Abstract
Microarrays of virus-specific oligonucleotides may provide a method of screening samples for the presence or absence of a large variety of viruses simultaneously. Influenza viruses are ideal for evaluating such microarrays because of their genetic and host diversity, and the availability of an extensive sequence database. A collection of 476 influenza virus-specific oligonucleotides was spotted onto glass slides as probes. Viral RNAs were reverse transcribed and amplified by PCR, and the products were labeled with cyanine dyes. The presence of viruses and their identities were determined by hybridization. The fluorescence intensities of oligonucleotide spots were highly reproducible within each slide and satisfactorily proportional between experiments. However, the intensities of probe spots completely complementary to target sequences varied from background to saturation. The variations did not correlate with base composition, nucleotide sequence, or internal secondary structures. Therefore, thresholds for determining whether hybridization to a spot should be judged as positive were assigned individually. Considering only positive spots from probes predicted to be monospecific for influenza virus species, subtype, host source, or gene segment, this method made correct identifications at the species, hemagglutinin subtype, and gene segment levels. Monospecific neuraminidase (NA) subtype probes were insufficiently diverse to allow confident NA subtype assignment. Incorporating positive spots from polyspecific probes into the identification scheme gave similar results. Overall, the results demonstrate the potential of microarray-based oligonucleotide hybridization for multiple virus detection.
A rapid and sensitive molecular detection and identification procedure for the presence of a wide variety of viruses is desired in a range of applications (4, 23), from diagnostics for clinical specimens through biodefense to environmental investigations. A DNA microarray of virus-specific oligo- or polynucleotides can, in theory, detect multiple viruses in a single hybridization. DNA microarrays have been widely used for the analysis of gene expression. Their potential as a generalized molecular diagnostic tool for specific pathogens (4) is being explored. Microarrays have been used to detect and identify soil, intestinal, and other bacterial populations (7, 14, 20, 24). Their use in molecular detection and identification of viruses has been limited to small numbers of probes (3, 8, 12, 13). Recently, the use of a large microarray of long oligonucleotides in the detection of respiratory viruses was reported (23). Since the specificity of oligonucleotide probes should increase with a decrease in length, we examined the use of short (ca. 21 nucleotides [nt]) oligonucleotides for the detection of, and discrimination among, influenza viruses.
Influenza viruses are ideal for evaluating such microarrays because of their genetic and host diversity and the availability of an extensive sequence database (16). The wide use of reverse transcription-PCR (RT-PCR) has resulted in the identification of a large number of influenza virus-specific oligonucleotides. The oligonucleotide sequences have been gathered in the VirOligo database (18), together with information about the oligonucleotides and their use in PCR or hybridization. Because of conserved oligonucleotide sequences at each of the 5′ and 3′ ends of all RNA segments (1, 17), the preparation of labeled cDNA targets from influenza viruses for microarray hybridization presents less of a challenge than from other viruses. The potential of microarray hybridization for influenza virus detection and identification has been discussed (6), and Li et al. (13) have shown the potential of this method for diagnosis of influenza. However, in the latter study, only 24 probes with an average length of 500 nt were used. A microarray with many more probes of shorter length should allow a better discrimination among closely related viruses. We report here the result of an evaluation of a microarray of more than 400 probes averaging 21 nt in length. Probes that were monospecific for influenza virus species, subtype, host source or segment made correct identifications at the species, hemagglutinin (HA) subtype, and segment levels. Overall, the results demonstrate the potential of microarray-based oligonucleotide hybridization for multiple virus detection.
MATERIALS AND METHODS
Oligonucleotide choice and microarray fabrication.
For fabrication of the microarray, influenza virus specific oligonucleotides were chosen from VirOligo (18), a web-interfaced relational database of published virus-specific oligonucleotides, created and maintained in our laboratory (http://viroligo.okstate.edu/). Omitting oligonucleotides of <17 and >29 nt and eliminating those that had a perfect match or a 1-nt mismatch with an organism other than influenza virus in BLASTn searches (2), we selected 463 oligonucleotides. A 32-nt equine influenza virus-specific oligonucleotide was also included. Fourteen oligonucleotide sequences specific for the HA segment of KY98 (see Table 1 for virus abbreviations) were obtained by using the OMIGA software (Accelrys, San Diego, Calif.) with default settings, except that the number of primer pairs to be generated was set at 10. The three homologous 20-nt HA-segment specific oligonucleotides S1, S2, and S3 for the viruses KY95, KY98, and MI63, respectively, were also synthesized. Oligonucleotide S0 was synthesized as the reverse of the HA-specific KY98 oligonucleotide sequence. It showed no similarity in BLASTn search to other sequences. A list of oligonucleotide identifiers and sequences is available as a table in supplemental material available online (referred to hereafter simply as the supplemental material [http://opbs.okstate.edu/∼melcher/ViSH/home.html]).
TABLE 1.
Influenza viruses used in this study
| Virus | Abbre- viation | Segment no. | GenBank or Influenza Sequence Database accession no. |
|---|---|---|---|
| A/Equine/Kentucky/1/98 (H3N8)a | KY98 | 4 | AF197241 |
| A/Equine/Miami/63 (H3N8)a | MI63 | 4 | M29257 |
| 5 | M22575 | ||
| 6 | L06580 | ||
| 7 | AF001674 | ||
| A/Panama/2007/99 (H3N2)b | Panama 99 | 4 | ISDNCDA001 |
| A/Aichi/2/68 (H3N2)c | HK68 | 1 | AF348170 |
| 2 | AF348172 | ||
| 3 | AF348170 | ||
| 4 | AF348176, J02090 | ||
| 5 | X15890 | ||
| 6 | U42630 | ||
| 7 | AF348198, M63515 | ||
| 8 | AF348198 | ||
| A/PR/8/34 (H1N1)c | PR8 | 1 | ISDN13419 |
| 2 | ISDN13420 | ||
| 3 | ISDN13421 | ||
| 4 | ISDN13422 | ||
| 5 | ISDN13423 | ||
| 6 | ISDN13424 | ||
| 7 | ISDN13425 | ||
| 8 | ISDN13426 | ||
| A/Equine/Kentucky/9/95 (H3N8)a | KY95 | 4 | AF197247 |
Obtained from T. Chambers.
Obtained from N. Cox.
Obtained from E. Kilbourne.
The oligonucleotides were synthesized with a 5′ C6 amino linker by the Laboratory for Microbial Genomics of the University of Oklahoma Health Sciences Center (Oklahoma City). To validate the synthesis, five pairs of oligonucleotides were tested by RT-PCR and found to yield PCR products of the expected size. In addition, randomly selected oligonucleotides were characterized by matrix-assisted laser desorption-ionization mass spectrometry at the OSU Recombinant DNA/Protein Resource Facility. These oligonucleotides had the predicted molecular masses.
