Abstract
Mammalian Ran-binding protein-1 (RanBP1) and its fission yeast homologue, sbp1p, are cytosolic proteins that interact with the GTP-charged form of Ran GTPase through a conserved Ran-binding domain (RBD). In vitro, this interaction can accelerate the Ran GTPase-activating protein–mediated hydrolysis of GTP on Ran and the turnover of nuclear import and export complexes. To analyze RanBP1 function in vivo, we expressed exogenous RanBP1, sbp1p, and the RBD of each in mammalian cells, in wild-type fission yeast, and in yeast whose endogenous sbp1 gene was disrupted. Mammalian cells and wild-type yeast expressing moderate levels of each protein were viable and displayed normal nuclear protein import. sbp1− yeast were inviable but could be rescued by all four exogenous proteins. Two RBDs of the mammalian nucleoporin RanBP2 also rescued sbp1− yeast. In mammalian cells, wild-type yeast, and rescued mutant yeast, exogenous full-length RanBP1 and sbp1p localized predominantly to the cytosol, whereas exogenous RBDs localized predominantly to the cell nucleus. These results suggest that only the RBD of sbp1p is required for its function in fission yeast, and that this function may not require confinement of the RBD to the cytosol. The results also indicate that the polar amino-terminal portion of sbp1p mediates cytosolic localization of the protein in both yeast and mammalian cells.
INTRODUCTION
Proteins to be transported across the eukaryotic nuclear membrane often contain sequence motifs that function as nuclear import or export signals (Nigg, 1997). Transport requires both insoluble components, associated with the nuclear pore, and soluble components that interact with the transport cargo and/or pore. Among the soluble factors are members of the karyopherin β family, the small GTPase Ran, Ran-binding protein-1 (RanBP1), and nuclear transport factor 2 (Mattaj and Englmeier, 1998). Components of the nuclear pore complex, termed nucleoporins, have been identified by biochemical analysis of partially purified pore complexes and by genetic analysis of nuclear transport in budding yeast (Fabre and Hurt, 1997; Ohno et al., 1998). A pore component that appears to play a central role in interactions with the soluble factors is RanBP2 (also known as Nup358) (Wu et al., 1995; Yokoyama et al., 1995).
Many models of nuclear transport have been proposed, and central to them all is the control exerted by Ran in regulating the association and dissociation of transport complexes, linked to the hydrolysis and turnover of Ran-bound guanine nucleotides (Koepp and Silver, 1996; Panté and Aebi, 1996; Rush et al., 1996; Görlich, 1997; Nakielny and Dreyfuss, 1997; Nigg, 1997; Yoneda, 1997; Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Melchior and Gerace, 1998; Moore, 1998). The intrinsic rates of GTP hydrolysis and guanine nucleotide release by Ran are slow and are accelerated in vivo by a Ran GTPase-activating protein (RanGAP) and a guanine nucleotide release protein, regulator of chromosome condensation 1 (RCC1), respectively (Bischoff and Ponstingl, 1991; Bischoff et al., 1994). RanGAP activity decreases the Ran ·GTP/Ran·GDP ratio, whereas RCC1 activity increases it. (GTP is at least 10 times more abundant than GDP in vivo.) RanGAP appears to be located in the cytosol and on the cytosolic face of nuclear pores (Mahajan et al., 1997; Saitoh et al., 1997; Matunis et al., 1998; Saitoh et al., 1998), whereas RCC1 appears to be located in the nucleus (Ohtsubo et al., 1989), so the Ran·GTP/Ran·GDP ratio is expected to be low in the cytosol and high in the nucleus.
This difference could explain directionality in the movement of transport complexes. For nuclear import, a member of the karyopherin β importin subfamily binds to the import substrate directly or via an adapter, such as karyopherin α, specific for import substrates containing a nuclear localization signal (NLS) sequence motif, and this complex docks at the cytosolic face of the nuclear pore (Görlich et al., 1994; Chi et al., 1995; Görlich et al., 1995; Imamoto et al., 1995; Radu et al., 1995; Pollard et al., 1996; Fridell et al., 1997). The docked complex is then transported through the nuclear pore. Once in the nucleus, the complex could be disrupted by Ran·GTP, which in vitro can bind to karyopherin β and displace the import substrate (Rexach and Blobel, 1995; Chi et al., 1996, 1997; Görlich et al., 1996; Izaurralde et al., 1997; Lounsbury and Macara, 1997; Siomi et al., 1997). Complex assembly would thus be favored in the cytosol (low [Ran·GTP]), and disassembly would be favored in the nucleus (high [Ran·GTP]). For nuclear export, members of the karyopherin β exportin subfamily bind to Ran·GTP and export substrates (Fornerod et al., 1997; Fukuda et al., 1997; Görlich et al., 1997; Stade et al., 1997). Formation of these exportin–Ran·GTP–export substrate complexes requires Ran·GTP, so assembly would be favored in the nucleus (high [Ran·GTP]). After the complexes traverse the nuclear pore, RanGAP-stimulated conversion of Ran·GTP to Ran·GDP in the cytosol could drive the complexes to disassemble (Görlich, 1997; Lounsbury and Macara, 1997). The same mechanism could explain the efficient recycling to the cytosol of karyopherin β importins as Ran·GTP complexes (Floer et al., 1997).
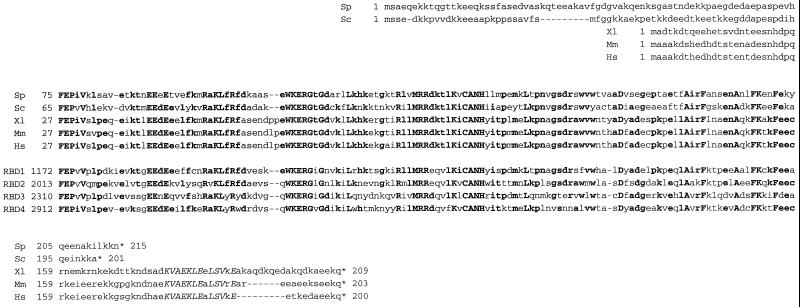
A limitation of this simple model is that Ran·GTP complexed to karyopherin β (importin or exportin) exhibits a decreased rate of nucleotide release and is almost completely resistant to RanGAP-stimulated GTP hydrolysis. This resistance could prevent efficient RanGAP-mediated disruption of Ran·GTP–importin and Ran·GTP–exportin–export substrate complexes in the cytosol. Two other cytosolic proteins, RanBP1 and RanBP2, appear to be critically important for efficient disassembly of these complexes. RanBP1 is a small hydrophilic protein present in both yeast and higher eukaryotes (Coutavas et al., 1993), and RanBP2 is a large nucleoporin (present in higher eukaryotes but absent in yeasts) located on cytoplasmic nuclear pore fibrils (Wilken et al., 1995; Wu et al., 1995; Yokoyama et al., 1995). Both proteins bind specifically to Ran·GTP but not Ran·GDP, via well-conserved Ran-binding domain (RBD) sequence motifs (Vetter et al., 1999). These RBD sequences are ∼130 aa long; RanBP1 has one and RanBP2 has four (Figure 1). Ran·GTP complexed to the RBDs of RanBP1 or RanBP2 exhibits a decreased rate of nucleotide release (Lounsbury et al., 1994) but an ∼10-fold increase in RanGAP-mediated GTP hydrolysis (Bischoff et al., 1995). In vitro, RanBP1 and RanBP2 interact with the karyopherin β complexes through Ran·GTP (Delphin et al., 1997) and stimulate dissociation of the complexes in one or more steps coupled to the hydrolysis of GTP on Ran (Bischoff and Görlich, 1997; Chi et al., 1996, 1997).
Figure 1.
Conserved sequences of RBDs. Alignment of the predicted sequence of S. pombe sbp1p (Sp, D86381) with S. cerevisiae Yrb1p (Sc, P41920), the RanBP1 proteins of Xenopus laevis (Xl, Y09128), mouse (Mm, X56046), and human (Hs, D38076), and RBDs 1–4 (RBD1, … , 4) of human RanBP2 (P49792). Asterisks indicate termination codons, and dashes indicate gaps introduced to maximize homology. Residues identical in five or more of the sequences are shown in boldface type, and residues identical in all nine sequences are shown in uppercase boldface type. The NES motif of the human, mouse, and Xenopus RanBP1 proteins is shown in uppercase italics.
The essential role of RanBP1 in nuclear–cytosolic transport may not be limited to disruption of export complexes. RanBP1 has been reported to stimulate the formation of a stable RanBP1–Ran·GDP–karyopherin β trimeric complex, even though Ran·GDP does not interact singly with either of these proteins (Chi et al., 1996, 1997). Such a complex may be an intermediate in the nuclear protein import pathway, a possibility supported by the observation that RanBP1 stimulates import in a digitonin-permeabilized cell assay supplemented with karyopherin α, karyopherin β, and Ran (Chi et al., 1996).
