enhancement of the sequence-specific DNA binding of the factors (3, 26, 28, 47, 48). These observations raise the possibility that P and the cellular proteins competitively interfere with the binding of each protein to HMGB1 in BDV-infected cells. Among these cellular factors, p53 may be a particularly interesting protein, because it has been demonstrated that p53 binds directly to HMGB1 only via the A-box domain (26). Therefore, we examined whether p53 and P competitively interfere with each other in the interaction with HMGB1 in transiently transfected cells (Fig. 5). We performed mammalian two-hybrid analysis to examine the interference between the proteins. To eliminate the effects of endogenous p53 in this experiment, we used a p53-deficient cell line, NCI-H1299, for the transfection. As shown above, efficient binding between HMGB1 and P was observed in the cells without transfection of p53. However, the luciferase activity in the cells significantly decreased when p53 was expressed in a dose-dependent fashion (Fig. 5A). A mutant p53 (pcD-Δp53) which lacks a domain interacting with HMGB1 was unable to inhibit the activation of luciferase by P, indicating p53-specific inhibition of P binding to HMGB1. In contrast, increased amounts of transfected P resulted in the recovery of the luciferase activity in the NCI-H1299 cell line even in the presence of p53 (Fig. 5B). These results suggested that P and p53 competitively interfere with the binding of each protein to HMGB1 in the cells.
FIG. 5.
BDV P competitively interferes with binding between p53 and HMGB1. p53-deficient NCI-H1299 cells were cotransfected with pG5luc (a luciferase reporter plasmid), VP16-P, and GAL4-HMGB1 plasmids with or without the p53 expression plasmid pcD-p53. Binding between BDV P and HMGB1 was monitored by luciferase activity. (A) Effect of p53 expression on binding of BDV P to HMGB1. VP16-P (0.5 μg) was cotransfected with various amounts of pcD-p53 plasmid into the cells. pcD-Δp53, p53 mutant plasmid lacking a domain interacting with HMGB1. +, present; −, absent. (B) Effect of BDV P on luciferase activity in p53-transfected NCI-H1299 cells. Various amounts of VP16-P plasmid were transfected with pcD-p53 plasmid (0.05 μg) into the cells. The total DNA transfected was normalized with the pcDNA3 plasmid. Luciferase activity in the cells was measured 36 h after transfection using the Dual-Luciferase Reporter Assay System. Values are expressed as means plus standard errors of the mean.
BDV P represses transcriptional activation of cyclin G by p53 and HMGB1.
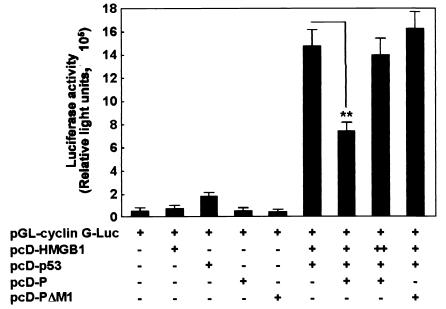
p53 is a transcriptional activator that binds to sequence-specific binding sites at the promoters of several cellular genes and activates their transcription (11, 32). Recent studies have indicated that HMGB1 enhances p53-mediated transactivation of the promoter by direct binding to p53 via its A-box domain (26). Therefore, to determine the effect of BDV P expression on p53-mediated transcriptional activation, a cyclin G-luciferase reporter that contains p53-specific binding sites was transfected into p53-null NCI-H1299 cells together with a p53, HMGB1, and/or P expression vector. As shown in Fig. 6, repeated experiments revealed that p53 dramatically induces activation of the cyclin G promoter in the cotransfection with HMGB1 compared with transfection alone. In addition, HMGB1 itself did not affect the promoter activity of cyclin G, indicating that the interaction between p53 and HMGB1 specifically up-regulates the transcriptional activity of p53. On the other hand, cyclin G promoter activity was significantly decreased by the expression of wild-type P but not by mutant P (pcD-PΔM1), which lacks the region responsible for binding to HMGB1 (Fig. 6). In contrast, overexpression of HMGB1 overcame P-mediated inhibition of p53-dependent transcriptional activation of the promoter (Fig. 6). These results suggested that BDV P specifically inhibits p53-mediated transcriptional activation through interference with the binding of HMGB1 to p53.