The synthesized oligonucleotide set was printed in quadruplicate as four side-by-side columns of 12×41 spots each on aldehyde-derivatized glass slides (CEL Associates, Inc.), without prior treatment of the slides, at a concentration of 7.5 g liter−1 in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Quill-type pins in a PixSys 5500 arrayer (Cartesian Technologies, Ann Arbor, Mich.) deposited approximately 1 to 2 nl in spots of 100 to 150 μm diameter over 20 h at room temperature in a chamber controlled to have 65% humidity. The slides were dried overnight in a covered box, processed (i.e., washed with 0.2% sodium dodecyl sulfate [SDS] for 1 min and then with water twice for 1 min each time, followed by treatment for 5 min with 0.1 M NaBH4 in 0.75× phosphate-buffered saline-25% ethanol and subsequent washing in 0.2% SDS for 1 min and in water for 1 min), and stored in a dark, humidity-free environment.
Target preparation.
Virus suspensions in phosphate-buffered saline with antibiotics were inoculated into 9- to 11-day-old fertilized chicken eggs. Allantoic fluid was harvested after 3 days at 37°C and clarified by centrifugation at 700 × g for 20 min. RNA isolation and reverse transcription was done according to the method of Lai and Chambers (11). PCR was carried out (94°C for 2 min and 25 cycles of 94°C for 1 min, 40°C for 2 min, and 72°C for 3 min, followed by a final 72°C for 10 min) in a 50-μl volume (3.5 U of Taq Long Plus [Stratagene], 0.16 mM amino allyl modified deoxynucleoside triphosphates [Sigma], 1× thermophilic DNA polymerase buffer [Promega], 2 mM MgCl2, and 4 μg of uni3 and uni5 universal primers ml−1 [see reference 17]). We also amplified a 1-kbp fragment of the HA segment of KY98 by using EH3-1 (11) and EH3-1061 (5′-TCTGATTTGCTTTTCTGGTA-3′) primers. PCR products were sonicated to yield DNA fragments of ca. 200 bp. Unincorporated amino allyl-deoxynucleoside triphosphates were removed by using Microcon YM-30 (Millipore Corp., Bedford, Mass.) spin columns, and the PCR products were conjugated to Cy5 or Cy3 cyanine dyes in 10 μl of 45 mM Na2CO3 buffer. The unreacted dyes were quenched by incubation for 15 min in the dark with 4.5 μl of 4 M hydroxylamine. The conjugated PCR products were recovered by using a QIAquick PCR Clean-Up kit (Qiagen, Valencia, Calif.).
Hybridization.
The labeled target was concentrated in a Speed Vac, resuspended in 2 μl of water, and denatured for 5 min at 100°C. It was snap-cooled on ice and mixed with 8 μl of preheated (65°C for 3 min) Unihyb hybridization buffer (Telechem International, Sunnyvale, Calif.). The 10-μl total volume was hybridized to the microarray under 22-by-22 mm coverslips at 22°C (or higher in selected experiments) for 1 h, followed by washing of slides (once in 2% SDS-2× SSC and once in 1× SSC) according to the manufacturer's instructions. Scanning was done (Packard BioScience Scanarray 3000; Perkin-Elmer, Boston, Mass.) at 100% laser and 95% photomultiplier tube (PMT) settings.
Data analysis.
The files of scanned intensities were transferred to GenePix Pro 4.0 (Axon Instruments, Union City, Calif.) for subsequent analysis. Pixel intensities were processed by GenePix to calculate the median pixel intensity of each spot and to subtract the median local background intensity (averaging 6% of saturation, ranging from 1 to 15%, as estimated by using Imagene software [Biodiscovery, Inc., Marina del Rey, Calif.]) from it. Outlier corrected median intensity values were identified as those whose difference from the mean of the middle pair of four values was more than 10 times the difference of the opposite extreme value from the mean. When outliers occurred, the remaining three corrected median intensity values were averaged by using Excel (Microsoft, Seattle, Wash.). The average values for each experiment are available in the supplemental material as item 2.
Normalization of pixel intensities.
Variations in experimental conditions from experiment to experiment could produce different mean median intensities for the same probe. To compensate for such variation, a correction for inequalities in the mean of median pixel intensities between experiments was applied. Seven experiments were selected as a training set (one experiment each for PR8, Panama 99, MI63, and KY98 and three experiments for HK68). Five oligonucleotide probes (oligonucleotides 127, 128, 131, 132, and 142; see supplemental material, item 1, for sequences) that resulted in high median pixel intensities with each of the training set were identified. The average over the seven experiments and the five probes of the mean median pixel intensities was designated Hstd. Similarly, Lstd was calculated as the average of the mean median pixel intensities for five probes (75, 77, 84, 88, and 93) that resulted in low median pixel intensities for every member of the training set. For each nontraining set target, we calculated the averages of the mean median pixel intensities for the two sets of five probes and designated them Hi and Li. A slope correction factor, m, was calculated as follows: m = (Hstd − Lstd)/(Hi − Li). An intercept correction factor, b, was calculated as b = Lstd − mLi. Finally, each median pixel intensity, Iraw, was adjusted by using the equation Inorm = b + mIraw to determine the normalized median pixel intensity. An Excel workbook that carries out these calculations is available as item 3 in the supplemental material.
Establishment of threshold intensities.
To analyze the results, we needed to determine whether a particular median pixel intensity value was indicative of hybridization. For this purpose, we defined threshold intensities as the values for normalized median pixel intensities above which hybridization was judged to be significant. First, we established sequenced-segment profiles by searching The Influenza Sequence Database (16) for each virus of the training set to determine which of its RNA segments had been sequenced. For this purpose, sequences of the A/Hong Kong/1/68 virus, derived from the A/Aichi/2/68 virus, were used for HK68 since the sequences of more segments were available for this derived virus. Then, for each probe, the BLASTn result table and the sequenced-segment profiles were used to classify the training set viruses into three groups: (i) those for which the probe target had been sequenced and the probe sequence was present; (ii) those for which the probe target had not been sequenced; and (iii) those for which the probe target had been sequenced but the probe sequence was absent. Two types of thresholds were calculated. The exclusive threshold was calculated first. The threshold was set to 110% of the maximum among the normalized median pixel intensities viruses in group 3. The inclusive threshold was set at 90% of the minimum of the normalized median pixel intensity values for group 1 viruses. The Excel spreadsheet used for these calculations is available in the supplemental material as item 3. A stringent threshold was defined as the lower of the exclude and include thresholds, whereas a relaxed threshold was defined as the higher threshold.
Oligonucleotide characterization by database search.
The results of BLASTn searches (expect value [E-value] of 100) of The Influenza Sequence Database (http://www.flu.lanl.gov) with each arrayed oligonucleotide were parsed to determine the type, source species, segment, and subtype of the retrieved sequences (these are available as supplemental material item 4). The numbers of retrieved sequences of each type (A, B, or C), each host species (human, swine, avian, equine, or other), each gene segment, each HA subtype, and each neuraminidase (NA) subtype were counted for each oligonucleotide by using Excel. The counts were entered in a BLASTn result table of oligonucleotides (available as supplemental material items 6 and 7).
Signature oligonucleotide analysis.
