Abstract
Porcine endogenous retroviruses (PERV) are discussed as putative infectious agents in xenotransplantation. PERV classes A, B, and C harbor different envelope proteins. Two different types of long terminal repeat (LTR) structures exist, of which both are present only in PERV-A. One type of LTR contains a distinct repeat structure in U3, while the other is repeatless, conferring a lower level of transcriptional activity. Since the different LTR structures are distributed unequally among the proviruses and, apparently, PERV is the only virus harboring two different LTR structures, we were interested in determining which LTR is the ancestor. Replication-competent viruses can still be found today, suggesting an evolutionary recent origin. Our studies revealed that the age of PERV is at most 7.6 × 106 years, whereas the repeatless LTR type evolved approximately 3.4 × 106 years ago, being the phylogenetically younger structure. The age determined for PERV correlates with the time of separation between pigs (Suidae, Sus scrofa) and their closest relatives, American-born peccaries (Tayassuidae, Pecari tajacu), 7.4 × 106 years ago.
Xenotransplantation involves the transfer of cells, tissues, or organs from one species to another for therapeutic reasons. While the technology is under development, it has the potential to overcome the significant shortage of human allotransplants (31). Using the pig (Sus scrofa; order, Suidae) as the favored donor animal presents a number of important microbiological safety issues (40) which may be solved regarding exogenous agents by proper handling of the animals, such as barrier breeding according to specific-pathogen-free conditions and application of vaccinations (38). However, porcine endogenous retroviruses (PERV) pose a more serious challenge, since they are present in almost every individual (26) and are transmitted vertically.
Endogenous retroviruses (ERV) are present in the genomes of all vertebrate species analyzed (8), and their general organization corresponds to exogenous retroviruses (8, 35, 41). Whereby multiple copies of ERV are present in their host genomes, most are truncated or mutated, rendering them replication incompetent. Nevertheless, some of the viral integration sites still display transcriptional activity and even produce viral particles by complementation in trans. Only a minority of the distinct proviruses are functional as reported for pigs (1, 2, 20, 25, 28, 29). While PERV belong to the gamma-retroviruses, other ERV also share homologies with beta- and delta-retroviruses but not with lentiviruses (12, 16, 28, 41).
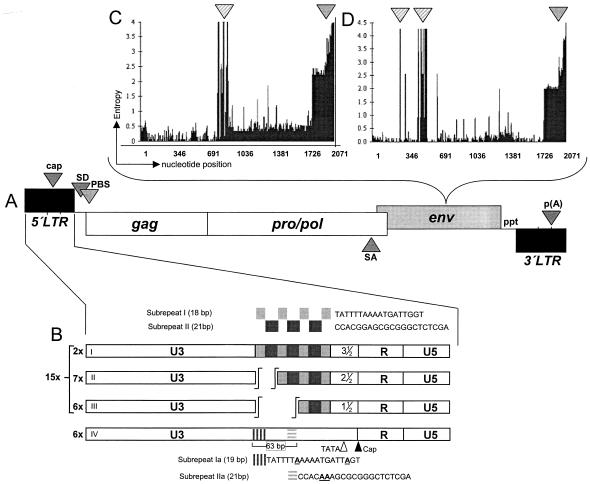
For PERV, three different classes, designated PERV-A, PERV-B, and PERV-C (1, 22), exist, whereby PERV-A and -B are polytropic and productively infect human cells in vitro, thus posing a serious risk in xenotransplantation and xenogeneic cell therapies. Ecotropic PERV-C (1) does not replicate on human cells and is therefore not included in this study. There are only minor genetic differences between the classes, which are most prominent in the receptor binding domain of the env protein (Fig. 1C and D). In addition, there are two different types of long terminal repeats (LTR) (Fig. 1B) that significantly affect the replication properties of single viruses (33) by instrumentalizing a special set of transcription factors (32). Both PERV-A and PERV-B proviruses demonstrate LTRs that harbor repeats in U3. On the other hand, PERV-A and PERV-C were found to display repeatless LTRs (33). LTRs displaying repeat structures and possible multimerization mechanisms were first described for murine retroviruses (23, 43). These retroviruses, along with other gamma-type retroviruses, share a common homology of approximately 60% among each other and with PERV. Furthermore, so as not to complicate the analysis, we excluded any of these viruses from this study and concentrated on determining the relative age of the two different PERV LTR types not found in any other virus. It was shown that PERV in cell culture actively adapt their LTR repeat structure, if present, to match the optimal but not the maximum replication performance in a given host cell (33). Conversely, the recombinant incorporation of an artificially created repeat structure, i.e., 10 times 39 bp, into U3 of the 5′ LTR of a molecular clone caused rapid cell death after transfection into susceptible cells (Scheef and Tönjes, unpublished data). While different env isoforms are present in a variety of retroviruses (7), only PERV is known to harbor two related but profoundly different LTR types in addition to three env classes.