FIG. 6.
BDV P inhibits p53-mediated transcriptional activation of cyclin G promoter. NCI-H1299 cells were transiently transfected with the pGL-cyclin G-Luc reporter plasmid (0.5 μg) and tested plasmids (pcD-HMGB1 [1.25 μg], pcD-p53 [0.3 μg], pcD-P [0.15 μg], and pcD-PΔM1 [0.5 μg]). pcD-PΔM1 contains a small deletion corresponding to the HMGB1-binding domain of BDV P amino acids 78 to 86. The total DNA transfected (3.0 μg) was normalized with the pcDNA3 plasmid. +, present; −, absent; ++, overexpression of pcD-HMGB1 (2.5 μg) in transfected cells. Twenty-four hours after transfection, the cell extracts were assayed for luciferase activity. Values are expressed as means plus standard errors of the mean. **, P < 0.01 (Student's t test).
Suppression of p21waf1 activation in cells expressing P and persistently BDV infected.
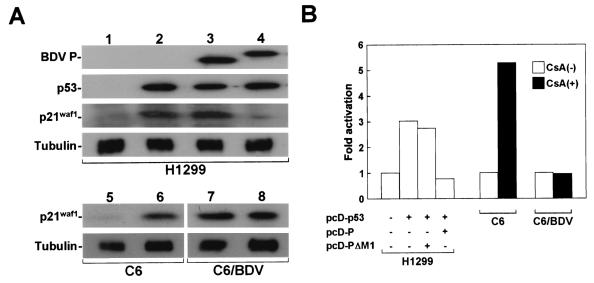
We investigated whether BDV P can influence p53 activity by interfering with endogenous HMGB1. Previous work has demonstrated that a cell cycle inhibitor, p21waf1, is a major target for p53-mediated transcriptional activation (11, 12). Thus, we estimated the expression level of p21waf1 protein in NCI-H1299 cells transfected with p53 expression plasmid. As shown in Fig. 7A, in the absence of BDV P, transduction of p53 could efficiently up-regulate the expression of p21waf1 in the transfected cells. On the other hand, coexpression of BDV P significantly inhibited the induction of p21waf1 expression in the cells, while no inhibition was found in the cells cotransfected with pcD-PΔM1, which lacks the domain for HMGB1 binding (Fig. 7A). This result indicated that P may repress up-regulation of p21waf1 by interference with endogenous HMGB1.
FIG. 7.
Activation of p21waf1 in p53-transduced and CsA-treated cell lines. (A) Western immunoblot analysis of BDV P, p53, and p21waf1 in total proteins extracted from transfected NCI-H1299 cells. pcD-p53 (0.5 μg) was cotransfected with pcD-P (1.5 μg) or pcD-PΔM1 (1.5 μg) into NCI-H1299 cells. The total DNA transfected (3.0 μg) was normalized with the pcDNA3 plasmid. Lanes: 1, pcDNA3 only; 2, pcDNA3 plus pcD-p53; 3, pcDNA3 plus pcD-p53 plus pcD-PΔM1; 4, pcDNA3 plus pcD-p53 plus pcD-P. p21waf1 expression was also demonstrated in the cell lysates prepared from uninfected and BDV-infected C6 cells before (lanes 5 and 7) and 24 h after (lanes 6 and 8) CsA treatment. As a control for protein input, the level of expression of tubulin was measured. (B) Activation of p21waf1 protein in BDV p53-induced NCI-H1299 cells showing P expression and BDV-infected C6 cells. The signal intensity shown in panel A was quantified by NIH Image software. Similar activation levels were obtained in at least three independent experiments.