The BLASTn result table was used to establish criterion tables. In these tables, one for each category of characteristics (type, source species, segment, HA subtype, and NA subtype), we identified signature oligonucleotides. When all hits within a category were for one characteristic, the oligonucleotide was judged positively diagnostic for that characteristic and called a signature oligonucleotide. When hits within a category occurred at more than one characteristic, the oligonucleotide was judged as nondiagnostic for that category. For each experiment, the number of signature oligonucleotides with normalized median pixel intensity values above threshold were totaled for each characteristic. Calculations can be accessed as supplemental material item 6.
Weighted analysis.
The results of hybridization to many oligonucleotides were ignored in the signature oligonucleotide analysis because many oligonucleotides recognized, in BLASTn searches, sequences belonging to more than one characteristic within a category. To utilize the information also in these nondiagnostic oligonucleotides, a weighting approach was used. For each characteristic, the number of BLASTn hits generated by all arrayed oligonucleotides was summed. The fraction of these that were generated by oligonucleotides whose normalized median pixel intensities were above threshold was designated the weighted score. The weighted scores for all characteristics in a category were totaled, and the score for each characteristic was expressed as a percentage of that total.
RESULTS
Influenza cDNA target preparation.
Use of universal primers in RT-PCR of extracted influenza virus RNA resulted in the amplification of all eight genome segments, as revealed by agarose gel electrophoresis. There were no major differences between experiments in the relative intensities of each of the bands corresponding to the RNA segments. In initial experiments, labeled HA segment-specific PCR products were used directly in hybridization. Only oligonucleotides that matched perfectly produced a detectable signal, and the signal was faint. When hybridization was performed with sheared labeled PCR products, the intensity of the perfect match spots increased approximately threefold. The S0 control spot (no matches) continued to show background levels of fluorescence. Thus, the signal-to-noise ratio was improved, and all further experiments were done with sheared PCR products.
Reproducibility.
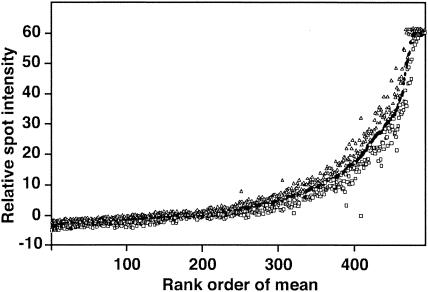
Labeled products for KY98, Panama 99, MI63, and HK68 were hybridized to the microarray, and the spot intensities were evaluated. As a control, RNA from chicken red blood cells (RBC) not exposed to virus was RT-PCR amplified, and the products were labeled. Only 44 spots reacted. For virus templates, 186 or more strongly fluorescent spots were observed. Visual inspection of pixel intensities generated from labeled influenza virus cDNAs revealed a general high reproducibility of the intensities for the quadruplicated spots of each oligonucleotide. To further examine the reproducibility within a microarray, the maximum, minimum, and mean intensities were plotted in rank order of the mean intensities (see example in Fig. 1). The plots confirmed an excellent reproducibility of spot intensity over the quadruplicate spots but also revealed occasional oligonucleotides with outlier intensities. As described in Materials and Methods, when the intensity was substantially different from the other three, the outlier was deleted from further analysis. A general spot-to-spot reproducibility within an array suggests that variables influencing the performance of individual microarray spots did not significantly affect the results.
FIG. 1.
Within-slide reproducibility of median pixel intensities. Influenza virus oligonucleotide microarray was hybridized with KY98 cDNA. Minimum (squares) and maximum (triangles) median intensities for quadruplicate spots plotted against the rank order of the average of the median intensities (line) for the four spots.
Normalization.
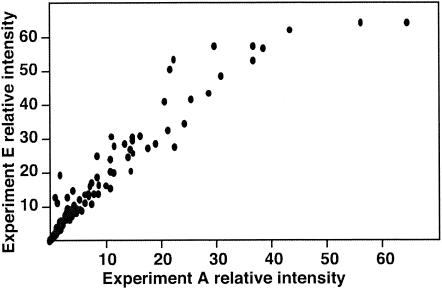
Comparisons of plots of rank-ordered pixel intensities, such as that of Fig. 1, from 30 different experiments revealed substantial between-microarray variation in the intensities of individual oligonucleotides. In some hybridization experiments, a plateau was reached at the high end of the distributions for a considerable number of oligonucleotides, because the detector had been saturated. In others, very few oligonucleotides had reached saturation of detection. Six hybridization experiments were performed with cDNA prepared from the RNA of the same virus, HK68. Two of these exhibited similar degrees of saturation. A plot of the intensity of oligonucleotides in one experiment against the intensity of the second experiment (Fig. 2) showed agreement between the two determinations of oligonucleotide reactivity. Scattering of the points beyond that seen in Fig. 1 was observed and might be due to variables occurring during preparation of the labeled targets, including the relative efficiency of cDNA amplification of the different gene segments, the effectiveness of shearing, the amount of cDNA available for hybridization, and the efficiency of the dye labeling reaction. As expected, comparisons of dissimilar samples had a much greater scatter of points (data not shown).
FIG. 2.
Experiment-to-experiment reproducibility of pixel intensities. Relative intensities (i.e., normalized median pixel intensity [10−3]) from influenza virus oligonucleotide microarray hybridized with HK68 target cDNA in one experiment (experiment A, Table 3) were plotted against the intensities obtained for the same spot with another target cDNA preparation made from the same virus (experiment E, Table 3).
The reproducibility between experiments (Fig. 2) suggested that a normalization procedure could allow a direct comparison from one experiment to another. The microarray contained 40 oligonucleotides that include the universal influenza virus 3′- or 5′-terminal sequence. These spots thus serve as positive controls for the success of the hybridization experiment. Examination of the rank order of the intensities indicated that these probes reacted similarly with the cDNA of every virus in the training set. Thus, normalization was done by using the results of hybridization of the test set to 10 oligonucleotides: five giving high-intensity results, but still in the responsive range of the detector (oligonucleotides 127, 128, 131, 132, and 142), and five whose hybridization was substantially less (oligonucleotides 75, 77, 84, 88, and 93). Normalization with these values was performed as described in Materials and Methods.
Spot intensity variation.
Figure 1 illustrates that oligonucleotide pixel intensities were continuously distributed from background to saturation values. Such distributions were seen with all targets used in these experiments. The lack of definite boundaries precluded any attempt to designate a cutoff value between significant hybridization and nonspecific background binding. As a possible control for background, hybridization to a labeled target cDNA derived from RBC RNA was used. Although a high percentage of oligonucleotides showed low levels of fluorescence, 22 displayed levels comparable to those obtained with the training set. Inspection of the sequences of the probes revealed that highly reactive oligonucleotides contain oligodeoxyribosylthymidylate sequences, allowing them to bind to double-stranded poly(A) tail-derived cDNA sequences. Subtraction of normalized RBC pixel intensities from the normalized intensities for samples was not done because the oligonucleotides used for normalization were not the ones that reacted with the RBC cDNA.
A possible explanation of the wide variation in spot intensities is that they may have different degrees of complementarity to the target. To address this possibility, a series of three probes (S1, S2, and S3; see supplemental material, item 1), for which the second differed from the first at a single base and the third differed at three positions, were compared qualitatively by hybridization to the target prepared from the HA segment of KY98. As expected, the signal of the perfect match was more intense than that of the one that had one mismatch. Hybridization to the multiple mismatch spot was barely detectable.