FIG. 1.
Genomic organization of proviral PERV. (A) PERV displays genes for group specific antigen (gag), protease/polymerase polyprotein (pro/pol), and envelope protein (env), flanked by LTR. Both LTR and env vary significantly between individual proviruses, with the LTR determining the transcriptional activity and the envelope protein determining the host range with two polytropic virus classes, designated A and B. The gag and pol genes show only minor variations between proviruses. (B) Two different PERV LTR structures exist, one type harboring different numbers of a distinct 39-bp repeat in U3 (I to III) which is composed of subrepeat I (18 bp) and subrepeat II (21 bp) (26, 33). The second LTR type has no repeat structure, although sequences homologous to the subrepeats (designated Ia and IIa) can be found scattered across U3. The numbers given for each LTR designate the presence of proviral PERV LTR found in 21 BAC clones of a genomic large white library (26, 30). (C and D) Entropy plots for PERV envelope genes (panel C, PERV-A; panel D, PERV-B). For calculating entropy, all env sequences (see Appendix) were aligned, and the number of nucleotide differences was determined for every position. This difference is plotted against the position, thus indicating regions of high sequence variation. The sequence variation at the 3′ end of both env genes is caused by two factors. Since sequences used for entropy analysis were taken unmodified from GenBank, some truncated sequences led to an overall increase in diversity. The more important fact, however, is a structural divergence of the R-peptide structure of various PERV. This divergence corresponds in large part, but not exclusively, with the LTR structure (see “Phylogenetic analysis of env” in the text for details). Broadly striped arrowhead, hot spot with very high sequence variation; finely striped arrow head, variations at the C terminus of Env; cap, cap site; SD, splice donor; PBS, primer binding site; SA, splice acceptor; ppt, polypurine tract; p(A), poly adenylation site.
In anthropological studies, human endogenous retroviruses (HERV) have been used successfully to designate the age of the proviruses in hominoids as well as the time points of species separation events and phylogenetic relationships of different hominoids (17, 18, 34). These studies had the benefit of comparing many different HERV from different hominoids, ranging from humans to apes to monkeys. To repeat such a study for PERV is complicated due to the present diversity of the Artiodactyla (27, 37). Therefore, the closest relatives to pigs, the American-born peccaries (Tayassuidae, Pecari tajacu; see below), were included in this study to analyze an archaeological fixed relationship between two species.
Besides determining the age of PERV, we were particularly interested in investigating which of the two LTR types is the phylogenetic predecessor.
We have analyzed the prevalence of six well-characterized full-length PERV, five of them being replication competent and four of them being chromosomally assigned (20, 25). These analyses revealed a heterogeneous distribution of PERV among individuals (26), and since no PERV is present in every pig, it seems feasible to generate pigs free of functional PERV by conventional breeding. In addition, specific proviruses show internal point mutations which significantly affect their replication capacities. Since there are two different types of PERV LTR structures (Fig. 1B) showing various levels of transcriptional capacity (33), an analysis of 21 distinct chromosomal locations revealed that PERV which harbor highly active LTR with repeat elements in U3 are dominant (26). In addition, the two polytropic envelope genes were assayed for sequence variations, displaying class-specific hot spots of variation, as well as variations in the R peptide region (Fig. 1C and D).
Sample acquisition.
In order to study the phylogenetics of PERV, we have analyzed the LTRs and env genes of 17 mostly intact and chromosomally localized proviruses (25, 30) by PCR and subsequent sequencing. These sequences were submitted to GenBank, and the accession numbers of those as well as of instrumental full-length proviral sequences are given in the Appendix. In addition, sequences of 27 envelope genes published in GenBank were analyzed. Their accession numbers are given in the Appendix as well.
Molecular clock analysis.