RanBP1 has also been implicated in cellular processes that are independent of nuclear transport (Dasso, 1995; Avis and Clarke, 1996; Pu and Dasso, 1997; Hughes et al., 1998). Overexpression of RanBP1 in either yeast or mammalian cells inhibits cell division. In the fission yeast Schizosaccharomyces pombe, overexpression results in a defective mitotic exit phenotype: cells undergo cell cycle arrest with condensed chromosomes, a wide medial septum, and a fragmented nuclear envelope (He et al., 1998). In mammalian cells, overexpression in G0 results in an inhibition of G1 progression, whereas overexpression at the G2–M boundary results in an inhibition of mitotic exit and a cell cycle arrest phenotype similar to fission yeast (Battistoni et al., 1997). An excess of RanBP1 in Xenopus egg extracts blocks assembly of replication- and import-competent nuclei (Nicolas et al., 1997; Pu and Dasso, 1997). All these data suggest that a proper balance between RanBP1 and other components of the Ran pathway may be required for normal function, but the molecular basis of the disruptions caused by excess RanBP1 remains unclear. It is also unclear how RanBP1 can have a constitutive role in nuclear–cytosolic transport yet be expressed predominantly in the S, G2, and M phases of the mammalian cell cycle (Battistoni et al., 1997; Guarguaglini et al., 1997).
Like endogenous protein, full-length exogenously expressed RanBP1 localizes predominantly to the cytosol. This localization appears to be an active process, because the isolated RBD of RanBP1 localizes predominantly to the nucleus and the presence of a leucine-rich nuclear export signal (NES) motif in the carboxyl-terminal portion of the full-length protein is required for its cytosolic localization (Richards et al., 1996; Zolotukhin and Felber, 1997). It is unclear whether the NES mediates normal shuttling of RanBP1 between nuclear and cytosolic compartments or whether it serves to ensure confinement of RanBP1 to the cytosol. However, ectopic nuclear expression of the RanBP1-RBD in mammalian cells or microinjection of full-length RanBP1 into the nuclei of Xenopus oocytes inhibits nuclear protein import and export, respectively (Richards et al., 1996; Izaurralde et al., 1997). Both the amount and localization of RanBP1 thus appear to be critical for its biological function(s).
RanBP1 homologues have been described for diverse eukaryotic species, including human, mouse, Xenopus laevis, budding yeast, and fission yeast (Coutavas et al., 1993; Bischoff et al., 1995; Ouspenski et al., 1995; Schlenstedt et al., 1995; Pu and Dasso, 1997; He et al., 1998). Given its essential and apparently well-conserved role in nuclear–cytosolic transport and other eukaryotic cellular processes, the substantial divergence in the primary structure of RanBP1 between vertebrates and yeasts is surprising (Figure 1). Aside from their overall lengths (200–215 aa) and their RBD sequences (∼50–60% identical), vertebrate and yeast RanBP1s are quite different. The yeast proteins (Yrb1p in S. cerevisiae and sbp1p in S. pombe) contain a polar amino-terminal segment not found in vertebrate proteins, whereas the vertebrate proteins contain a carboxyl-terminal sequence including the leucine-rich NES motif not found in yeast.
To better define the roles of specific domains of RanBP1 in nuclear protein import and other cellular functions, we have studied the effects of exogenous full-length RanBP1 proteins and isolated RBDs on viability and nuclear protein import in mammalian and S. pombe cells. We have also tested the abilities of these proteins to rescue S. pombe cells whose endogenous sbp1 gene was disrupted. Our results suggest that only the well-conserved RBD of mammalian RanBP1 or yeast sbp1p is required for yeast cell viability, that the RBDs of RanBP2 can substitute for RanBP1, and that these RBDs can localize predominantly to the cell nucleus without disrupting cell function.
MATERIALS AND METHODS
Plasmid Construction
Mammalian Cell Expression Plasmids.
ORFs encoding full-length RanBP1 (mouse, aa 1–203) and sbp1p (S. pombe RanBP1 homologue, aa 1–215), and the RBDs of RanBP1 (aa 38–159) and sbp1p (aa 75–215), were amplified by PCR and subcloned in frame downstream of a T7 epitope tag under the control of a cytomegalovirus promoter. To accomplish this, the pCGTVP16ΔC plasmid (Wilson et al., 1997) was digested with XbaI and BamHI to delete the VP16 insert, which was replaced with the PCR fragment.
Fission Yeast Expression Plasmids.
ORFs were expressed under the control of the thiamine-repressible promoter of the S. pombe nmt1 gene. PCR products encoding T7-tagged full-length RanBP1 (mouse) and sbp1p (S. pombe), and the RBD of mouse RanBP1 were subcloned into the NdeI and BamHI sites of plasmids pREP1 (nmt full-strength promoter), pREP41 (nmt* medium-strength promoter), and pREP81 (nmt** low-strength promoter) (Basi et al., 1993). Plasmids pSLF173 (nmt full-strength promoter), pSLF273 (nmt* medium-strength promoter), and pSLF373 (nmt** low strength-promoter) (Forsburg and Sherman, 1997) were modified to have a leu2+ marker instead of the ura4+ marker (pSLFleu173, pSLFleu273, and pSLFleu373, respectively). These plasmids allow the expression of a protein fused to a triple HA epitope tag under the control of different-strength nmt promoters. The RBD of sbp1p and RBDs 2 (aa 2013–2142) and 4 (aa 2912–3040) of Nup 358/RanBP2 were PCR amplified and cloned into the SalI–BglII sites of the pSLFleu plasmids.
Construction of the GST-GFP-NLS Protein Import Reporter Gene
Plasmid pEGFP-C3 (Clontech, Palo Alto, CA) was used as a template to amplify by PCR a green fluorescent protein (GFP) ORF followed by the NLS of SV40 large T antigen (aa 126–132, PKKKRKV) (Kalderon et al., 1984). A 5′ end primer with an NdeI site hybridizing to the amino-terminal region of GFP and a set of two overlapping primers encoding the carboxyl-terminal region of GFP fused to the SV40 large T antigen NLS with a BamHI site were used to amplify a GFP-NLS PCR product. The GST coding sequence (plasmid pGEX-5X-1; Pharmacia, Piscataway, NJ) was amplified with a PstI site added to its 5′ end and a NdeI site added to its 3′ end and then ligated in frame to the amino-terminus of the GFP-NLS coding sequence. The vector pART1 (McLeod et al., 1987) was modified to replace the leu2+ marker by an ade2+ marker, and the new pARTade vector was digested with PstI and BamHI and ligated to the two PCR products to obtain the GST-GFP-NLS gene under the control of the adh promoter. To express the same protein in mammalian cells, the pSVL vector (Pharmacia) digested with XhoI and BamHI was ligated to the GST gene amplified with an XhoI site at its 5′ end (instead of a PstI site), and the GFP-NLS sequence was generated with restriction enzyme sites as described above. As a control, a GST-GFP protein without the NLS was also constructed in both vectors (pARTade and pSVL).
Mammalian Cell Transfection and Immunostaining
Mammalian cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. Cells were transfected with LipofectAMINE reagent (Life Technologies, Gaithersburg, MD) following the manufacturer’s protocol. Approximately 48 h after transfection, cells were fixed with 3% paraformaldehyde in PBS for 15 min at room temperature, washed three times with PBS plus 0.1 M glycine, permeabilized in 0.2% Triton X-100 in PBS for 15 min, and incubated for 30 min in blocking buffer (2% BSA in PBS). Cells were then incubated 1 h with a 1:500 dilution of anti-T7 tag mAb (Novagen, Madison, WI) in blocking buffer, washed three times with blocking buffer, incubated 1 h with a 1:700 dilution of TRITC-conjugated goat anti-mouse IgG (Sigma, St. Louis, MO) in blocking buffer, washed three times in blocking buffer, incubated for a few seconds with 1 μg/ml DAPI (Sigma) in PBS, washed three times with PBS, mounted with an antibleaching agent (FITC-Guard; Testog, Chicago, IL), and examined using an Axiophot microscope (Carl Zeiss, Thornwood, NY).
Yeast Strains and Cell Culture
Yeast strains were grown by standard procedures (Moreno et al., 1991), and transformations were carried out by the lithium acetate method (Okazaki et al., 1990). Total yeast DNA for Southern analysis was prepared by a standard method (Alfa et al., 1993). The haploid strain h90 leu1-32 ura4-D18 ade6-M216 was used for overexpression of RanBP1 (mouse), sbp1p (S. pombe), and the RBD of each, under the control of nmt promoters of various strengths (Maundrell, 1990; Basi et al., 1993). Thiamine was added to Edinburgh minimal medium to a concentration of 5 μg/ml to repress the nmt promoter.