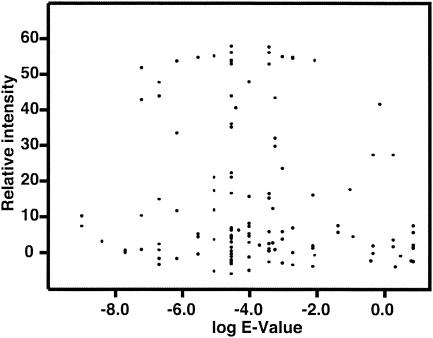
Nevertheless, the probe-dependent variation in normalized pixels did not reflect the degree of complementarity of the probe with the target. The microarray included 128 oligonucleotides that detected PR8 sequences in the Influenza Virus Database by using BLASTn searching with an E-value cutoff of 100. The normalized intensity values for PR8 hybridization to these oligonucleotides were plotted against the logarithm of the E-values of the BLASTn search as a measure of their complementarity (Fig. 3). No correlation was apparent. Indeed, oligonucleotides with the lowest and the highest E-values were among those that barely showed positive pixel intensities. Of the 42 oligonucleotides that gave normalized pixel intensities of <2,000 27 had E-values of <0.001, and E-values of >0.001 were scored by 4 of 25 oligonucleotides that produced normalized pixel intensities of >40,000.
FIG. 3.
Independence of oligonucleotide normalized median pixel intensities from complementarity to template as measured by BLASTn E-values. Influenza virus oligonucleotide microarray was hybridized with labeled cDNA derived from PR8. Logarithms of E-values were plotted against relative intensity (i.e., normalized median pixel intensity [10−3]).
Because several laboratories have designed similar PCR primers, VirOligo, and consequently the microarray, contained 32 oligonucleotides that overlapped one another in 15 sets of sequences (Table 2). In addition, a series of six overlapping oligonucleotides complementary to the HA segment of KY98 had been designed by the OMIGA primer design program. The variation in pixel intensities seen with VirOligo-derived oligonucleotides was also noted with this set of oligonucleotides. In four sets the normalized median intensities of all probes were <10% of saturation, and in two others the values were within 50% of one another. However, dramatic differences did occur. In one case, a mismatch at the 3′ terminus resulted in a >2-fold reduction in intensity. In several pairs, additional nucleotides at the 5′ end resulted in dramatic increases in hybridization, whereas similar additions at the 3′ end had little or no effect. However, not all 5′ additions had this effect. Some of the high-intensity producing oligonucleotides had features that violated rules previously proposed for the design of effective oligonucleotides for hybridization (15), such as that the oligonucleotides should have fewer than 10 A's or 10 T's and should contain fewer than 6 G's or 6 C's.
TABLE 2.
Variation in normalized pixel intensities for similar oligonucleotidesa
| Segment | Sequence | E-value | Pixel intensity |
|---|---|---|---|
| 1 | AATCTAATGTCGCAGTCTC | −4.0 | 0 |
| 1 | CTGATGTCGCAGTCTCGCAC | −2.7 | 7,283 |
| 1 | CTGTCAGTAAGTATGCTAGAGTCCC | −7.2 | 43,161 |
| 1 | GGCTGTCAGTAAGTATGCTA | −4.5 | 7,289 |
| 2 | TTGAATGGATGTCAATCCGA | −4.5 | 0 |
| 2 | GAATGGATGTCAATCCGACC | −4.5 | 0 |
| 4 | TTTCTAATATCCACAAAATGAAGGC | 0.5 | 0 |
| 4 | ACCAAAATGAAGGCAAACC | −4.0 | 7,769 |
| 4 | CAGATGCAGACACAATATGT | −4.5 | 3,314 |
| 4 | CAGATGCAGACACAATATGTATAGG | −7.2 | 1,341 |
| 4 | AAACCGGCAATGGCTCCAAA | −4.5 | 4,842 |
| 4 | ACCGGCAATGGCTCCAAA | −3.4 | 1,581 |
| 4 | GATGCAGACACAATATGTATAGG | −6.2 | 0 |
| 4 | CAGATGCAGACACAATATGT | −4.5 | 3,314 |
| 4 | CAGATGCAGACACAATATGTATAGG | −7.2 | 1,341 |
| 4 | AAAGCAGGGGAAAATAAAAACAACC | −7.2 | 52,256 |
| 4 | GCAGGGGAAAATAAAAACAACC | −5.5 | 52 |
| 4 | GCAGGGGAAAATAAAAGCCAC | −2.0 | 0 |
| 5 | TGCGGGGAAGGATCCTAAGAAAAC | −4.3 | 6,709 |
| 5 | GGGAAAGATCCTAAGAAAAC | −4.5 | 884 |
| 5 | TCCTCTGCATTGTCTCCG | −3.4 | 58,051 |
| 5 | CTCTGCATTGTCTCCGAAG | −4.0 | 48,211 |
| 7 | GTCAGCATCCACAGCACTCTGCTGTTCC | −9.0 | 10,645 |
| 7 | TCGTCAGCATCCACAGCA | −3.4 | 56,454 |
| 7 | GACCAGCACTGGAGCTAGGA | −4.5 | 36,488 |
| 7 | GACCAGCACTGGAGCTAGGG | −4.0 | 15,976 |
| 7 | GGCAAGTGCACCAGCAGAATAAC | −6.2 | 33,891 |
| 7 | GGCAAGTGCACCAGCAGAATAACT | −6.7 | 48,119 |
| 7 | AGCGTAGACGCTTTGTC | −3.0 | 0 |
| 7 | CTGCAGCGTAGACGCTTTGTCCAAAATG | −9.0 | 7,788 |
| 8 | CCCATTCTCATTACTGCTTC | −4.5 | 54,028 |
| 8 | ATAATGTTTTTCTCATTACT | 0.9 | 1,731 |
| KY | ATTGACCCCTAACCCACG | 30,727 | |
| KY | CTGCCTGATTGACCCCTAACC | 60,471 | |
| KY | TATCCTGCCTGATTGACC | 18,603 | |
| KY | TTATCCTGCCTGATTGACC | 4,997 | |
| KY | CTTATCCTGCCTGATTGACC | 24,744 | |
| KY | TGCTTATCCTGCCTGATTGACC | 9,453 |
Values reported with segment numbers are from hybridization of the influenza virus oligonucleotide microarray with PR8 cDNA target, whereas those identified as “KY” are from hybridization of the microarray with KY98 cDNA target. Values are normalized mean net median pixel intensities for quadruplicate spots. Negative values (due to spot intensities being insignificantly less that background intensities) are reported as zeros.
Besides using targets amplified by universal primers, we amplified a specific region of the HA gene from KY98 and used the product as a target for hybridization with the microarray. As expected, the number of spots showing hybridization was dramatically less as only one viral segment was used. Spot intensities observed with this target for software-designed oligonucleotides were proportional to those observed with a target derived from universal amplification of all segments. This suggested that the labeled cDNAs from other gene segments did not enhance or interfere with the hybridization of HA-specific oligonucleotides.