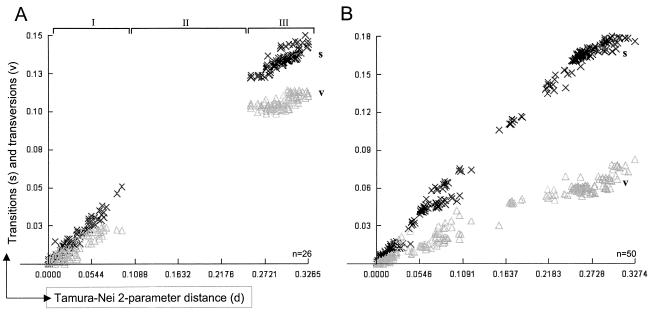
In order to evaluate the clocklike behavior of PERV proviruses, distinct trees were constructed using the datasets of LTR and env sequences, proving that the molecular clock (5, 24, 45, 46) is the best approximation and could not be rejected assuming a constant rate of evolution (Table 1). When both data sets were tested for substitution saturation using the DAMBE software package (44), no saturation could be observed when transitions (s) and transversions (v) were plotted versus the evolutionary distance (Fig. 2). The plot shows a linear increase of s and v values based on increasing divergence between different PERV. Since s occurs much more often than v, s should increase faster than v. In case of substitution saturation, where multiple substitutions have occurred at each site, the phylogenetic signal is essentially lost, and its effect is detectable because v gradually outnumbers s. Thus, the established graphs (Fig. 2) indicate that no substitution saturation has occurred in the investigated data sets of PERV LTR (Fig. 2A) and env (Fig. 2B) sequences, suggesting the reliability of the chronological dating based on the molecular clock.
TABLE 1.
Maximum-likelihood test for the molecular clock hypothesisa
| Data set | Log likelihood
|
2(In ML − In MLK) | χ2 (0.95) | Statistical significance level (P) | Rejection of the molecular clock hypothesis | |
|---|---|---|---|---|---|---|
| ML | MLK | |||||
| LTR | −1,968.13 | −2,727.04 | 1,517.81 | 23 | 0.001 | No |
| env | −7,334.43 | −10,017.67 | 5,366.47 | 35 | 0.001 | No |
Calculation of the statistical significance of rejecting the molecular clock hypothesis using the χ2 test comparing maximum likelihood and maximum likelihood with enforced molecular clock. The hypothesis that PERV does evolve according to a molecular clock is rejected, if the significance p is larger than 0.05. ML, maximum likelihood method; MLK, likelihood method enforcing a molecular clock using the ML topology. The calculation was performed using the software package DAMBE (44) according to the instructions given in reference 39.
FIG. 2.
Transitions and transversions of PERV sequences plotted versus Timura-Nei′s two-parameter genetic distance. Nucleotide sequences of LTRs (A) and env (B) used to determine the evolutionary age of PERV were analyzed with the program DAMBE (44) for their transitions (× and s) and transversions (Δ and v) ratio employing the Tamura-Nei substitution model (36). Transitions are nucleotide changes T↔C or A↔G and should happen more frequently than transversions, which are changes encompassing T↔A, T↔G, C↔A, or C↔G. While the s/v plot of env sequences (B) shows a linear increase of both transitions and transversions, with transitions progressing much faster, the s/v plot of LTR sequences (A) shows a gap (II) based on LTRs with different numbers of repeats, leading to a stepwise increase in sequence variations. When comparing viruses with different LTR types, each repeat-harboring LTR type increases genetic distance by 39 bp, while the repeatless LTRs deviate considerably from the repeat-harboring LTRs on the basis of pairwise comparison, thus producing the gap in genetic distance that is observed in the LTR s/v plot. The significant dominance of transitions and no crossing of the different symbols, representing transitions and transversions, suggest no substitution saturation and proof of a constant rate of evolution, therefore allowing a determination of age by molecular clock. Each × or Δ in the plot represent a transition or transversion event at a given genetic distance, respectively. n, number of sequences analyzed for each plot (see Appendix).
The gap in the s/v plot of the LTR data set (Fig. 2A, II) is due to the repeat structure present in U3 of the PERV LTR (Fig. 1B). The comparison of LTRs with a different repeat number results in this steplike increase of genetic distance as insertions or deletions of 39 nucleotides happen at once. The separation of env sequences into two subpopulations according to their class (A or B) results in similar s/v plots, as observed for the combined sequence set (data not shown), suggesting similar rates of evolution for both env classes.
Phylogenetic analysis of LTR sequences.
The clocklike evolution of PERV allowed an attempt of dating by sequence comparison of the 5′ and 3′ LTR of distinct proviruses. Since the number of fully sequenced PERV is not large enough to allow for phylogenetic analyses based on intact and defect proviral sequences, we amplified the LTRs of distinct chromosomally assigned PERV present in a genomic pig library constructed in bacterial artificial chromosomes (BAC) (30). Because every BAC statistically harbors only one provirus (checked by PCR; data not shown), we could amplify 5′ and 3′ LTRs of different PERV from this quasigenomic template without interference with LTRs of unrelated proviruses.