Protein Extracts of S. pombe
Proteins were extracted in lysis buffer (1% Triton X-100, 0.1% SDS, 4 μg/ml leupeptin, and 1 mM PMSF in PBS) using glass beads, as described by Moreno et al. (1991). Expression of epitope-tagged proteins was analyzed by immunoblotting of protein extracts separated in 15% SDS-PAGE. T7- and hemagglutinin (HA)-tagged proteins were detected with anti-T7 mAb (1:10,000 dilution; Novagen) and anti-HA antibody (1:1000 dilution; Babco, Richmond, CA), respectively, in 1% nonfat dry milk and 0.1% Tween 20 in PBS plus peroxidase-coupled anti-mouse Ig (1:5000 dilution; Amersham, Arlington Heights, IL) in the same buffer. Chemiluminescent detection was carried out with the LumiGLO kit (Kirkegaard & Perry, Gaithersburg, MD) following the manufacturer’s instructions.
Disruption of sbp1 and Rescue of sbp1− Mutant Cells
A 5.2-kb HindIII fragment of S. pombe genomic DNA that spanned the sbp1 gene was cloned from an S. pombe genomic DNA library (from Dr. David Frendeway, New York University School of Medicine). To disrupt the sbp1 gene, the 300-bp XbaI–PstI fragment that extends from the first intron of the gene into its second exon (He et al., 1998) was replaced with the ura4 gene, and this construct was used to transform diploid h90/h90 leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M210/ade6-M216 S. pombe cells. ura+ transformants were analyzed by Southern blotting to identify ones heterozygous for sbp1 disruption (h90/h90 sbp1::ura4+/sbp1 leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M210/ade6-M216), and lethality of the disruption was confirmed by random spore analysis. Heterozygous diploid cells were transformed with pREP41 and pSLFleu constructs (Okazaki et al., 1990). Leu+ ura+ transformants were sporulated, digested with Zymolase 20-T (ICN, Costa Mesa, CA), and plated on Edinburgh minimal medium supplemented with adenine.
S. pombe Immunostaining and Fluorescence Microscopy
Epitope-tagged proteins were localized in paraformaldehyde-fixed cells following the protocol of Alfa et al. (1993), except that cell walls were digested with 0.25 mg/ml lysing enzymes from Trichoderma harzianum (Sigma) plus 0.3 mg/ml Zymolase 20-T for ∼4 min at 37°C. Monoclonal anti-T7 and anti-HA antibodies were used at dilutions of 1:250 and 1:50, respectively, and FITC-conjugated anti-mouse IgG was used at a dilution of 1:250. After staining, cells were mounted in Vectashield mounting medium (Vector Laboratories, Burlingame, CA) plus antifade and DAPI (Alfa et al., 1993) and examined using an Axiophot microscope. GST-GFP-NLS localization was carried out in methanol-fixed cells (Alfa et al., 1993).
RESULTS
Exogenously Expressed RanBP1 and sbp1p Localize to the Cytosol of Transfected Mammalian Cells, Whereas Their RBDs Localize Predominantly to the Nucleus
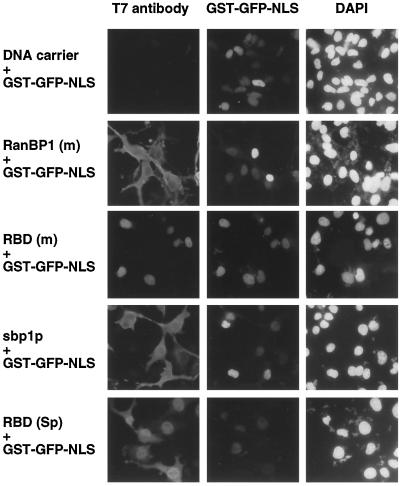
Full-length RanBP1 (mouse) is known to localize to the cytoplasm, but its RBD localizes predominantly to the nucleus (Richards et al., 1996). To compare the protein sequence motifs responsible for subcellular localization of sbp1p (S. pombe) and mammalian RanBP1, COS cells were transfected with constructs encoding full-length mouse or S. pombe proteins or the RBD of each. The RBD of mouse RanBP1 consisted of amino acids 38–159. The sbp1p RBD construct consisted of amino acids 75–204, the region homologous to vertebrate RBDs (Figure 1), plus the carboxyl-terminal 11 amino acids of the protein (aa 205–215). Each of these proteins was subcloned with a T7 epitope tag in frame at its amino terminus (see MATERIALS AND METHODS). Consistent with previous studies (Richards et al., 1996; Zolotukhin and Felber, 1997), cells transfected with a construct expressing T7-tagged full-length mouse RanBP1 showed largely cytoplasmic staining (Figure 2, left panels). In contrast, in cells expressing the T7-tagged mouse RBD, which lacks the NES motif (Figure 1), staining was largely nuclear (Figure 2, left panels). Likewise, full-length and RBD forms of sbp1p localized mostly to the cytoplasm and nuclei, respectively (Figure 2, left panels), despite the lack of any discernible NES motif in the intact S. pombe protein (Figure 1). The same subcellular distributions were observed for all four proteins in transfected HeLa cells (our unpublished results). These results suggest that the amino-terminal 74 residues of the S. pombe RanBP1 homologue include a sequence that mediates cytoplasmic localization in mammalian cells and that is distinct from the leucine-rich NES of mouse RanBP1.
Figure 2.
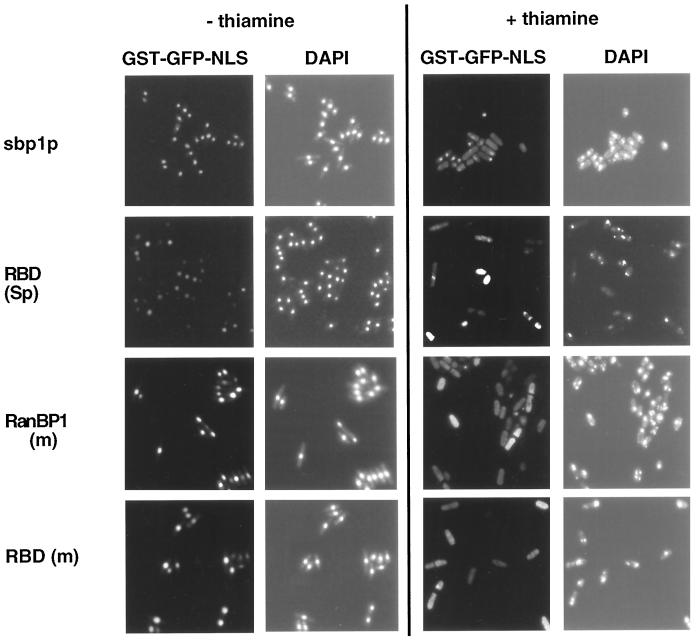
Localization of RanBP1 (m), sbp1p, and their RBDs and effect on nuclear protein import in transiently transfected COS cells. COS cells grown in 35-mm dishes were cotransfected with ∼0.5 μg pSVL.GST-GFP-NLS (a plasmid encoding GST-GFP-NLS protein) and with 0.5 μg of either DNA carrier (salmon sperm DNA), or plasmids encoding T7-tagged RanBP1 (mouse, aa 1–203), sbp1p (S. pombe RanBP1 homologue, aa 1–215), the RBD of RanBP1 (mouse, aa 38–159), or the RBD of sbp1p (S. pombe, aa 75–215). Approximately 48 h after transfection, cells were fixed, and RanBP1 proteins were detected by immunofluorescence using a monoclonal anti-T7 epitope antibody and a TRITC-conjugated secondary antibody. Cells were examined under the microscope using filters specific for TRITC (exogenous RanBP1 proteins, left panels), GFP (GST-GFP-NLS, center panels), and DAPI (DNA, right panels).
Exogenously Expressed RanBP1, sbp1p, or Their RBDs Do Not Block NLS-mediated Nuclear Protein Import in Mammalian Cells
Previous studies had suggested that mislocalization of the RBD of RanBP1 to the nucleus blocked NLS-mediated nuclear protein import (Richards et al., 1996). To determine the effects of our constructs on nuclear protein import, COS cells were cotransfected with a construct encoding a GST-GFP-NLS chimeric protein and with constructs encoding full-length mouse (m) RanBP1 or S. pombe (Sp) sbp1p or the RBD of each. As shown in Figure 2, center panels, the NLS protein localized efficiently to nuclei in all cases. These results suggest that full-length RanBP1 and sbp1p and their respective RBDs do not interfere with NLS-mediated nuclear import in mammalian cells, even though the RBDs are predominantly mislocalized to the nucleus.