Analysis of signature oligonucleotides.
We used the oligonucleotide sequences to search the Influenza Virus Database (16) using BLASTn with an E-value cutoff of 100. This approach identified target sequences of a length expected to contribute to hybridization. The BLASTn result table was processed to identify, for each hit, the oligonucleotide, and the following virus characteristics: the virus type, the source host, the RNA segment and, if applicable, its HA and NA subtypes.
For the purposes of discussion, “characteristics” refers here to the identifications within a category (type A, type B, or type C; human, swine, avian, equine, or other; etc.). Signature oligonucleotides were defined as the oligonucleotides that recognized only one of the possible characteristics for a category. There were signature oligonucleotides for each viral species (Table 3; 274, 32, and 1 for types A, B, and C, respectively). However, for the viral host, signature oligonucleotides were only available for avian, equine, and human hosts (Table 4; 2, 3, and 31, respectively). For any specific gene segment, there were 4 to 122 oligonucleotides available as specific oligonucleotides (Table 5). HA subtypes 2, 6, 8, and 10 through 15, since they are not well characterized, had no signature oligonucleotides (Table 6). Of the nine NA subtypes (Table 7), only subtypes 1 and 2 had signature oligonucleotides (3 and 17 oligonucleotides, respectively). Subtypes H1 and H3 had the most signature oligonucleotides (23 and 53 oligonucleotides, respectively), since they have been well studied. One oligonucleotide each for H4, H7, and H9 (oligonucleotides 607, 285, and 105, respectively) and two for H5 (oligonucleotides 61 and 489) were signature oligonucleotides.
TABLE 3.
Identification of influenza virus type by microarray hybridization
| Virus (expt)a | Scoreb
|
||
|---|---|---|---|
| Type A | Type B | Type C | |
| KY98* | 80.3 (55) | 14.4 (0) | 5.3 (0) |
| MI63* | 83.2 (69) | 12.0 (0) | 4.7 (0) |
| Panama 99* | 80.8 (88) | 13.6 (0) | 5.5 (0) |
| PR8* | 86.1 (54) | 8.5 (0) | 5.3 (0) |
| HK68 (A)* | 89.6 (70) | 7.8 (0) | 2.6 (0) |
| HK68 (B)* | 82.5 (70) | 11.4 (0) | 6.1 (0) |
| HK68 (C)* | 80.8 (79) | 11.7 (0) | 7.5 (0) |
| HK68 (D) | 75.8 (47) | 20.8 (1) | 3.4 (0) |
| HK68 (E) | 89.4 (63) | 7.9 (0) | 2.7 (0) |
| HK68 (F) | 91.5 (60) | 8.5 (0) | 0.0 (0) |
| Available signature oligonucleotides | (274) | (32) | (1) |
Hybridization with HK68 cDNA was performed in six experiments: experiments B and C were done at 42 and 32°C, respectively, while all others were done at 22°C. A high concentration of cDNA was used for experiment D. F was scanned at lower laser (95%) and PMT (90%) settings; all others were at 100 and 95% laser and PMT settings, respectively. *, Experiments included in training set.
The scores are from the weighted oligonucleotide calculation (see Materials and Methods). Values in parentheses are the numbers of signature oligonucleotides that registered positive.
TABLE 4.
Identification of influenza virus source host by microarray hybridization
| Virus (expt)a | Scoreb
|
||||
|---|---|---|---|---|---|
| Human | Swine | Avian | Equine | Other | |
| KY98* | 40.1 (1) | 13.9 | 26.0 (0) | 8.1 (1) | 12.0 |
| MI63* | 42.0 (1) | 16.8 | 23.3 (0) | 9.0 (2) | 8.9 |
| Panama 99* | 46.0 (1) | 15.6 | 23.5 (0) | 5.3 (0) | 9.6 |
| PR8* | 39.7 (0) | 14.0 | 28.7 (0) | 3.7 (0) | 13.9 |
| HK68 (A)* | 40.5 (1) | 15.9 | 26.1 (0) | 6.1 (0) | 11.3 |
| HK68 (B)* | 40.7 (0) | 13.9 | 26.7 (0) | 7.0 (0) | 11.6 |
| HK68 (C)* | 41.1 (0) | 14.4 | 25.8 (0) | 6.7 (0) | 12.0 |
| HK68 (D) | 46.3 (2) | 12.0 | 23.2 (0) | 5.8 (0) | 12.7 |
| HK68 (E) | 40.1 (1) | 14.4 | 25.6 (0) | 6.4 (0) | 13.5 |
| HK68 (F) | 41.8 (1) | 14.6 | 25.9 (0) | 5.9 (0) | 11.8 |
| Available signature oligonucleotides | (31) | (0) | (2) | (3) | (0) |
See Table 3 for the conditions of experiments A to E. *, Experiments included in training set.
The scores are from the weighted oligonucleotide calculation (see Materials and Methods). Values in parentheses are the numbers of signature oligonucleotides that registered positive.
TABLE 5.
Identification of influenza virus segments by microarray hybridization
| Virus (expt)a | Scoreb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Segment 1 | Segment 2 | Segment 3 | Segment 4 | Segment 5 | Segment 6 | Segment 7 | Segment 8 | |
| KY98* | 1.6 (2) | 6.9 (1) | 3.8 (1) | 26.1 (5) | 13.4 (12) | 17.3 (5) | 16.4 (12) | 14.5 (9) |
| MI63* | 2.4 (4) | 5.4 (3) | 6.1 (1) | 31.7 (6) | 9.7 (9) | 12.8 (6) | 17.2 (21) | 14.8 (9) |
| Panama 99* | 2.1 (5) | 2.9 (2) | 6.0 (3) | 27.0 (7) | 9.3 (12) | 16.4 (11) | 22.9 (29) | 13.4 (13) |
| PR8* | 1.6 (2) | 6.2 (2) | 1.6 (0) | 27.0 (4) | 13.2 (10) | 7.3 (1) | 29.1 (22) | 13.9 (8) |
| HK68 (A)* | 2.7 (2) | 4.2 (2) | 4.3 (2) | 21.5 (6) | 11.6 (13) | 14.4 (4) | 27.6 (24) | 13.6 (11) |
| HK68 (B)* | 2.7 (3) | 1.9 (3) | 8.0 (2) | 25.1 (1) | 12.1 (12) | 10.0 (7) | 26.7 (22) | 13.4 (12) |
| HK68 (C)* | 3.2 (3) | 1.7 (3) | 6.0 (2) | 26.7 (1) | 10.4 (13) | 9.9 (7) | 27.2 (29) | 15.0 (14) |
| HK68 (D) | 0.8 (1) | 1.9 (0) | 7.3 (2) | 28.2 (2) | 11.5 (10) | 18.7 (6) | 15.5 (8) | 16.1 (9) |
| HK68 (E) | 3.8 (2) | 4.5 (1) | 3.9 (2) | 23.5 (7) | 10.4 (12) | 11.5 (3) | 27.4 (20) | 15.1 (11) |
| HK68 (F) | 3.2 (3) | 3.6 (3) | 2.8 (1) | 28.4 (3) | 10.4 (12) | 12.8 (4) | 29.2 (19) | 9.7 (8) |
| Available signature oligonucleotides | (7) | (4) | (5) | (122) | (25) | (22) | (41) | (20) |
See Table 3 for the conditions of experiments A to E. *, Experiments included in training set.