We also examined the activation of p21waf1 in BDV-infected C6 cells by treatment with CsA, which is known to be able to efficiently induce p53 in C6 cells (35). Twenty-four hours after the treatment, induction of p53 was detected in both the infected and uninfected C6 cells (data not shown). As shown in Fig. 7A, CsA treatment significantly induced p21waf1 expression in the uninfected C6 cells (lane 6). Intriguingly, despite the fact that there was no difference between the abilities for cell growth and the viabilities of the infected and uninfected cells, expression of p21waf1 was up-regulated in the infected cells even under normal culture conditions. Several different signaling pathways have been identified in p21waf1 activation with or without p53 involvement. We demonstrated, however, that CsA at least failed to increase p21waf1 up-regulation in the infected cells by 24 h after treatment (Fig. 7A, lane 8). The quantification analysis of the signal intensities of the protein indicated that both NCI-H1299 cells showing p53-induced P expression and BDV-infected C6 cells had p21waf1 activation ratios reduced to almost 70 and 80% of those of control cells, respectively (Fig. 7B). Similar activation levels were obtained in at least three independent experiments. These observations suggested that BDV infection may decrease the chance of interaction between p53 and HMGB1 in cells by the expression of P, resulting in the repression of p53-mediated transcriptional activity in infected cells.
DISCUSSION
The 24-kDa phosphoprotein of BDV is abundant in infected cells. Although the precise role of the protein in the viral life cycle has not yet been elucidated, it is assumed that P plays a pivotal role in viral transcription and replication in the nucleus in cooperation with the viral pol protein. We have recently demonstrated that P specifically binds to HMGB1 and inhibits its functions in vivo. In this study, we identified those amino acid regions of BDV P and HMGB1 required for the interaction (Fig. 1 to 4). Furthermore, we showed that BDV P interferes with p53 binding to HMGB1 and inhibits p53 activity in transiently transfected cells (Fig. 5 and 6). We were also able to demonstrate that activation of p21waf1 expression is repressed in cells showing p53-induced P expression and in BDV-infected cell lines (Fig. 7). These results suggest that BDV P may repress p53 transcriptional activity by interfering with HMGB1. This strategy of P may be involved in unique features of BDV infection in CNS cells, such as noncytolytic replication and persistent infection.
Increasing evidence has demonstrated that HMGB1 is multifunctional in neuronal cells. As we described in a previous report, some roles of HMGB1 are conducted by secretion from the cell plasma membrane. Surface coating of culture plates with HMGB1 directly mediated neurite outgrowth in neuronal cell lines and primary rat brain cells via interaction with the extracellular moiety of RAGE (23, 36). In addition, activation of RAGE by HMGB1 appeared to promote cell survival through increased expression of the anti-apoptotic protein Bcl-2 (13, 24). In accordance with these observations, HMGB1 is likely to be critical in the maturation or construction of the CNS, as well as in network formation of neuronal cells in the developing or injured brain. Such extracellular functions of HMGB1 have been the prime focus of recent studies of the protein. The extracellular functions of HMGB1 should provide an important paradigm for understanding the neuropathogenesis of BDV in various animals.
Beyond its extracellular roles, HMGB1 also has intranuclear functions, which have been extensively studied. The HMG box (A and B) domains in HMGB1 allow the protein to bind to DNAs exclusively through the minor groove and to modify the structure of DNA (4, 18, 45). This feature may be relevant to the biological functions of HMGB1 in the nucleus, such as DNA repair, recombination, replication, and transcription. In addition, HMGB1 can interact via the HMG boxes with a broad range of proteins, from nuclear proteins to virus components. Interaction with HMGB1 has been described in several transcription factors (p53, Hox, Pou, Oct, steroid hormone receptors, and TATA-binding protein) (3, 5, 17, 26, 28, 47, 48), viral proteins (adeno-associated virus Rep) (7), and the recombination activation gene proteins (RAG1 and -2) (1). In general, HMGB1 increases the DNA binding affinities of these factors and shows either a negative or positive effect on transcription. A recent study using phage display analysis has identified peptide motifs that are recognized by HMGB1 (10). The study revealed that the peptide sequences selected by the assay are very variable and that HMGB1 may not have a preferred interaction sequence (10). Our results also support this finding. The HMGB1-binding region of P, K77LVTELAENS86, has only slight homology to the peptide identified in the work mentioned above, suggesting high complexity in the sequences for HMGB1 binding. On the other hand, however, it was postulated that the sequences binding to HMGB1 may be grouped into at least two classes by their amino acid distributions: proline-rich (PxxPxP) and tryptophan-rich (WxxW) motifs (10). In fact, several HMGB1-binding proteins, including p53, progesterone, and Groucho-related gene protein 1 (Grg1), contain a consensus proline-rich motif in their sequences (10). Although the HMGB1-binding region of P contains neither a proline-rich nor a tryptophan-rich region, interestingly, P has two consensus proline-rich motifs in both the N and C termini of the sequence (R25PGSPRP31 and L174PSHPAP181). This might indicate the possibility that P recognizes HMGB1 with more than a region in the sequence. As we clearly demonstrated, amino acids 77 to 86 of P must be essential for interaction with HMGB1, while a cooperative role of the proline-rich motifs may also exist in the secure binding between P and HMGB1. Also note that our experiment could not rule out the possibility that the amino acid sequence between positions 87 and 143 of P is also required for interaction with HMGB1.