Analyses of primate ERV showed that these ERV accumulate mutations with a rate of approximately 2.3 × 10−9 to 5.0 × 10−9 substitutions per nucleotide per year (17). Assuming the same mutation rate and with a mean LTR length of 700 bp, this translates to 250.000 to 650.000 years per mutation for PERV, with a mean of 450.000 years used for the calculations in Table 2. The fact that transcriptionally active viruses exist (11, 19, 25) favors a relatively recent phylogenetic origin of PERV. While the results of this attempt of chronological dating are given in Table 2 and the actual estimation of time may be improved in the future, it is clear that the repeatless LTR (Fig. 1B) is phylogenetically younger than the repeat-harboring LTR (Fig. 1B).
TABLE 2.
Estimation of PERV age by comparison of differences in the 5′ and 3′ LTRsa
| LTR type | No. | Ø no. of mutations | Approx age
|
|
|---|---|---|---|---|
| Min | Max | |||
| w/o repeats | 6 | 4 ± 3 | 400,000 | 3.1 × 106 |
| 1 1/2 repeats | 6 | 12 ± 5 | 1.1 × 106 | 7.6 × 106 |
| 2 1/2 repeats | 7 | 9 ± 6 | 900,000 | 6.7 × 106 |
| 3 1/2 repeats | 2 | 2 ± 1 | 100,000 | 1.3 × 106 |
The 5′ and 3′ LTRs of 21 chromosomally assigned proviral clones were amplified and sequenced, thus revealing nucleotide differences in the corresponding LTRs. With estimated mutation rates of approximately 2.3 × 10−9 to 5.0 × 10−9 substitutions per nucleotide per year, any substitution translates to 250,000 and 650,000 years of evolution, respectively. The mean of 450,000 years was used for calculating the age of PERV, multiplying this value with the average number of mutations per LTR type. The appearance of very low or very high mutation numbers in a single copy of an LTR giving rise to high standard deviation was corrected for the final estimation of age using the median. No. occurrences of the respective LTR type in the 21 BAC clones assayed; Ø no. of mutations, mean number of mutations found in all LTRs of a given type, given as mean number with standard deviation; approx age, the estimation of age is given in years with the minimum (Min) and maximum (Max) times since the proviral integrations into the host genome have occurred. w/o, without.
A possible mechanism could be an insertional mutation in an LTR with 1.5 repeats, disrupting this motif, since highly homologous but spatially not organized subrepeat fragments can be found in the LTRs that are devoid of 39-bp repeats (Fig. 1B, subrepeats Ia and IIa). An insertional mutation only needs to happen in the 3′ LTR, since the replication mechanism of retroviruses will copy that mutation to the 5′ LTR as well. Unfortunately, the stretch of DNA inserted into the LTR is too small to allow tracing of its origin confidently in the databases, producing matches with many genes of different species. The repeatless LTRs show a significantly lower level of transcriptional activity because the binding site of the major transcription factor NF-Y is located directly at the junction of the 18-bp and 21-bp-repeat and, therefore, is missing in the repeatless LTRs (33). We assume that repeatless LTRs are an adaptation to the acquired endogenous replication cycle of PERV, since this LTR structure largely reduces the transcriptional activity (33), which is in line with observations that artificially elongated LTRs kill their host cells rather rapidly (see above) and that replication-competent viruses dynamically adjust their repeat numbers to host cells, favoring lower copy numbers of these repeats in U3 (33). Furthermore, adaptations reducing the replicative performance have been described for a number of virus families, such as endogenously replicating members like MLV or HERV (10). While PERV are discussed as major obstacles to xenotransplantation applications, since they productively infect human cells in vitro, some of our results indicate that viruses carrying repeatless LTRs, though capable of minor replication in vitro, are not able to cross the species barrier on their own. Proviruses isolated from PERV-infected human cell line 293-PERV-PK (29) all showed a repeat-harboring LTR, and no integrated repeatless LTR could be identified in genomic DNA of that cell line by PCR (11, 25).
Phylogenetic analysis of env sequences.