The scores given are from the weighted oligonucleotide calculation (see Materials and Methods). Values in parentheses are the numbers of signature oligonucleotides that registered positive.
TABLE 6.
Identification of influenza virus HA subtype by microarray hybridization
| Virus (expt)a | Scoreb (by HA subtype)
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subtype 1 | Subtype 2 | Subtype 3 | Subtype 4 | Subtype 5 | Subtype 6 | Subtype 7 | Subtype 8 | Subtype 9 | Subtype 10 | Subtype 11 | Subtype 12 | Subtype 13 | Subtype 14 | Subtype 15 | |
| KY98* | 5.5 (0) | 0.8 | 40.8 (2) | 5.5 (0) | 10.7 (0) | 6.5 | 5.6 (0) | 0.0 | 4.4 (0) | 0.0 | 7.4 | 0.0 | 12.9 | 0.0 | 0.0 |
| MI63* | 8.8 (0) | 1.1 | 36.0 (6) | 3.3 (0) | 7.0 (0) | 3.3 | 10.3 (0) | 5.3 | 3.1 (0) | 3.3 | 3.8 | 0.0 | 8.0 | 0.0 | 6.6 |
| Panama 99* | 5.1 (0) | 0.9 | 48.4 (7) | 5.6 (0) | 4.3 (0) | 4.4 | 2.7 (0) | 0.0 | 2.7 (0) | 0.0 | 13.7 | 0.0 | 12.2 | 0.0 | 0.0 |
| PR8* | 69.6 (2) | 0.0 | 1.2 (0) | 0.0 (0) | 8.2 (0) | 0.0 | 0.0 (0) | 17.8 | 0.0 (0) | 0.0 | 3.2 | 0.0 | 0.0 | 0.0 | 0.0 |
| HK68 (A)* | 4.6 (0) | 0.8 | 44.0 (6) | 5.7 (0) | 4.6 (0) | 4.0 | 2.4 (0) | 0.0 | 2.4 (0) | 7.9 | 12.5 | 0.0 | 11.1 | 0.0 | 0.0 |
| HK68 (B)* | 8.6 (0) | 1.4 | 25.8 (1) | 7.6 (0) | 7.6 (0) | 7.1 | 4.3 (0) | 0.0 | 3.9 (0) | 0.0 | 12.2 | 0.0 | 21.4 | 0.0 | 0.0 |
| HK68 (C)* | 8.0 (0) | 1.5 | 19.2 (1) | 8.8 (0) | 6.7 (0) | 6.8 | 4.2 (0) | 0.0 | 4.2 (0) | 0.0 | 21.5 | 0.0 | 19.2 | 0.0 | 0.0 |
| HK68 (D) | 6.1 (0) | 1.0 | 11.8 (0) | 7.0 (0) | 5.3 (0) | 16.4 | 5.5 (0) | 0.0 | 3.2 (0) | 4.9 | 21.1 | 0.0 | 17.7 | 0.0 | 0.0 |
| HK68 (E) | 10.0 (1) | 0.8 | 41.5 (6) | 5.3 (0) | 4.4 (0) | 3.7 | 2.3 (0) | 0.0 | 2.3 (0) | 7.5 | 11.8 | 0.0 | 10.5 | 0.0 | 0.0 |
| HK68 (F) | 6.6 (0) | 1.1 | 32.2 (1) | 7.7 (0) | 3.4 (0) | 2.2 | 5.7 (0) | 0.0 | 2.1 (0) | 20.2 | 13.4 | 0.0 | 5.4 | 0.0 | 0.0 |
| Available signature oligonucleotides | (23) | (53) | (1) | (2) | (1) | (1) | |||||||||
See Table 3 for conditions of experiments A to E. *, Experiments included in training set.
Scores given are from the weighted oligonucleotide calculation (see Materials and Methods). Values in parentheses are the numbers of signature oligonucleotides that registered positive.
TABLE 7.
Identification of influenza virus NA subtype by microarray hybridization
| Virus (expt)a | Scoreb (by NA subtype)
|
|||||
|---|---|---|---|---|---|---|
| Subtype 1 | Subtype 2 | Subtype 3 | Subtype 5 | Subtype 6 | Subtype 7 | |
| KY98* | 0.0 (0) | 100 (5) | 0.0 | 0.0 | 0.0 | 0.0 |
| MI63* | 2.9 (0) | 72.5 (6) | 0.0 | 0.0 | 12.3 | 12.3 |
| Panama 99* | 4.3 (0) | 86.9 (10) | 0.0 | 0.0 | 8.8 | 0.0 |
| PR8* | 97.2 (1) | 2.8 (0) | 0.0 | 0.0 | 0.0 | 0.0 |
| HK68 (A)* | 0.0 (0) | 100 (4) | 0.0 | 0.0 | 0.0 | 0.0 |
| HK68 (B)* | 0.0 (0) | 100 (7) | 0.0 | 0.0 | 0.0 | 0.0 |
| HK68 (C)* | 0.0 (0) | 100 (7) | 0.0 | 0.0 | 0.0 | 0.0 |
| HK68 (D) | 16.9 (1) | 83.1 (5) | 0.0 | 0.0 | 0.0 | 0.0 |
| HK68 (E) | 0.0 (0) | 100 (3) | 0.0 | 0.0 | 0.0 | 0.0 |
| HK68 (F) | 0.0 (0) | 100 (4) | 0.0 | 0.0 | 0.0 | 0.0 |
| Available signature oligonucleotides | (3) | (17) | (0) | (0) | (0) | (0) |
See Table 3 for the conditions of experiments A to E. *, Experiments included in training set.
The scores given are from the weighted oligonucleotide calculation (see Materials and Methods). Values in parentheses are the numbers of signature oligonucleotides that registered positive. There were no NA-specific oligonucleotides that reacted with N4, N8, or N9 subtypes in BLASTn searches.
In all experiments, at least 47 of 247 signature oligonucleotides correctly identified the viruses tested as type A (Table 3). Oligonucleotide 29, in one of six experiments, misidentified HK68 as type B. For virus host, as expected from the ecology of influenza viruses, identification was more problematic. None of 31 signature oligonucleotides identified PR8 as a human virus, and only four of six experiments correctly identified HK68 as a human virus (Table 4). Although the two equine viruses had signature oligonucleotides to identify them as equine viruses, there was also a positive reaction with human virus-specific oligonucleotide 180. For all except one experiment, the presence of all eight RNA segments could be verified by at least one segment-specific signature oligonucleotide (Table 5). The exception was for PR8 segment 3. In all except one of six experiments with HK68 did signature oligonucleotides identify the correct HA or NA subtype (Table 6). In that experiment, none of the signature oligonucleotides for any subtype reacted positively. In another experiment, oligonucleotide 29 misidentified the HK68 subtype as H1, whereas the six other signature oligonucleotides correctly identified it as H3. Identification of NA subtype was less satisfactory. The correct N2 subtype was identified for viruses Panama 99 (six oligonucleotides) and HK68 (three to five oligonucleotides). Similarly, the N1 subtype was identified for PR8. In one of the HK68 experiments, however, although five signature oligonucleotides made the correct identification, one misidentified it as the N1 subtype. The equine influenza viruses KY98 and MI63 (H3N8) reacted incorrectly positively with five and six, respectively, signature oligonucleotides for the N2 subtype.