We determined that the HMGB1 A box is necessary for binding to P. The A box is also known to be involved in interactions with several cellular proteins, including p53, Hox, steroid hormone receptors, and RAGs (1, 3, 26, 47). These observations indicate the possibility that the presence of P in BDV-infected cells competitively interferes with the binding of these cellular proteins to HMGB1 and affects their functions. Among them, p53 may be a particularly interesting protein, because it has been revealed that p53 directly binds to HMGB1 only via the A box, while in other proteins, either the A or B box is sufficient for the interactions. We could clearly demonstrate that the transcriptional activity of p53 is significantly reduced when P is present in the transfected cells, suggesting that P could efficiently block p53 binding to the HMGB1 A box in the nucleus.
The possibility of competitive inhibition of p53 activity by P allows us to speculate about the role of P in the infected CNS. Activation of p53 gives rise to a variety of cellular responses, most notably cell cycle arrest and apoptosis, through the activation of numerous cellular genes, such as Mdm2, cyclin G, p21waf1, Bax, and Apaf1 (11, 12, 15, 32). These cellular responses led by p53 could be part of the host defense mechanisms against several cellular stresses, such as viral infection (2, 9). Many viruses have been shown to interfere with the function of p53, suggesting that alteration of p53 activity is likely to be important for viral replication or survival in infected cells. It has been demonstrated that the LANA protein of human herpesvirus 8 contributes to viral persistence through its ability to promote cell survival by altering p53 function (16). BDV shows noncytolytic replication and easily establishes persistent infection in various cultured cells and brain cells. The long-lasting persistent infection is frequently found in infected animal brains (22). Recent studies have indicated that induction of immune tolerance or T-cell ignorance may play a major role in BDV persistence in the brain (19, 21). Indeed, it has been demonstrated that T lymphocytes from the brains of acutely BDV-infected rats, but not those from persistently infected rats, could lyse BDV-infected target cells (39). Although evasion of host immune response must be essential for the establishment of persistence in the CNS, the viral strategy that disturbs host surveillance for viral replication could also be required for maintenance of the persistent infection. It has been demonstrated that the expression patterns of BDV mRNAs do not shift from acute to persistent stages of infection in the brain and that the viral proteins are abundantly expressed throughout the infection (21). These observations strongly suggest that BDV replication itself protects infected cells against host defense mechanisms. Our results demonstrated the possibility that the modulation of p53 activity by P may perturb p53-dependent cell defense responses. The expression of P may prolong cell survival through the reduction of p53-mediated apoptosis, resulting in the maintenance of BDV persistence in the CNS. Although neuronal cell loss is found in specific regions of BDV-infected rat brains (20, 46), most cells appear to survive and express the viral antigens during persistent infection. Recently, it has been demonstrated that HMGB1 has the potential to cell- and promoter-specifically down- or up-regulate in vivo transcriptional activities of different members of the p53 family (p53 and p73α/β) (42). This finding suggests that BDV P may show cell-type- or region-specific modulation of p53 transcriptional activity in the brain. On the other hand, we could demonstrate that a persistently BDV-infected cell line exhibits up-regulation of p21waf1 even under normal culture conditions, although induction of p53 by CsA failed to increase the level of p21waf1 in the cells (Fig. 7). This observation suggested that p53-independent responses may play key roles in cellular defense in BDV-infected cells. However, our experiment could not exclude the possibility that levels of p21waf1 had already reached their peaks in the infected cells and that the lack of further activation in response to CsA is not related to the inhibition of p53 activity by P. It should be also noted that alterations of other cellular factors, the functions of which are mediated by HMGB1 binding, by P are also involved in the persistence and neuropathogenesis of BDV. Further study will be needed to understand the effect of P expression on BDV survival in CNS cells.