Since the reliability of the molecular clock was proven for PERV LTR and env sequences (Table 1 and Fig. 2), an alternative method for phylogenetic dating is the comparison of coding sequences generating phylogenetic trees based on Env (Fig. 3). In general, the homologies between PERV-A and PERV-C are approximately 85%, while the similarities between PERV-B and both PERV-A and PERV-C barely exceed 70%. This and the occurrence of repeatless LTRs in both PERV-A and PERV-C but not in PERV-B (see above) lead to the assumption of a common evolutionary origin of these two classes, both seeming younger than PERV-B. Unfortunately, there are not enough PERV-C sequences available currently to conduct a thorough analysis. The exclusion of PERV-C sequences may present a weak point. There is the possibility that PERV-C recombined with PERV-A, which is evident by A/C env recombinants (13, 42). Therefore, a PERV-A population with mixed LTR sequences is conceivable. PERV-C obviously was not able to spread in the porcine genome (initial results of screening several porcine genomic libraries suggest a very limited number of PERV-C sequences in the pig genome; Preuβ and Tönjes, unpublished data); however, PERV-A with both types of LTR did. We admit that if more PERV-C sequences become available the study must be extended. While the observed hot spots of variation specific for each class most likely are due to different selective pressures (Fig. 1C and D), there is a principle sequence difference between both classes in the receptor binding region, while other regions display high levels of homology. When using such sequences for computational analysis, this may lead to intrinsic bias. Therefore, both subpopulations were assayed separately. However, the comparison of coding sequences, in contrast to the comparison of LTR sequences, lacks an internal reference and must be calibrated with an externally generated fixed time point, most preferably related to archaeological data. Since those data are limited for pigs, an appropriate analysis includes study of whether peccary (Tayassuidae) genomes harbor PERV sequences. Peccaries are the closest evolutionary relatives of pigs, live isolated in America, and were separated from the Suidae approximately 7.4 × 106 years ago (18). Analysis of different peccary genomic DNA samples failed to identify any PERV-specific signals by PCR using degenerate primers (13a) suitable for amplification of gamma-type retroviral sequences or PERV specific primers (11); however, both approaches produced faint, unspecific signals (data not shown). We are currently investigating whether these unspecific signals belong to any unknown gamma-type retrovirus, which would not be surprising, since ERV sequences were found in any vertebrate species (14). These results are in line with a recent publication which identified several new PERV-related sequences in pigs and peccaries but also did not detect any PERV sequences in peccaries (28). The absence of PERV sequences in peccaries indicate that the age of PERV cannot exceed the calculated 7.4 × 106 to 7.6 × 106 years. According to calculations based on the LTR data, the repeatless LTR evolved between 400,000 and 3.1 × 106 years ago (Table 2). We assume that the appearance of that LTR which shows a low transcriptional activity enabled the virus to pursue an endogenous replication cycle more easily, since the low replication level does not damage the host cell while the virus still retains its ability to replicate outside its host.
FIG. 3.
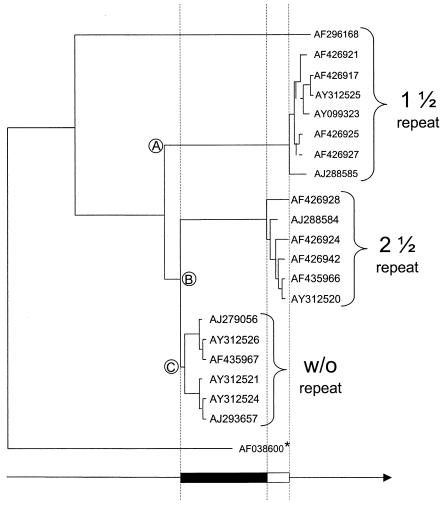
Phylogenetic tree of PERV-A envelope protein sequences aligned for evolutionary time, calculated as in Table 2. Env sequences were shortened to amino acid position 654 to avoid any bias due to R-peptide variations or truncation (see the text and Fig. 1C and D for details). The analyzed PERV-A Env sequences cluster into three groups, with their branching points indicated by letters. The most divergent group (A) is the oldest one, the second group (B) being only slightly younger. This behavior correlates with LTR types with 1 1/2 and 2 1/2 repeats, respectively. The less-divergent group (C) is the youngest one, with all members belonging to proviruses with repeatless LTRs. The distance between cluster A and B is approximately one-fourth of the distance between cluster C and cluster A. When assuming a difference of approximately 0.9 million years between LTRs with 1 1/2 and 2 1/2 repeats (white box) (see Table 2), these two groups are about 3.5 millions years older than groups C (black box) (compare table 2). *, PERV-C (1) was chosen as the outgroup because of its close relationship to PERV-A sequences. The arrow indicates increasing divergence and therefore increasing evolutionary age. Please note that no absolute age is indicated, since an external reference is missing (see the text), and that only relative age can be determined from the graph.