Expression of RanBP1 (m), sbp1p, or RBD (Sp) Can Inhibit Cell Division in S. pombe
To examine the effects of overexpression of these proteins in S. pombe, thiamine-repressible expression vectors based on the nmt promoter (nmt = no message in thiamine) for fission yeast were used (Maundrell, 1990; Basi et al., 1993). The TATA box of the nmt promoter has been mutated to yield promoters that have three different strengths in the absence of thiamine: nmt (full strength), nmt* (medium strength), and nmt** (low strength) (Basi et al., 1993). Full-length RanBP1 (mouse) and sbp1p, and the mouse RBD were expressed with an amino-terminal T7 epitope tag, whereas the S. pombe RBD was expressed with an amino-terminal influenza HA epitope tag (see MATERIALS AND METHODS) (Forsburg and Sherman, 1997).
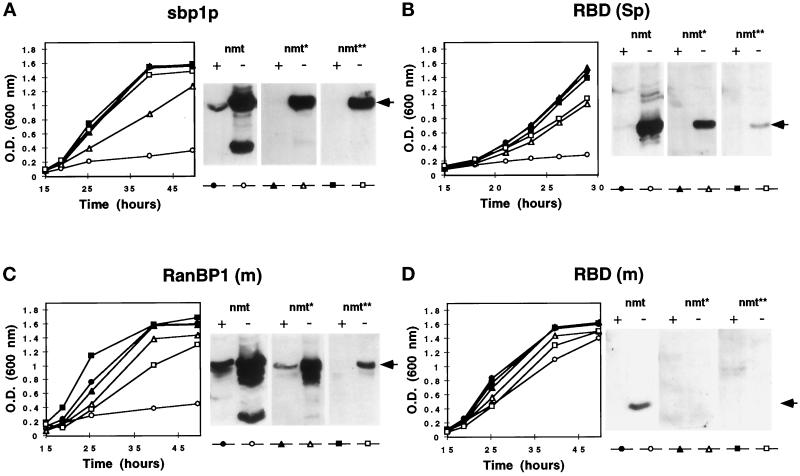
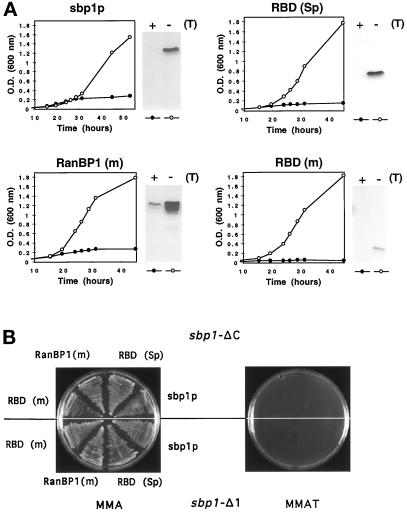
Transformed S. pombe cells were grown in liquid media in the presence (repressed) or absence (derepressed) of thiamine to determine the effect of exogenous RanBP1 protein expression on cell growth (Figure 3). Division of cells transformed with sbp1p, RanBP1 (m) and RBD (Sp) constructs in full-strength (nmt) plasmids was strongly inhibited in the absence of thiamine (Figure 3, A–C), whereas transformation of cells with medium-strength (nmt*) and low-strength (nmt**) promoter plasmids had little or no effect. Growth inhibition was correlated with high levels of protein detected by immunoblotting (Figure 3, A–C). Relatively low amounts of exogenous protein were observed in cells transformed with RBD (m) even under control of the full-strength nmt promoter (Figure 3D), which may explain the normal growth of these cells irrespective of the expression vector.
Figure 3.
Cell growth and expression of RanBP1 proteins in S. pombe. S. pombe cells transformed with the full-strength (nmt, filled and empty circles), medium-strength (nmt*, filled and empty triangles), or low-strength (nmt**, filled and empty squares) nmt promoter plasmids expressing full-length sbp1p (A), RBD (Sp) (B), RanBP1 (m) (C), or RBD (m) (D) were grown in Edinburgh minimal medium in the presence of 5 μg/ml thiamine (+, filled symbols) or in its absence (−, empty symbols). The optical density at 600 nm was monitored as a function of time. Ten micrograms of protein extracts from cells grown to midlog phase in the presence of thiamine or for an equal amount of time in its absence were analyzed by immunoblotting using mAbs against the T7 or HA epitope tags. Arrows indicate full-length proteins.
The lethality associated with high levels of sbp1p in S. pombe cells is consistent with the results of previous studies of sbp1p, including overexpression, and studies of defects in other elements of the spi1p GTPase system (S. pombe Ran homologue). Growth-arrested cells exhibit the characteristic phenotype of condensed chromosomes, fragmentation of the nuclear envelope, and an increase in the number of septated cells (Demeter et al., 1995; Matynia et al., 1996; He et al., 1998). Our data show that this phenotype is also associated with high levels of full-length mouse RanBP1 or sbp1p RBD.
Exogenously Expressed sbp1p and RanBP1 Localize to the Cytosol of S. pombe, Whereas RBD (Sp) and RBD (m) Localize Predominantly to the Nucleus
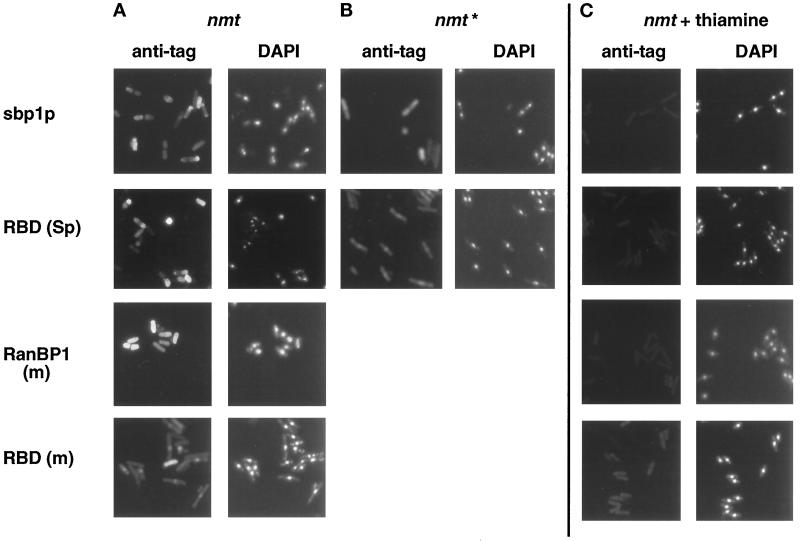
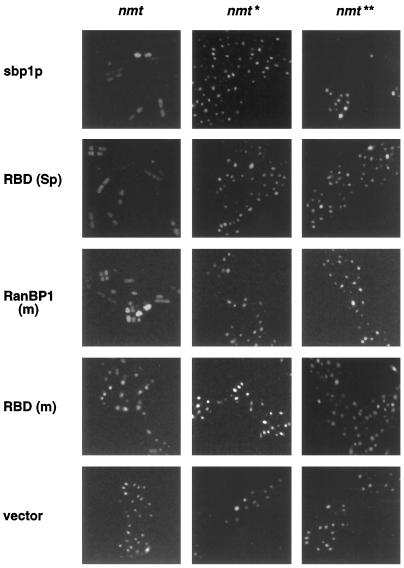
To determine the subcellular distribution of exogenous RanBP1 proteins in transformed S. pombe cells, we performed indirect immunofluorescence using either anti-T7 or anti-HA epitope tag mAbs. When T7-tagged sbp1p or RanBP1 (m) or HA-tagged RBD (Sp) was expressed under the full-strength nmt promoter (growth-inhibiting conditions; Figure 3, A–C), staining was dispersed throughout the cells, and DAPI staining tended to be diffuse (Figure 4A, nmt). When T7-tagged RBD (m) was expressed under the full-strength nmt promoter (normal growth; Figure 3D), staining was mostly nuclear (Figure 4A), and DAPI staining was more restricted. In control cells grown in the presence of thiamine (repressed conditions), no immunofluorescence signal was detected after treatment with anti-epitope mAbs (Figure 4C).
Figure 4.
Localization of exogenous RanBP1 (m), sbp1p, and their RBDs in transformed S. pombe by indirect immunofluorescence. S. pombe cells transformed with full-strength (nmt) or medium-strength (nmt*) plasmids were grown to midlog phase in the presence of thiamine (C) or for an equal amount of time in its absence (A and B). Cells were fixed with paraformaldehyde and permeabilized, and tagged proteins were detected by immunofluorescence using a 1:250 dilution of anti-T7 [(sbp1p, RanBP1 (m), and RBD (m)] or a 1:50 dilution of anti-HA [RBD (Sp)] mAbs, followed by a 1:250 dilution of FITC-labeled anti-mouse secondary antibody. Cells were stained with DAPI before mounting.