BDV P may be a unique inhibitor of p53. Almost all viral proteins known to modify p53 activity have been shown to directly interact with p53 (14). Furthermore, many viruses encoding p53-binding proteins are known to be oncogenic, e.g., adenovirus, papillomavirus, retrovirus, and herpesvirus. The features of BDV, such as indirect modification of p53 via interference with its cellular coactivator and nononcogenesis, would make BDV a unique p53-inhibiting virus. In tumor viruses, control of the cell cycle and apoptosis by p53 inhibition, as well as the production of viral oncogenes, would be necessary for the transformation of infected cells. In BDV, the modification of p53, as well as inhibition of neurite outgrowth or cell migration, by interfering with HMGB1 may be linked to the successful persistence and unique neuropathogenesis of the virus in the CNS. Studies using experimental animals to understand the roles of interference with HMGB1 in BDV-induced neuronal disturbance and neurobehavioral disorders are in progress.
Acknowledgments
We thank H. Nojima (Research Institute for Microbial Diseases, Osaka University, Osaka, Japan) for generously providing the expression plasmid carrying full-length human p53 cDNA, Y. Nakanishi (Kyushu University, Fukuoka, Japan) and A. F. Gazdar (Hamon Cancer Center, University of Texas Southwestern Medical Center, Dallas) for NCI-H1299 cells, and M. Oren (Institute of Science, Rehovot, Israel) for the pGL-cyclin G-Luc reporter plasmid.
This study was supported in part by Special Coordination Funds for Science and Technology and by Grants-in-Aid, both from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by the Japan Society for the Promotion of Science (JSPS).
REFERENCES
- 1.Aidinis, V., T. Bonaldi, M. Beltrame, S. Santagata, M. E. Bianchi, and E. Spanopoulou. 1999. The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol. 19:6532-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barber, G. N. 2001. Host defense, viruses and apoptosis. Cell Death Differ. 8:113-126. [DOI] [PubMed] [Google Scholar]
- 3.Boonyaratanakornkit, V., V. Melvin, P. Prendergast, M. Altmann, L. Ronfani, M. E. Bianchi, L. Taraseviciene, S. K. Nordeen, E. A. Allegretto, and D. P. Edwards. 1998. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol. 18:4471-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butteroni, C., M. De Felici, H. R. Scholer, and M. Pesce. 2000. Phage display screening reveals an association between germline-specific transcription factor Oct-4 and multiple cellular proteins. J. Mol. Biol. 304:529-540. [DOI] [PubMed] [Google Scholar]
- 6.Carbone, K. M. 2001. Borna disease virus and human disease. Clin. Microbiol. Rev. 14:513-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costello, E., P. Saudan, E. Winocour, L. Pizer, and P. Beard. 1997. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 16:5943-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cubitt, B., C. Oldstone, and J. C. de la Torre. 1994. Sequence and genome organization of Borna disease virus. J. Virol. 68:1382-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Laurenzi, V., and G. Melino. 2000. Evolution of functions within the p53/p63/p73 family. Ann. N. Y. Acad. Sci. 926:90-100. [DOI] [PubMed] [Google Scholar]
- 10.Dintilhac, A., and J. Bernues. 2002. HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. J. Biol. Chem. 277:7021-7028. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry, W. S. 1998. p21/p53, cellular growth control and genomic integrity. Curr. Top. Microbiol. Immunol. 227:121-137. [DOI] [PubMed] [Google Scholar]
- 12.El-Deiry, W. S. 1998. Regulation of p53 downstream genes. Semin. Cancer Biol. 8:345-357. [DOI] [PubMed] [Google Scholar]
- 13.Fages, C., R. Nolo, H. J. Huttunen, E. Eskelinen, and H. Rauvala. 2000. Regulation of cell migration by amphoterin. J. Cell Sci. 113:611-620. [DOI] [PubMed] [Google Scholar]