Due to missing external references, protein sequences could be employed only for dating the relative age, thereby reinforcing the LTR analysis and indicating that the repeatless LTRs are in fact phylogenetically younger (data not shown).
Conclusion.
A number of authors have pointed out that molecular clock calibrations are subject to a wide margin of error (3, 4, 15). Therefore, the calculations given in Table 2 are only rough estimates of absolute time, but they are nonetheless useful for comparing the relative ages of different PERV. In summary, we have shown that the repeatless PERV LTR evolved from the repeat-harboring LTR, most likely by insertional mutation. This can be proven by employing two different sets of data (LTR and env/Env). PERV is calculated to be approximately 7.4 million years old. This age correlates with the separation of pigs and their closest relatives, the peccaries, approximately 7.6 × 106 years ago, and the event that led to the generation of the repeatless type of LTR occurred between 400,000 and 3 million years ago (Table 2). Upon integration into the germ line, both LTR variants will have coexisted exo- and endogenously for some time. It is evident from observations of other endogenous retroviruses (especially HERV) that these ERV acquire mutations which reduce and finally abolish their replicative performance. The acquisition of a weakly performing promoter may help the virus to do no damage to its host cell while retaining some levels of replication competence. However, the generation of repeatless LTRs may reflect an adaptation process of the virus, switching from an exogenous to an endogenous life cycle. The repeatless LTR is present in the relatively closely related PERV-A and PERV-C proviruses, while the more distant PERV-B proviruses exclusively display repeat-harboring LTRs.
Acknowledgments
This study was supported by a grant from the German Ministry of Health (Bundesministerium für Gesundheit), Bonn, Germany.
APPENDIX
Specific PERV loci isolated from the BAC library (30) are grouped according to their respective PERV subgroup, while the accession numbers are given for 5′ LTR, 3′ LTR, and env for the respective clone. PERV-A: 242D4 (AY312539, AY312555, AY312523); 141G12 (AY312537, AY312553, AY312521); 305F5 (AY312542, AY312558, AY312526); 258A11 (AY312541, AY312557, AY312525); 135E5 (AY312536, AY312552, AY312520); 253B6 (AY312540, AY312556, AY312524); 383E10 (AY312543, AY312559, AY312527); and 1079D9 (AY312535, AY312551, AY312519). PERV-B: 161B7 (AY312538, AY312554, AY312522); 484G4 (AY312544, AY312560, AY312528); 498D8 (AY312545, AY312561, AY312529); 534G4 (AY312546, AY312562, AY312530); 647G4 (AY312547, AY312563, AY312531); 667G4 (AY312548, AY312564, AY312532); 783D7 (AY312549, AY312565, AY312533); 80H6 (AY312550, AY312566, AY312517); and 1058D6 (AY312534, AY312567, AY312518). PERV-C: none.
In addition, we have used full-length sequences of previously described proviruses isolated in our laboratory as well as sequences available in the international databases. PERV-A: AY099323 (6); AJ288585, AJ288584 (9), AJ133817 (11); AJ293656 (20); AF426917, AF426921, AF426924, AF426925, AF426927, AF426928, AF426942, (21); AJ279056, AJ279056, AF435966 (25); and AF296168 (Chang et al., unpublished data). PERV-B: AY099324 (6); AJ288589, AJ288592, AJ288591, AJ288590, AJ288586, AJ288588 (9); AJ133816, AJ133818 (11); AJ293657 (20); AF426916, AF426940, AF426937, AF426936, AF426935 AF426933, AF426946, (21); Y12239 (22); and AJ293656 (25). PERV-C: AF038600 (1).
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 72:4503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. S. Potterfield, and A. T. DeMadrid. 1971. C-type virus particles in pig kidney cell lines. J. Gen. Virol. 10:195-198. [DOI] [PubMed] [Google Scholar]
- 3.Arnason, U., A. Gullberg, and A. Janke. 1998. Molecular timing of primate divergences as estimated by two nonprimate calibration points. J. Mol. Evol. 47:718-727. [DOI] [PubMed] [Google Scholar]
- 4.Avise, J. C. 1994. Molecular markers, natural history and evolution. Chapman & Hall, New York, N.Y.