Analysis by oligonucleotide weighting.
The identification capacity of signature oligonucleotides was limited by the sparsity of signature oligonucleotides in some categories, particularly the HA and NA subtypes. However, several probes were predicted to react with targets of the missing subtypes, as well as with those of other subtypes. We therefore considered a scoring system based not on an “all-or-none” criterion (e.g., signature oligonucleotides) but on the proportion of available BLASTn hits indicated as consonant with each characteristic. This approach might provide further useful information. The weighting system is described in Materials and Methods, and the results are displayed in Tables 3 through 7. For identification of species, non-influenza A viruses scored less than 25% of the sum of the weighted scores (Table 3). Equine influenza viruses received 8.1 and 9.0% of the sum of weighted scores for the source host (Table 4), whereas the percentages for human influenza viruses ranged from 3.7 to 7.0, with a mean of 5.9 ± 1.1. The values for segments varied considerably (Table 5), even among the six experiments with the same virus. This variation may be due to subtle variation in the efficiency of amplification of different segments of the same virus. Using this approach, MI63, KY98, Panama 99, and A/Puerto Rico/8/34 and, in three of six experiments, HK68, had their HA subtypes correctly identified (Table 6). In the other HK68 experiments, the rare subtypes 11 and 13 appeared falsely to be significant. Oligonucleotides predicted to react with these rare subtypes also should react with almost all subtypes, perhaps accounting for the misidentification. For example, of 23 H13-reacting probes, 13 also should react with eight or more other HA subtypes. The performance of scoring for the NA subtype by using this approach produced more consistent results for HK68 than scoring for HA subtype. However, the method was not able to identify that the equine viruses were of N8 subtype because no NA-specific oligonucleotides were expected to react with N8 cDNAs.
Effect of experimental variables on detection.
The effect of hybridization temperature on microarray performance was tested with HK68. Increasing the hybridization temperature from 22 to 32°C affected the results for some characteristics (Tables 3 through 7; experiments A, B, and C). However, a further increase to 42°C did not affect results significantly. The increase in temperature resulted in increases in the proportions of weighted scores for misidentified virus species. The single diagnostic oligonucleotide that at 22°C implicated a human host no longer appeared positive at the elevated temperatures. Raising the temperature from 22 to 42°C increased the number of signature oligonucleotides positive for segments 1, 2, 6, 7, and 8, but decreased that number for the HA segment. On raising the temperature, segment 3 increased more than 50% in its percentage of the weighted scores, whereas segment 2 decreased by a similar amount. The increase in hybridization temperature correlated with a decreased ability to identify the H3 subtype, favoring the rare 11 and 13 subtypes. For NA subtyping, the temperature change did not drastically affect the values leading to the correct identification but did increase the values for N1, while decreasing those for N4 and N6.
The effect of quantity of labeled cDNA on the ability of the analysis method to identify viruses was also examined, again with HK68 (Tables 3 through 7; experiments A, D, E, and F). The mean percent saturations of pixel intensity of the five oligonucleotides used as medium intensity standards in normalization for experiments A, D, E, and F were, respectively, 11.9, 92.3, 5.5, and 30.2%. Dramatic differences from experiment A results were only noticed at the highest percent saturation (experiment D). The changes noted from experiment A to experiment D were similar, but not identical, to the effects of raising the temperature. The increased misidentification of HK68 virus species was concentrated more on type B than on type C. The effect on virus host identification was not distinguishable from that of increased hybridization temperature. The changes in the scores for the eight segments were noticeably different. Scores for segments 1, 2, and 7 decreased at the higher concentration, whereas those for segments 3 and 6 increased. The effects of the higher DNA concentration on subtyping were similar to those of elevated temperature during hybridization. Overall, these experimental variations had little negative effect on the detection and identification of the viruses.
DISCUSSION
Our results demonstrate the potential of the use of short oligonucleotide microarrays in accurate typing of influenza virus species, distinction between human and equine hosts, and detection of the presence of each of the eight viral gene segments. The method, as tested here, can distinguish H1 from H3 and N1 from N2 subtypes. The ability to make these distinctions was relatively independent of the quantity of labeled cDNA used and only slightly affected by elevated hybridization temperatures. Clinical application of the method will require validation experiments, such as sensitivity determinations, testing for interference by substances likely to be in clinical material, and blind sample studies.
Our experiments revealed that not all oligonucleotides are equally useful in making distinctions among influenza viruses. The intensities of hybridization for perfectly complementary oligonucleotides to their targets varied from none to very high. Such variation can be noted in results obtained by others in virus identification (23) and other assays (14, 19, 22, 24). “Rules-of-thumb” for oligonucleotide design have been promulgated to minimize this variation (15, 19, 24). Although these recommendations are consistent with a low reactivity of some oligonucleotides, they do not account for many of the anomalies. One possibility for the nonreactivity of some oligonucleotides is that the viruses used in the experiments did not retain the sequence deposited in the databases. It is well known that influenza viruses have high mutation rates. Mutations may account for a few of the discrepancies. However, they are unlikely to account for most of them since the oligonucleotides were mostly chosen via VirOligo (18), which is derived from PCR literature, and had been demonstrated to be able to amplify the targeted viruses. Another possible contributor to the variation in reactivity is the availability of the target for hybridization. Formation of secondary structure by target cDNAs would inhibit hybridization (21). Consistent with this view is the observation that shearing of cDNA targets (20) was essential to obtain satisfactory hybridization. Shearing prevents the formation of intramolecular base pairing of distant sequences. However, at an average length of 200 nt for the sheared fragments, intramolecular base pairing between closely spaced sequences may still occur. To improve the ability to identify influenza virus and other pathogens, these factors need to be considered further. Design of probes solely based on sequence uniqueness and melting temperature (10, 25) will not suffice efficiently to produce an effective set of probes.
Very few of the oligonucleotides misidentified the virus type, source species, or HA subtype. Misidentifications are not due to nonspecific interactions, since the same oligonucleotide was not misdiagnostic for all viruses and did not react with negative control (RBC) cDNA. Since influenza viruses exist as quasispecies, some minor components of the target cDNA may hybridize to the incorrect probe.