- 14.Fazakerley, J. K., and T. E. Allsopp. 2001. Programmed cell death in virus infections of the nervous system. Curr. Top. Microbiol. Immunol. 253:95-119. [DOI] [PubMed] [Google Scholar]
- 15.Fortin, A., S. P. Cregan, J. G. MacLaurin, N. Kushwaha, E. S. Hickman, C. S. Thompson, A. Hakim, P. R. Albert, F. Cecconi, K. Helin, D. S. Park, and R. S. Slack. 2001. APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J. Cell Biol. 155:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 17.Ge, H., and R. G. Roeder. 1994. The high mobility group protein HMG1 can reversibly inhibit class II gene transcription by interaction with the TATA-binding protein. J. Biol. Chem. 269:17136-17140. [PubMed] [Google Scholar]
- 18.Hardman, C. H., R. W. Broadhurst, A. R. Raine, K. D. Grasser, J. O. Thomas, and E. D. Laue. 1995. Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry 34:16596-16607. [DOI] [PubMed] [Google Scholar]
- 19.Hausmann, J., W. Hallensleben, J. C. de la Torre, A. Pagenstecher, C. Zimmermann, H. Pircher, and P. Staeheli. 1999. T cell ignorance in mice to Borna disease virus can be overcome by peripheral expression of the viral nucleoprotein. Proc. Natl. Acad. Sci. USA 96:9769-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornig, M., H. Weissenbock, N. Horscroft, and W. I. Lipkin. 1999. An infection-based model of neurodevelopmental damage. Proc. Natl. Acad. Sci. USA 96:12102-12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hornig, M., T. Briese, and W. I. Lipkin. 2001. Bornavirus tropism and targeted pathogenesis: virus-host interactions in a neurodevelopmental model. Adv. Virus Res. 56:557-582. [DOI] [PubMed] [Google Scholar]
- 22.Hornig, M., M. Solbrig, N. Horscroft, H. Weissenbock, and W. I. Lipkin. 2001. Borna disease virus infection of adult and neonatal rats: models for neuropsychiatric disease. Curr. Top. Microbiol. Immunol. 253:157-177. [DOI] [PubMed] [Google Scholar]
- 23.Huttunen, H. J., C. Fages, and H. Rauvala. 1999. Receptor for advanced glycation end products (RAGE)-mediated neurite outgrowth and activation of NF-κB require the cytoplasmic domain of the receptor but different downstream signaling pathways. J. Biol. Chem. 274:19919-19924. [DOI] [PubMed] [Google Scholar]
- 24.Huttunen, H. J., J. Kuja-Panula, G. Sorci, A. L. Agneletti, R. Donato, and H. Rauvala. 2000. Coregulation of neurite outgrowth and cell survival by amphoterin and S100 proteins through receptor for advanced glycation end products (RAGE) activation. J. Biol. Chem. 275:40096-40105. [DOI] [PubMed] [Google Scholar]
- 25.Ikuta, K., M. S. Ibrahim, T. Kobayashi, and K. Tomonaga. 2002. Borna disease virus and infection in humans. Front. Biosci. 7:D470-D495. [DOI] [PubMed] [Google Scholar]
- 26.Imamura, T., H. Izumi, G. Nagatani, T. Ise, M. Nomoto, Y. Iwamoto, and K. Kohno. 2001. Interaction with p53 enhances binding of cisplatin-modified DNA by high mobility group 1 protein. J. Biol. Chem. 276:7534-7540. [DOI] [PubMed] [Google Scholar]
- 27.Jayaraman, L., K. G. Murthy, C. Zhu, T. Curran, S. Xanthoudakis, and C. Prives. 1997. Identification of redox/repair protein Ref-1 as a potent activator of p53. Genes Dev. 11:558-570. [DOI] [PubMed] [Google Scholar]
- 28.Jayaraman, L., N. C. Moorthy, K. G. Murthy, J. L. Manley, M. Bustin, and C. Prives. 1998. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 12:462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan, I., and W. I. Lipkin. 2001. Borna disease virus. Rev. Med. Virol. 11:37-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamitani, W., Y. Shoya, T. Kobayashi, M. Watanabe, B. J. Lee, G. Zhang, K. Tomonaga, and K. Ikuta. 2001. Borna disease virus phosphoprotein binds a neurite outgrowth factor, amphoterin/HMG-1. J. Virol. 75:8742-8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig, H., and L. Bode. 2000. Borna disease virus: new aspects on infection, disease, diagnosis and epidemiology. Rev. Sci. Technol. 19:259-288. [DOI] [PubMed] [Google Scholar]