The localization of these proteins expressed from the medium-strength promoter constructs (Figure 4B, nmt*) was also analyzed, but we only detected a specific staining when T7-tagged sbp1p or HA-tagged RBD (Sp) was expressed. Although sbp1p was mainly cytosolic, the RBD (Sp) was predominantly nuclear (Figure 4B, nmt*). The localization of the RanBP1 (m) and RBD (m) expressed from the medium- and low-strength nmt promoters could not be determined because of a signal below the level of detection by immunofluorescence, even though RanBP1 (m) was readily detectable by immunoblotting (Figure 3C).
The subcellular distribution of these proteins was also analyzed by direct fusion of GFP to the N-terminal region of each protein and direct fluorescence analysis of cells fixed by paraformaldehyde (our unpublished results). In agreement with the T7 and HA immunofluorescence data, the two RBDs expressed from the medium-strength promoter were predominantly present in the nucleus, although not completely excluded from the cytoplasm, and an increased amount of RBD (Sp) in the cytoplasm was detected when this protein was expressed with the full-strength promoter. Full-length sbp1p and RanBP1 (m) fused to the GFP and expressed from the medium-strength promoter were mainly cytoplasmic, but at higher levels of expression the nuclei were not excluded. These results suggest that the amino-terminal 74 residues of sbp1p mediate cytoplasmic localization in S. pombe cells and that when isolated mouse or S. pombe RBDs are expressed at nontoxic levels, they localize predominantly to S. pombe cell nuclei as they do in mammalian cells. When these proteins are produced at toxic levels their diffuse localization probably reflects the disorganized phenotype described for overexpression of sbp1p or rna1p (RanGAP homologue in S. pombe) in which the nuclear envelope integrity is lost (Matynia et al., 1996; He et al., 1998).
Exogenously Expressed RanBP1, sbp1p, or Their RBDs Do Not Block NLS-mediated Nuclear Protein Import in S. pombe
To further analyze the phenotype of overexpression of RanBP1 proteins in S. pombe, cells were transformed with a plasmid expressing the GST-GFP-NLS protein under the constitutive adh promoter (see MATERIALS AND METHODS). The SV40 large T-antigen NLS is functional in fission yeast (Shiozaki and Yanagida, 1992), and its fusion to GST-GFP causes efficient nuclear import of this normally cytoplasmic protein. Figure 5 shows the distribution of the GST-GFP-NLS when S. pombe cells are expressing full-length sbp1p, RanBP1 (m), or their RBDs under different-strength nmt promoters. Nuclear import proceeds normally, as reflected by nuclear GFP staining, when proteins are expressed from medium- and low-strength nmt promoters (nmt* and nmt**, respectively). The signal remains nuclear when the RBD (m) is expressed from the full-strength nmt promoter (nmt), but in the presence of high levels of full-length sbp1p, RanBP1 (m), or RBD (Sp), the NLS protein is distributed throughout the entire cell, presumably a result of fragmentation of the nuclear envelope (He et al., 1998).
Figure 5.
Nuclear protein import of GST-GFP-NLS in S. pombe expressing sbp1p, RanBP1 (m), or their RBDs. S. pombe cells were cotransformed with a GST-GFP-NLS–expressing construct and with nmt promoter plasmids expressing the indicated RanBP1 or RBD proteins (nmt, full-; nmt*, medium-; and nmt**, low-strength promoter). Cells were grown in the absence of thiamine and fixed by methanol, and the NLS protein was visualized by fluorescence microscopy. Vector indicates control transformations using nmt plasmids lacking RanBP1 or RBD inserts.
RanBP1 (m), RBD (m), and RBD (Sp) Can All Rescue an S. pombe sbp1 Null Mutant
The results of our studies on RanBP1 and RBDs in wild-type S. pombe indicated that strong overexpression is lethal, whereas moderate overexpression is not, even though much of the exogenous RBD protein is mislocalized to the nucleus. To examine these findings more rigorously, as well as to test RBD functionality in vivo, we constructed an S. pombe mutation in which the carboxyl-terminal portion of sbp1 was replaced by ura4+. As expected from the work of He et al. (1998), this sbp1-ΔC mutation was lethal in haploid cells and could be rescued by moderate-level (nmt*) but not high-level (nmt) expression of plasmid-borne wild-type sbp1 (Figure 6). Under repressed conditions (+thiamine), sbp1-ΔC cells containing the nmt* sbp1 plasmid showed a phenotype of condensed chromosomes and disrupted nuclei, indistinguishable from that associated with sbp1 overexpression in wild-type cells (Figure 4).
Figure 6.
Cell growth and expression of RanBP1 proteins in the rescued S. pombe sbp1-ΔC genomic mutant. (A) Growth of S. pombe haploid sbp1-ΔC genomic mutant cells transformed with nmt* plasmids encoding T7-tagged sbp1p, HA-tagged RBD (Sp), T7-tagged RanBP1 (m), or T7-tagged RBD (m) in liquid media in the presence (closed circles) or absence (open circles) of thiamine. Cell samples (20 μg of protein) collected at midlog phase when grown in the absence of thiamine or for an equal amount of time in its presence were analyzed by immunoblotting using anti-T7 or anti-HA epitope mAbs. (B) Growth of S. pombe haploid sbp1-ΔC and sbp1-Δ1 genomic mutants transformed with the plasmids encoding sbp1p, RBD (Sp), RanBP1 (m), or RBD (m) on Edinburgh minimal medium agar plates supplemented with adenine in the absence (MMA) or presence (MMAT) of thiamine and cultivated at 30° for 3 d.
We then tested the ability of full-length mouse RanBP1 and of the RBDs of the S. pombe and mouse proteins to rescue the sbp1-ΔC mutant. The heterozygous mutant diploid strain was transformed with plasmids bearing the appropriate ORF under the control of the medium-strength nmt* promoter, ura+ leu+ transformants were sporulated, and spores were grown in selective medium. The full-length mouse protein and both mouse and S. pombe RBDs were capable of rescuing the sbp1 knockout mutant (Figure 6), suggesting that RanBP1 function is conserved between mouse and S. pombe and showing that the part(s) of that function required for viability in S. pombe are contained within the RBD. (Others have shown that full-length human RanBP1 can also rescue an S. cerevisiae YRB1 disruptant [Noguchi et al., 1997]). When transcription of the nmt promoter was turned off by the presence of thiamine in the medium, growth was inhibited, confirming that rescue was dependent on the expression of each of these proteins (Figure 6). The levels of expression of RanBP1 and sbp1p proteins in the four rescued strains were measured by immunoblotting of whole-cell extracts, and as noted previously, the RBD of mouse RanBP1 was reproducibly expressed at lower levels than the full-length sbp1p, RanBP1 (m), or RBD (Sp) proteins (Figure 6A).
Our strategy for disruption of the sbp1 gene replaced only the carboxyl-terminal region of the sbp1 ORF with ura4. It was therefore possible that rescued S. pombe sbp1-ΔC cells were viable because of transcomplementation between a chromosomally encoded sbp1p amino-terminal sequence and a plasmid-encoded RBD. To examine this possibility, we tested the previously described sbp1 genomic mutant sbp1-Δ1 (a generous gift from Dr. S. Sazer, Baylor College of Medicine; He et al., 1998), to see whether it could also be rescued with the same plasmids. sbp1-Δ1 was constructed by replacing the amino-terminal two-thirds of the ORF with the ura4 gene. As shown in Figure 6B, ura+ leu+ sbp1-Δ1 haploid spores that expressed plasmid-borne sbp1p, RanBP1 (m), RBD (Sp), or RBD (m) were viable in the absence of thiamine. This result supports the conclusion that each of these constructs can rescue a functional null mutation in sbp1.