- 5.Ayala, F. J. 1986. On the virtues and pitfalls of the molecular evolutionary clock. J. Hered. 77:226-235. [DOI] [PubMed] [Google Scholar]
- 6.Bartosch, B., R. A. Weiss, and Y. Takeuchi. 2002. PCR-based cloning and immunocytological titration of infectious porcine endogenous retrovirus subgroup A and B. J. Gen. Virol. 83:2231-2240. [DOI] [PubMed] [Google Scholar]
- 7.Battini, J. L., O. Danos, and J. M. Heard. 1995. Receptor-binding domain of murine leukemia virus envelope glycoproteins. J. Virol. 69:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke, J. D., and J. P. Stoye. 1997. Retrotransposons, endogenous retroviruses and the evolution of retroelements, p. 343-436. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 9.Bosch, S., C. Arnauld, and A. Jestin. 2000. Study of full-length porcine endogenous retrovirus genomes with envelope gene polymorphism in a specific-pathogen-free Large White swine herd. J. Virol. 74:8575-8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffin, J. M. 1984. Endogenous viruses, p. 1109-1203. In R. Weiss, N. Teich, H. Varmus, and J. Coffin (ed.), RNA tumor viruses, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Czauderna, F., N. Fischer, K. Boller, R. Kurth, and R. R. Tönjes. 2000. Establishment and characterization of molecular clones of porcine endogenous retroviruses replicating on human cells. J. Virol. 74:4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ericsson, T., B. Oldmixon, J. Blomberg, M. Rosa, C. Patience, and G. Andersson. 2001. Identification of novel porcine endogenous betaretrovirus sequences in miniature swine. J. Virol. 75:2765-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ericsson, T. A., Y. Takeuchi, C. Templin, G. Quinn, S. F. Farhadian, J. C. Wood, B. A. Oldmixon, K. M. Suling, J. K. Ishii, Y. Kitagawa, T. Miyazawa, D. R. Salomon, R. A. Weiss, and C. Patience. 2003. Identification of receptors for pig endogenous retrovirus. Proc. Natl. Acad. Sci. USA 100:6759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Heneine W., A. Tibell, W. M. Switzer, P. Sandstrom, G. V. Rosales, A. Mathews, O. Korsgren, L. E. Chapman, T. M. Folks, and C. G. Groth. 1998. No evidence of infection with porcine endogenous retrovirus in recipients of porcine islet-cell xenografts. Lancet 352:695-699. [DOI] [PubMed]
- 14.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hillis, D. M., C. Moritz, and B. K. Mable (ed). 1996. Molecular systematics, p. 321-341. Sinauer Associates, Sunderland, Mass.
- 16.Horwitz, M. S., M. T. Boyce-Jacino, and A. J. Faras. 1992. Novel human endogenous sequences related to human immunodeficiency virus type 1. J. Virol. 66:2170-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, W. E., and J. M. Coffin. 1999. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl. Acad. Sci. USA 96:10254-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, H. S., O. Takenaka, and T. J. Crow. 1999. Isolation and phylogeny of endogenous retrovirus sequences belonging to the HERV-W family in primates. J. Gen. Virol. 80:2613-2619. [DOI] [PubMed] [Google Scholar]
- 19.Krach, U., N. Fischer, F. Czauderna, R. Kurth, and R. R. Tönjes. 2000. Generation and testing of a highly specific anti-serum directed against porcine endogenous retrovirus nucleocapsid. Xenotransplantation 7:221-229. [DOI] [PubMed] [Google Scholar]
- 20.Krach, U., N. Fischer, F. Czauderna, and R. R. Tönjes. 2001. Comparison of replication-competent molecular clones of porcine endogenous retrovirus class A and class B derived from pig and human cells. J. Virol 75:5465-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, J. H., G. C. Webb, R. D. Allen, and C. Moran. 2002. Characterizing and mapping porcine endogenous retroviruses in Westran pigs. J. Virol. 76:5548-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeTissier, P., J. Stoye, Y. Takeuchi, C. Patience, and R. A. Weiss. 1997. Two sets of human-tropic retrovirus. Nature 389:681-682. [DOI] [PubMed] [Google Scholar]
- 23.Martiney, M. J., K. Rulli, R. Beaty, L. S. Levy, and J. Lenz. 1999. Selection of reversions and suppressors of a mutation in the CBF binding site of a lymphomagenic retrovirus. J. Virol. 73:7599-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan, G. J. 1998. Emile Zuckerkandl, Linus Pauling, and the molecular evolutionary clock, 1959-1965. J. Hist. Biol. 31:155-178. [DOI] [PubMed] [Google Scholar]
- 25.Niebert, M., C. Rogel-Gaillard, P. Chardon, and R. R. Tönjes. 2002. Characterization of chromosomally assigned replication-competent gamma porcine endogenous retroviruses derived from a large white pig and expression in human cells. J. Virol. 76:2714-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niebert, M., and R. R. Tönjes. 2003. Analyses of prevalence and polymorphisms of six replication-competent and chromosomally assigned porcine endogenous retroviruses in individual pigs and pig subspecies. Virology 313:427-434. [DOI] [PubMed]
- 27.Nowak, R. M. 1991. Walker′s mammals of the world, 5th ed. John Hopkins University Press, Baltimore, Md.