Two methods were used to make identifications. The signature oligonucleotide method used exclusivity of BLASTn hits to determine specificity. It is unambiguous but ignores the information in oligonucleotides that react with multiple characteristics. Of the 424 oligonucleotides used that were not eliminated from consideration for the reasons cited above, 307 were type specific and 246 were segment specific. However, only 36 were source host specific, 81 were HA subtype specific, and 21 were NA subtype specific. Therefore, for some source hosts (swine, and species other than human, avian, and equine) and many HA and NA subtypes there were no uniquely diagnostic probes. To compensate for the lack of signature oligonucleotides, an attempt was made to use the overrepresentation of some categories among BLASTn hits of the oligonucleotides and the underrepresentation of others to aid in the characterization. However, such weighting did not yield more information than obtained from the signature oligonucleotides.
Oligonucleotides designed by available software or by VirOligo provided oligonucleotides with comparable abilities to detect influenza virus, even though the requirements for PCRs differ from those for hybridization. Mismatches in the 3′-most nucleotides in a PCR primer prevent amplification, while they interfere minimally with hybridization. Conversely, the mismatches of an oligonucleotide probe that most dramatically reduce hybridization to a target are located in the middle of the sequence. These mismatches have less effect on use of the oligonucleotide as a PCR primer.
The experiments reported here, in combination with those reported by Wang et al. (23), demonstrate the tremendous potential of microarray hybridization for the detection and identification of viruses. The important role played by this method in the rapid characterization of the recently identified coronavirus associated with severe acute respiratory syndrome testifies to the utility of this method (5). Obviously, the present microarray has several deficiencies. However, they are not inherent in the method but rather stem from weaknesses in the choice of probes, which can be improved by better probe selection and addition of more probes. Thus, these studies open the door to designing a universal viral signature hybridization chip that can be applied to address the questions of whether there is a virus in a given sample and, if so, what that virus is. Although the studies described here used the fluorescence of hybridized target DNA molecules for detection of successful hybridization, the experience gained is relevant to any method that relies on the formation of hybrids between nucleic acids derived from samples to short oligonucleotide probes. Methods currently under development in other laboratories (9) have the potential of providing greater sensitivity than was achieved by the method used here (4).
Acknowledgments
This study was supported by the Centers for Water Research and for Sensors and Sensor Technology of Oklahoma State University, by the EPSCoR program of the National Science Foundation, and by the Oklahoma Agricultural Experiment Station, whose director approved the study for publication.
We thank J. Malayer and R. Prade for careful review of the manuscript. The assistance of the OSU Microarray Core Facility, the OSU Recombinant DNA/Protein Resource Facility, and the OU Health Sciences Center Laboratory for Microbial Genomics is gratefully acknowledged.
REFERENCES
- 1.Adeyefa, C., K. Quayle, and J. McCauley. 1994. A rapid method for the analysis of influenza virus genes: application to the reassortment of equine influenza virus genes. Virus Res. 32:391-399. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Cho, N. H., H. J. An, J. K. Jeong, S. W. Kang, J. W. Kim, Y. T. Kim, and T. K. Park. 2003. Genotyping of 22 human paillomavirus types by DNA chip in Korean women: comparison with cytologic diagnosis. Am. J. Obstet. Gynecol. 188:56-62. [DOI] [PubMed] [Google Scholar]
- 4.Delpech, M. 2000. Les puces a ADN. Ann. Biol. Clin. 58:29-38. (In French.) [PubMed]
- 5.Elias, P. 2003. Gene chip helps identify cause of SARS. Government tech-nology. [Online.] http://www.govtech.net/news/news.phtml?docid=2003.04.07-46078.
- 6.Ellis, J. S., and M. C. Zambon. 2002. Molecular diagnosis of influenza. Rev. Med. Virol. 12:375-389. [DOI] [PubMed] [Google Scholar]
- 7.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang, T. S., J. K. Jeong, M. Park, H. S. Han, H. K. Choi, and T. S. Park. 2003. Detection and typing of HPV genotypes in various cervical lesions by HPV oligonucleotide microarray. Gynecol. Oncol. 90:51-56. [DOI] [PubMed] [Google Scholar]
- 9.Jenison, R., M. Rihanek, and B. Polisky. 2001. Use of a thin film biosensor for rapid visual detection of PCR products in a multiplex format. Biosens. Bioelectron. 16:757-763. [DOI] [PubMed] [Google Scholar]
- 10.Kaderali, L., and A. Schliep. 2002. Selecting signature oligonucleotides to identify organisms using DNA arrays. Bioinformatics 18:1340-1349. [DOI] [PubMed] [Google Scholar]
- 11.Lai, A. C., and T. M. Chambers. 1995. Rapid protocol for sequencing RNA virus using delta Taq version 2.0 DNA polymerase. BioTechniques 19:704-706. [PubMed] [Google Scholar]
- 12.Lapa, S., M. Mikheev, S. Shchelkunov, V. Mikhailovich, A. Sobolev, V. Blinov, I. Babkin, A. Guskov, E. Sokunova, A. Zasedatelev, L. Sandakhchiev, and A. Mirzabekov. 2002. Species-level identification of orthopoxviruses with an oligonucleotide microchip. J. Clin. Microbiol. 40:753-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, J., S. Chen, and D. H. Evans. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, W. T., A. D. Mirzabekov, and D. A. Stahl. 2001. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol. 3:619-629. [DOI] [PubMed] [Google Scholar]
- 15.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 16.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection, p. 103-106. In A. D. M. E. Osterhaus, N. Cox, and A. W. Hampson (ed.), Options for the control of influenza, vol. IV. Elsevier Science, Amsterdam, The Netherlands.
- 17.Offringa, D. P., V. Tyson-Medlock, Z. Ye, and R. A. Levandowski. 2000. A comprehensive systematic approach to identification of influenza A virus genotype using RT-PCR and RFLP. J. Virol. Methods 88:15-24. [DOI] [PubMed] [Google Scholar]
- 18.Onodera, K., and U. Melcher. 2002. VirOligo: a database of virus-specific oligonucleotides. Nucleic Acids Res. 30:203-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Relogio, A., C. Schwager, A. Richter, W. Ansorge, and J. Valcarcel. 2002. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 30:e51. [DOI] [PMC free article] [PubMed]
- 20.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sohail, M., S. Akhtar, and E. M. Southern. 1999. The folding of large RNAs studied by hybridization to arrays of complementary oligonucleotides. RNA. 5:646-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohail, M., H. Hochegger, A. Klotzbucher, R. L. Guellec, T. Hunt, and E. M. Southern. 2001. Antisense oligonucleotides selected by hybridisation to scanning arrays are effective reagents in vivo. Nucleic Acids Res. 29:2041-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, D., L. Coscoy, M. Zylberberg, P. C. Avila, H. A. Boushey, D. Ganem, and J. L. DeRisi. 2002. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA 99:15687-15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, R., M. Beggs, L. Robertson, and C. Cerniglia. 2002. Design and evaluation of oligonucleotide-microarray method for the detection of human intestinal bacteria in fecal samples. FEMS Microbiol. Lett. 213:175-182. [DOI] [PubMed]
- 25.Zhang, Z., R. C. Willson, and G. E. Fox. 2002. Identification of characteristic oligonucleotides in the bacterial 16S ribosomal RNA sequence dataset. Bioinformatics 18:244-250. [DOI] [PubMed] [Google Scholar]