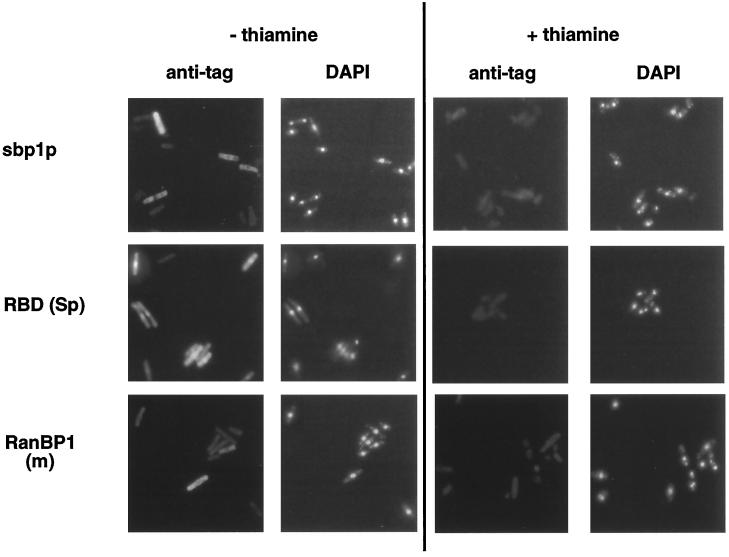
To compare the subcellular distribution of the RanBP1 proteins in rescued null mutant cells with those found when these proteins were expressed in wild-type S. pombe (Figure 4), haploid sbp1-ΔC genomic mutants expressing T7 or HA epitope-tagged forms of the four proteins were grown to midlog phase in the absence of thiamine or for an equal amount of time in its presence and examined by immunofluorescence microscopy (Figure 7). Full-length mouse and S. pombe proteins were concentrated in the cytoplasm, whereas the S. pombe RBD was predominantly localized to cell nuclei. We could not detect the distribution of RBD (m) because of its low level of expression. Cells grown in repressed conditions (+thiamine) showed no specific antibody staining, and the DAPI staining revealed condensed chromosomes (Figure 7).
Figure 7.
Localization of plasmid-encoded RanBP1, sbp1p, and RBD (Sp) in S. pombe sbp1-ΔC genomic mutant cells. Cells transformed with the indicated nmt* promoter-expressing constructs were grown to midlog phase in the absence of thiamine or for the same period in its presence. Cells were fixed with paraformaldehyde, permeabilized, and stained with monoclonal anti-T7 antibody [sbp1p, RanBP1 (m)] or anti-HA antibody [RBD (Sp)], plus FITC-conjugated goat anti-mouse Ig. Cellular DNA was detected by DAPI staining.
The wild-type growth properties of S. pombe sbp1-ΔC genomic mutant cells rescued with each of the four constructs (Figure 6) implied that macromolecular traffic between nuclear and cytoplasmic compartments in these cells was normal. To examine the one aspect of this traffic that can readily be assayed directly in S. pombe, NLS-mediated nuclear protein import was monitored by transforming the rescued S. pombe sbp1-ΔC genomic mutants with a plasmid encoding GST-GFP-NLS under the constitutive adh promoter. Cells grown to midlog phase in the absence of thiamine or for an equal amount of time in its presence were examined by fluorescence microscopy to determine the subcellular localization of the GST-GFP-NLS reporter protein (Figure 8). In the absence of thiamine (derepressed conditions), the reporter protein was consistently localized to cell nuclei. In the presence of thiamine (repressed conditions), cells were growth arrested with a large fraction of septated, chromatin-condensed cells, and the reporter protein was diffusely localized. This mislocalization of the NLS protein, when the plasmid-driven expression of sbp1p, RanBP1 (m), RBD (Sp), or RBD (m) is repressed, as noted previously, is a consequence of the loss of the nuclear envelope integrity (He et al., 1998).
Figure 8.
NLS nuclear protein import in rescued S. pombe sbp1-ΔC genomic mutant cells. sbp1-ΔC mutant cells rescued with the indicated nmt* constructs were transformed with a GST-GFP-NLS expressing construct. Cells were grown to midlog phase in the absence of thiamine or for an equal amount of time in its presence, fixed in methanol, and stained with DAPI before mounting. The GFP signal was analyzed by direct fluorescence microscopy.
The RBDs of Mammalian RanBP2 Can Also Rescue an S. pombe sbp1 Null Mutant
As discussed in INTRODUCTION, mammalian RanBP2, a component of the cytoplasmic fibrillar extensions of the nuclear pore complex, contains four RBDs (Figure 1): RBDs 1 and 2 interact strongly with Ran·GTP, whereas the interaction of RBDs 3 and 4 is weaker (Yokoyama et al., 1995). To test whether any of these RBDs can also functionally substitute for sbp1p in S. pombe, the sbp1ΔC genomic mutant was transformed with a plasmid bearing HA-tagged RBD2 or RBD4 (strong and weak interaction with Ran·GTP, respectively) under the control of nmt promoters. Although cells expressing moderate levels of these RBDs, under the control of the medium-strength nmt* promoter, were inviable, cells expressing high levels of each protein driven by the full-strength nmt promoter were rescued (Figure 9A). Immunoblot analysis of sbp1-ΔC and wild-type S. pombe cells transformed with the HA-tagged RBD2, RBD4, and RBD (Sp) under the control of the full-strength nmt promoter revealed that the levels of expression of all three proteins detected with the HA-epitope mAb were similar (Figure 9A; our unpublished results). These Western blot analyses confirmed that larger amounts of RBD2 and RBD4 proteins were required to rescue the sbp1-ΔC deletion mutant than were required of the RanBP1, sbp1p, RBD (m), and RBD (Sp) proteins (Figures 6A and 9A). The sbp1-Δ1 mutant (He et al., 1998) was also rescued with full-strength nmt expression of the RBD2 or RBD4 of RanBP2 (our unpublished results).
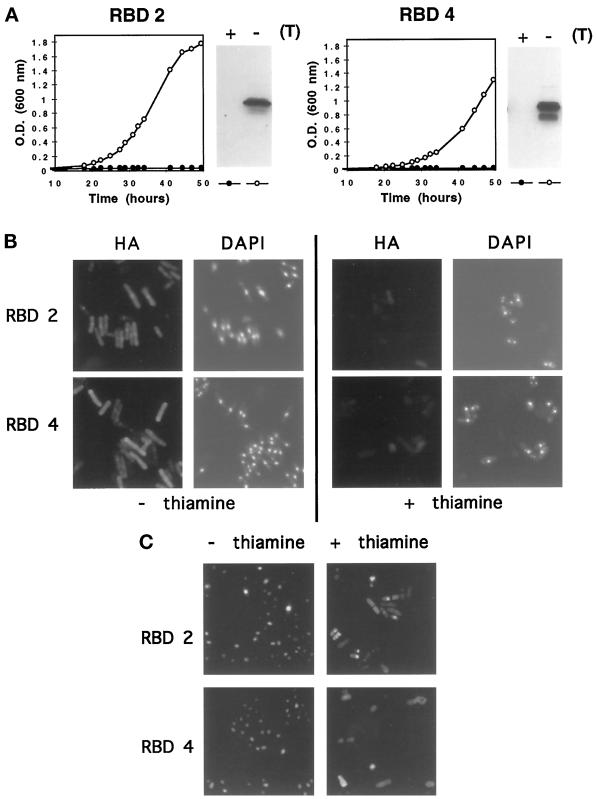
Figure 9.
Cell growth, protein expression, and nuclear import in the S. pombe sbp1-ΔC genomic mutant rescued with RBDs 2 and 4 of RanBP2. (A) Growth in liquid media in the presence (repressed, closed circles) or absence (derepressed, open circles) of thiamine and detection by immunoblotting of HA-tagged proteins present in 20 μg of total cell extracts. sbp1-ΔC was rescued with HA-tagged RBD2 or RBD4 expressed from full-strength nmt promoter plasmids. (B) Immunodetection of HA-tagged RBD2 and RBD4 of RanBP2, and DAPI staining of cellular DNA, in cells grown in the presence or absence of thiamine. (C) GST-GFP-NLS localization in sbp1-ΔC cells rescued by RBD2 or RBD4, grown in the presence or absence of thiamine, and analyzed as described in the legend to Figure 8.
The growth of cells rescued with RBD2 of RanBP2 was comparable to that of cells rescued with the RBDs of RanBP1 (m) or sbp1p (Figures 6A and 9A). Cultures of cells rescued with RBD4 grew more slowly but to the same final density. The distribution of RBD2 and RBD4 and their effects on nuclear import of an NLS reporter protein were also assayed. In the absence of thiamine (derepressed conditions), RBD2 (detected with HA epitope antibody) was mostly localized in the nucleus, whereas RBD4 was dispersed over the cytoplasm and nucleus (Figure 9B). Under these conditions, both rescued genomic mutants exhibit efficient transport of the NLS reporter protein to cell nuclei (Figure 9C). As expected, in the presence of thiamine (repressed conditions) the NLS protein was distributed throughout the cell. These results show that two RanBP2 RBDs, despite their different affinities for Ran·GTP, can complement the genomic disruption of sbp1, although very high levels of each protein are required. It is possible that the lower Ran·GTP binding affinity of RBD4 is responsible for the different distribution of RBD4 compared with RBD2 and thus affects the growth of rescued cells.