- 28.Patience, C., M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patience, C., Y. Takeuchi, and R. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 30.Rogel-Gaillard, C., N. Bourgeaux, A. Billault, M. Vaiman, and P. Chardon. 1999. Construction of a swine BAC library: application to the characterization and mapping of porcine type C endoviral elements. Cytogenet. Cell. Genet. 85:205-211. [DOI] [PubMed] [Google Scholar]
- 31.Sachs, D. H., M. Sykes, S. C. Robson, and D. K. Cooper. 2001. Xenotransplantation. Adv. Immunol. 79:129-223. [DOI] [PubMed] [Google Scholar]
- 32.Scheef, G., N. Fischer, E. Flory, I. Schmitt, and R. R. Tönjes. 2002. Transcriptional regulation of porcine endogenous retroviruses released from porcine and infected human cells by heterotrimeric protein complex NF-Y and impact of immunosuppressive drugs. J. Virol. 76:12553-12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheef, G., N. Fischer, U. Krach, and R. R. Tönjes. 2001. The number of a U3 repeat box acting as an enhancer in long terminal repeats of polytropic replication-competent porcine endogenous retroviruses dynamically fluctuates during serial virus passages in human cells. J. Virol. 75:6933-6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steinhuber, S., M. Brack, G. Hunsmann, H. Schwelberger, M. P. Dierich, and W. Vogetseder. 1995. Distribution of human endogenous retrovirus HERV-K genomes in humans and different primates. Hum. Genet. 96:188-192. [DOI] [PubMed] [Google Scholar]
- 35.Stoye, J. P. 2001. Endogenous retroviruses: still active after all these years? Curr. Biol. 11:R914-R916. [DOI] [PubMed] [Google Scholar]
- 36.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 37.Thenius, E. 1990. Even-toed ungulates, p. 1-15. In S. P. Parker (ed.), Grzimek′s encyclopedia of mammals, vol. 5. McGraw-Hill, New York, N.Y.
- 38.Tucker, A., C. Belcher, B. Moloo, J. Bell, T. Mazzulli, A. Humar, A. Hughes, P. McArdle, and A. Talbot. 2002. The production of transgenic pigs for potential use in clinical xenotransplantation: microbiological evaluation. Xenotransplantation 9:191-202. [DOI] [PubMed] [Google Scholar]
- 39.Van Dooren, S., E. Gotuzzo, M. Salemi, D. Watts, E. Audenaert, S. Duwe, H. Ellerbrok, R. Grassmann, E. Hagelberg, J. Desmyter, and A. M. Vandamme. 1998. Evidence for a post-Columbian introduction of human T-cell lymphotropic virus [type I] [corrected] in Latin America. J. Gen. Virol. 79:2695-2708. [DOI] [PubMed] [Google Scholar]
- 40.Weiss, R. A., S. Magre, and Y. Takeuchi. 2000. Infection hazards of xenotransplantation. J. Infect. 40:21-25. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson, D. A., D. L. Mager, and J. A. Leong. 1994. Endogenous human retroviruses, p. 465-535. In J. A. Levy (ed.), The retroviridae, vol. 1. Plenum Press, New York, N.Y.
- 42.Wilson, C. A., S. Wong, J. Muller, C. E. Davidson, T. M. Rose, and P. Burd. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolgamot, G., and A. D. Miller. 1999. Replication of Mus dunni endogenous retrovirus depends on promoter activation followed by enhancer multimerization. J. Virol. 73:9803-9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]
- 45.Zuckerkandl, E. 1976. Evolutionary processes and evolutionary noise at the molecular level. II. A selectionist model for random fixations in proteins. J. Mol. Evol. 7:269-311. [DOI] [PubMed] [Google Scholar]
- 46.Zuckerkandl, E. 1987. On the molecular evolutionary clock. J. Mol. Evol. 26:34-46. [DOI] [PubMed] [Google Scholar]