- 32.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293-299. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura, Y., H. Takahashi, Y. Shoya, T. Nakaya, M. Watanabe, K. Tomonaga, K. Iwahashi, K. Ameno, N. Momiyama, H. Taniyama, T. Sata, T. Kurata, J. C. de la Torre, and K. Ikuta. 2000. Isolation of Borna disease virus from human brain tissue. J. Virol. 74:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osaki, S., Y. Nakanishi, K. Takayama, X. H. Pei, H. Ueno, and N. Hara. 2000. Alteration of drug chemosensitivity caused by the adenovirus-mediated transfer of the wild-type p53 gene in human lung cancer cells. Cancer Gene Ther. 7:300-307. [DOI] [PubMed] [Google Scholar]
- 35.Pyrzynska, B., M. Serrano, A. C. Martinez, and B. Kaminska. 2002. Tumor suppressor p53 mediates apoptotic cell death triggered by cyclosporin A. J. Biol. Chem. 277:14102-14108. [DOI] [PubMed] [Google Scholar]
- 36.Rauvala, H., H. J. Huttunen, C. Fages, M. Kaksonen, T. Kinnunen, S. Imai, E. Raulo, and I. Kilpelainen. 2000. Heparin-binding proteins HB-GAM (pleiotrophin) and amphoterin in the regulation of cell motility. Matrix Biol. 19:377-387. [DOI] [PubMed] [Google Scholar]
- 37.Rott, R., and H. Becht. 1995. Natural and experimental Borna disease in animals. Curr. Top. Microbiol. Immunol. 190:17-30. [DOI] [PubMed] [Google Scholar]
- 38.Schwemmle, M., C. Jehle, S. Formella, and P. Staeheli. 1999. Sequence similarities between human Bornavirus isolates and laboratory strains question human origin. Lancet 354:1973-1974. [DOI] [PubMed] [Google Scholar]
- 39.Sobbe, M., T. Bilzer, S. Gommel, K. Noske, O. Planz, and L. Stitz. 1997. Induction of degenerative brain lesions after adoptive transfer of brain lymphocytes from Borna disease virus-infected rats: presence of CD8+ T cells and perforin mRNA. J. Virol. 71:2400-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staeheli, P., C. Sauder, J. Hausmann, F. Ehrensperger, and M. Schwemmle. 2000. Epidemiology of Borna disease virus. J. Gen. Virol. 81:2123-2135. [DOI] [PubMed] [Google Scholar]
- 41.Stitz, L., T. Bilzer, and O. Planz. 2002. The immunopathogenesis of Borna disease virus infection. Front. Biosci. 7:D541-D555. [DOI] [PubMed] [Google Scholar]
- 42.Stros, M., T. Ozaki, A. Bacikova, H. Kageyama, and A. Nakagawara. 2002. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J. Biol. Chem. 277:7157-7164. [DOI] [PubMed] [Google Scholar]
- 43.Tomonaga, K., T. Kobayashi, and K. Ikuta. 2002. Molecular and cellular biology of Borna disease virus infection. Microbes Infect. 4:491-500. [DOI] [PubMed] [Google Scholar]
- 44.Waltrip, R. W., II, R. W. Buchanan, W. T. Carpenter, Jr., B. Kirkpatrick, A. Summerfelt, A. Breier, S. A. Rubin, and K. M. Carbone. 1997. Borna disease virus antibodies and the deficit syndrome of schizophrenia. Schizophr. Res. 23:253-257. [DOI] [PubMed] [Google Scholar]
- 45.Weir, H. M., P. J. Kraulis, C. S. Hill, A. R. Raine, E. D. Laue, and J. O. Thomas. 1993. Structure of the HMG box motif in the B-domain of HMG1. EMBO J. 12:1311-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weissenbock, H., M. Hornig, W. F. Hickey, and W. I. Lipkin. 2000. Microglial activation and neuronal apoptosis in Bornavirus infected neonatal Lewis rats. Brain Pathol. 10:260-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zappavigna, V., L. Falciola, M. Helmer-Citterich, F. Mavilio, and M. E. Bianchi. 1996. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 15:4981-4991. [PMC free article] [PubMed] [Google Scholar]
- 48.Zwilling, S., H. Konig, and T. Wirth. 1995. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 14:1198-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]