DISCUSSION
The small cytosolic Ran binding protein, RanBP1, is as ubiquitous as Ran, although not as well conserved. As discussed in INTRODUCTION, the precise role of RanBP1 in overall Ran function has not been elucidated unambiguously, but most data strongly suggest that RanBP1 is required in the cytosol both to disrupt Ran·GTP–karyopherin β (importin and exportin) complexes, and to promote RanGAP-stimulated GTP hydrolysis. The proper directionality of nuclear import and export processes would appear to depend on such RanBP1 function being restricted to the cytosol, and it has been suggested that nuclear mislocalization of RanBP1 would interfere significantly, perhaps lethally, with nuclear-cytosolic trafficking. In this communication we demonstrate that, at least in the case of S. pombe, this is clearly not the case.
In both transfected mammalian cells and transformed S. pombe, exogenous RanBP1 (m) and sbp1p proteins localized to the cytosol, whereas their RBDs localized to the nucleus. In previous studies of mammalian RanBP1 proteins, a carboxyl-terminal leucine-rich NES has been identified, distinct from the protein’s RBD (residues 178–189 and 27–158, respectively; Figure 1), which mediates the cytosolic localization of RanBP1 (Richards et al., 1996; Zolotukhin and Felber, 1997). The RanBP1 NES is well conserved among vertebrate proteins but is absent from the homologous yeast proteins (Figure 1). Despite its lack of a canonical NES, however, exogenous full-length sbp1p localized predominantly to the cytosol of both mammalian and S. pombe cells (Figures 2 and 4). In contrast, the isolated RBD of sbp1p localized predominantly to the nuclei of both cell types. This result suggests that the polar, leucine-free 74-residue amino-terminal portion of sbp1p mediates the cytosolic localization of the protein, via a mechanism that operates in both yeast and mammalian cells. This result also suggests that the carboxyl-terminal 11 residues of sbp1p, which appear not to form part of its RBD based on sequence homology, do not play a role in subcellular localization of the protein in either yeast or mammalian cells, inasmuch as they were retained in our sbp1p-RBD construct (Figure 1). Mutagenesis and protein–protein interaction studies will be needed to identify the protein sequence motif(s) responsible for this novel localization process and to work out its mechanism. S. pombe cells are nevertheless capable of recognizing the canonical NES motif, as shown by the predominantly cytosolic localization of full-length mouse RanBP1, and the predominantly nuclear localization of its NES-free RBD in transformed S. pombe cells (Figure 4).
Expression of high levels of exogenous sbp1p inhibited cell proliferation in S. pombe (Figure 3), consistent with the previous result of He et al. (1998). Expression of high levels of exogenous RanBP1 (m) and RBD (Sp) also inhibited cell proliferation (Figure 3). The toxicity of mouse RanBP1 is consistent with the hypothesis that RanBP1 function is well conserved. The effects of high levels of exogenous mouse RBD could not be tested, because none of our expression constructs yielded high steady-state levels of this polypeptide in yeast cells. The epitope-tagged mouse RBD polypeptide may be less efficiently translated or less stable than its yeast homologue or either full-length protein.
The phenotype associated with overexpression of RanBP1, sbp1p, or RBD (Sp) in S. pombe cells (Figures 4 and 5) was indistinguible from that described previously for S. pombe cells in which the endogenous pim1 (S. pombe Ran guanine nucleotide exchange factor) gene had been inactivated or in which the sbp1 or rna1 (RanGAP) genes had either been inactivated or overexpressed (Demeter et al., 1995; Matynia et al., 1996; He et al., 1998). In contrast, expression of moderate levels of exogenous RanBP1 (m), sbp1p, or their RBDs had no measurable effect on either viability or NLS-mediated nuclear protein import in mammalian or S. pombe cells (Figures 2, 3, and 5). This result was unexpected, inasmuch as previous studies had suggested that mislocalization of RanBP1 to the nucleus in mammalian and Xenopus assay systems disrupted trafficking of macromolecules between the nucleus and the cytosol (Richards et al., 1996; Izaurralde et al., 1997). Interpretation of these results, however, may be complicated by the presence of endogenous RanBP1 or sbp1p.
Elimination of functional endogenous RanBP1 in the sbp1-ΔC mutant strain of S. pombe allowed us to circumvent these complications. Haploid mutant cells were inviable but could be rescued by expression of moderate levels of wild-type sbp1p, by expression of moderate levels of full-length mouse RanBP1 (m) or RBD (m) or RBD (Sp), or by expression of high levels of RBD2 or -4 of the human nucleoporin RanBP2 (Figures 6 and 9). In all cases, rescued S. pombe cells exhibited wild-type levels of nuclear protein import. sbp1− mutant cells rescued with RBDs or with full-length mouse RanBP1 grew to the same density as did sbp1+ cells or mutant cells rescued with full-length sbp1p, and all cultures except those rescued with RBD4 of human RanBP2 grew at rates similar to wild type. These results suggest that all functions of sbp1p essential for the viability of S. pombe can be mediated by the isolated RBD of the protein. The results also suggest that these functions are evolutionarily well conserved, inasmuch as RBDs of human RanBP2 can also substitute for sbp1p to restore wild-type function.
In the rescued sbp1− mutant cells, exogenous RBDs localized predominantly to cell nuclei, and exogenous full-length proteins localized predominantly to the cytosol, the same patterns as were observed when the proteins were expressed in sbp1+ S. pombe cells (Figures 4, 7, and 9; our unpublished results). These results are consistent with the previous predictions that RanBP1/RBD function is carried out in the cytosol, inasmuch as rescued S. pombe cells contained significant amounts of exogenous protein in that compartment. It remains necessary to explain how the cells can tolerate high nuclear levels of a protein that is normally confined to the cytoplasm and that, when present in the nucleus either as an isolated RBD or in its full-length form, disrupts nuclear–cytoplasmic trafficking (Richards et al., 1996; Izaurralde et al., 1997). Although we cannot exclude the possibility that nuclear RBD is somehow inactivated in our expression studies in yeast and mammalian cells, a more plausible hypothesis is that nuclear RanBP1/sbp1p or their RBDs might disrupt several processes, by different mechanisms and with different toxic concentration thresholds. In the nucleus, full-length RanBP1 or RBD might compete for factors such as Ran·GTP required for nuclear protein export, might perturb guanine nucleotide turnover on Ran, or might perturb the turnover of transport complexes. Assessing the effects of these processes on the successful rescue of sbp1-ΔC S. pombe and the reported functional disruption of Xenopus and mammalian nuclei will require additional work in each case to determine the functional states of the exogenous proteins, their concentrations, and their localizations within the nucleus. At the same time, our results are difficult to reconcile with models that require the maintenance of a fixed ratio of cytosolic to nuclear RanBP1 and would appear to be inconsistent with the specific hypothesis that the function of RanBP1/RBD in nuclear protein import (or any other essential cell process) requires it to be excluded from the cell nucleus.
Indeed, RanBP1/sbp1p may normally enter cell nuclei. Human RanBP1 proteins in which single leucine or valine residues within the carboxyl-terminal NES are replaced by alanine accumulate in the cell nucleus (Richards et al., 1996; Zolotukhin and Felber, 1997), suggesting that RanBP1 can readily enter the cell nucleus, and that an efficient export mechanism is required to maintain its predominantly cytoplasmic localization. The localization of mammalian RanGAP/yeast Rna1p to the cytoplasm appears likewise to be dynamic. Both mammalian and yeast proteins contain NES and NLS motifs, and Rna1p accumulates in the nuclei of Crm1p (exportin)-deficient S. cerevisiae cells (Matunis et al., 1998; Feng et al., 1999). The NES motifs in both RanGAP and RanBP1 proteins could simply facilitate the efficient purging of these molecules from the nucleus when the nuclear membrane reforms at the end of mitosis. Even in yeast cells, whose nuclear membranes remain grossly intact during mitosis, such purging might be needed to restore high-level nuclear–cytoplasmic trafficking at the onset of interphase. At the same time, the interphase nucleus contains multiple functionally distinct subregions (Lamond and Earnshaw, 1998). It is possible that in some of these, presumably well separated from nuclear pores, controlled amounts of RanBP1 and RanGAP generate local high concentrations of Ran·GDP.
Besides RanBP1 and RanBP2, other mammalian and yeast proteins that contain RBDs have been described: S. pombe hba1p, budding yeast Yrb2p, and human RanBP3. All three proteins are predominantly nuclear and share a sequence motif with limited but significant similarities to the RBD motif of RanBP1 and RanBP2. In assays in vitro, RanBP3 and Yrb2p bind Ran·GTP. The function of hba1p is unknown, but it is essential for cell viability in S. pombe. The budding yeast protein Yrb2p interacts in vivo with the budding yeast homologues of RanGAP (Rna1p) and RCC1 (Ran guanine nucleotide exchange factor, Prp20p) and appears to have a role in nuclear protein export (Turi et al., 1996; Noguchi et al., 1997; Taura et al., 1997, 1998; Mueller et al., 1998). However, two results suggest indirectly that Ran may not be the target GTPase of hba1p/Yrb2p/RanBP3. First, interactions between RanBP3 family proteins and Ran·GTP are weak and may not occur to a significant extent in vivo (Turi et al., 1996; Mueller et al., 1998). Second, in budding yeast, mutations in the YRB2 gene were deleterious but not lethal and had no measurable effects on nuclear protein import or mRNA export. The defects in yrb2-mutant cells were not affected by expression of a chimeric Yrb2 protein containing the RBD of Yrb1p (budding yeast RanBP1) (Noguchi et al., 1997; Taura et al., 1997). It is thus not surprising that hba1+ function cannot substitute for sbp1p in sbp1− S. pombe cells and unlikely that hba1+ function could contribute partially to the rescue of mutant S. pombe cells transformed with exogenous yeast and mammalian RBD constructs.
It is possible, however, perhaps even likely, that the rescue of sbp1− S. pombe by isolated RBDs is incomplete. Analysis of cell growth under conditions of stress or genome-wide scans for differences in patterns of gene expression (Brown and Botstein, 1999) may well reveal important physiological differences between cells using full-length sbp1p and cells using various RBDs. Indeed, a search for such differences could provide a useful systematic complement to mutational analysis of the amino-terminal region of sbp1p in studies to characterize the full range of functions of this protein in S. pombe.
ACKNOWLEDGMENTS
We thank Drs. Shelley Sazer, Eric Chang, S.L. Forsburg, David Frendeway, and J. Wu for gifts of strains, plasmids, and reagents and Dr. Elias Coutavas for isolating S. pombe sbp1 DNA clones. This work was supported by National Science Foundation grant MCB-9630675. I.N. was supported by a postdoctoral fellowship from the Ministerio de Educación y Cultura, Spain. DNA and protein sequence analyses were done at the Research Computer Resource of New York University Medical Center, supported by National Science Foundation grant DIR-8908095.
Abbreviations used:
- GFP
green fluorescent protein
- HA
hemagglutinin
- NES
nuclear export signal
- NLS
nuclear localization signal
- RanBP
Ran-binding protein
- RanGAP
Ran GTPase-activating protein
- RBD
Ran-binding domain
- RCC1
regulator of chromosome condensation 1
REFERENCES
- Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast. A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Avis JM, Clarke PR. Ran, a GTPase involved in nuclear processes: its regulators and effectors. J Cell Sci. 1996;109:2423–2427. doi: 10.1242/jcs.109.10.2423. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Battistoni A, Guarguaglini G, Degrassi F, Pittoggi C, Palena A, Di Matteo G, Pisano C, Cundari E, Lavia P. Deregulated expression of the RanBP1 gene alters cell cycle progression in murine fibroblasts. J Cell Sci. 1997;110:2345–2357. doi: 10.1242/jcs.110.19.2345. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Görlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Coactivation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Adam SA. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Adam SA. Different binding domains for Ran-GTP and Ran-GDP/RanBP1 on nuclear import factor p97. J Biol Chem. 1997;272:6818–6822. doi: 10.1074/jbc.272.10.6818. [DOI] [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Visser GD, Adam SA. RanBP1 stabilizes the interaction of Ran with p97 in nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutavas E, Ren M, Oppenheim JD, D’Eustachio P, Rush MG. Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature. 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- Dasso M. The role of the Ran GTPase pathway in cell cycle control and interphase nuclear functions. Prog Cell Cycle Res. 1995;1:163–172. doi: 10.1007/978-1-4615-1809-9_13. [DOI] [PubMed] [Google Scholar]
- Delphin C, Guan T, Melchior F, Gerace L. RanGTP targets p97 to RanBP2, a filamentous protein localized at the cytoplasmic periphery of the nuclear pore complex. Mol Biol Cell. 1997;8:2379–2390. doi: 10.1091/mbc.8.12.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter J, Morphew M, Sazer S. A mutation in the RCC1-related protein pim1 results in nuclear envelope fragmentation in fission yeast. Proc Natl Acad Sci USA. 1995;92:1436–1440. doi: 10.1073/pnas.92.5.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Hurt E. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu Rev Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- Feng W, Benko AL, Lee J-H, Stanford DR, Hopper AK. Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J Cell Sci. 1999;112:339–347. doi: 10.1242/jcs.112.3.339. [DOI] [PubMed] [Google Scholar]
- Floer M, Blobel G, Rexach M. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for Leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Sherman DA. General purpose tagging vectors for fission yeast. Gene. 1997;191:191–195. doi: 10.1016/s0378-1119(97)00058-9. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Truant R, Thorne L, Benson RE, Cullen BR. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Görlich D. Nuclear protein import. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- Görlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kostka S, Kraft R, Dingwall C, Laskey RA, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Laskey RA, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Guarguaglini G, Battistoni A, Pittoggi C, Di Matteo G, Di Fiore B, Lavia P. Expression of the murine RanBP1 and Htf9-c genes is regulated from a shared bidirectional promoter during cell cycle progression. Biochem J. 1997;325:277–286. doi: 10.1042/bj3250277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Hayashi N, Walcott NG, Azuma Y, Patterson TE, Bischoff FR, Nishimoto T, Sazer S. The identification of cDNAs that affect the mitosis-to-interphase transition in Schizosaccharomyces pombe, including sbp1, which encodes a spi1p-GTP-binding protein. Genetics. 1998;148:645–656. doi: 10.1093/genetics/148.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M, Zhang C, Avis JM, Hutchison CJ, Clarke PR. The role of the Ran GTPase in nuclear assembly and DNA replication: characterization of the effects of Ran mutants. J Cell Sci. 1998;111:3017–3026. doi: 10.1242/jcs.111.20.3017. [DOI] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Kose S, Takao T, Tachibana T, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 1995;368:415–419. doi: 10.1016/0014-5793(95)00699-a. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Silver PA. A GTPase controlling nuclear trafficking: running the right way or walking RANdomly? Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lounsbury KM, Beddow AL, Macara IG. A family of proteins that stabilize the Ran/TC4 GTPase in its GTP-bound conformation. J Biol Chem. 1994;269:11285–11290. [PubMed] [Google Scholar]
- Lounsbury KM, Macara IG. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin beta and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin beta. J Biol Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matynia A, Dimitrov K, Mueller U, He X, Sazer S. Perturbations in the spi1p GTPase cycle of Schizosaccharomyces pombe through its GTPase-activating protein and guanine nucleotide exchange factor components result in similar phenotypic consequences. Mol Cell Biol. 1996;16:6352–6362. doi: 10.1128/mcb.16.11.6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- McLeod M, Stein M, Beach D. The product of the mei3+ gene expressed under control of the mating type locus, induces meiosis and sporulation in fission yeast. EMBO J. 1987;6:729–736. doi: 10.1002/j.1460-2075.1987.tb04814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Gerace L. Two-way trafficking with Ran. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- Moore MS. Ran and nuclear transport. J Biol Chem. 1998;273:22857–22860. doi: 10.1074/jbc.273.36.22857. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Mueller L, Cordes VC, Bischoff FR, Ponstingl H. Human RanBP3, a group of nuclear RanGTP binding proteins. FEBS Lett. 1998;427:330–336. doi: 10.1016/s0014-5793(98)00459-1. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Nuclear export of proteins and RNAs. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- Nicolas F, Zhang C, Hughes M, Goldberg M, Watton S, Clarke P. Xenopus ran-binding protein 1: molecular interactions and effects on nuclear assembly in Xenopus egg extracts. J Cell Sci. 1997;110:3019–3030. doi: 10.1242/jcs.110.24.3019. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Noguchi E, Hayashi N, Nakashima N, Nishimoto T. Yrb2p, a Nup2p-related yeast protein, has a functional overlap with Rna1p, a yeast Ran-GTPase-activating protein. Mol Cell Biol. 1997;17:2235–2246. doi: 10.1128/mcb.17.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouspenski II, Mueller UW, Matynia A, Sazer S, Elledge SJ, Brinkley BR. Ran-binding protein-1 is an essential component of the Ran/RCC1 molecular switch system in budding yeast. J Biol Chem. 1995;270:1975–1978. doi: 10.1074/jbc.270.5.1975. [DOI] [PubMed] [Google Scholar]
- Panté N, Aebi U. Toward the molecular dissection of protein import into nuclei. Curr Opin Cell Biol. 1996;8:397–406. doi: 10.1016/s0955-0674(96)80016-0. [DOI] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Pu RT, Dasso M. The balance of RanBP1 and RCC1 is critical for nuclear assembly and nuclear transport. Mol Biol Cell. 1997;8:1955–1970. doi: 10.1091/mbc.8.10.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Richards SA, Lounsbury KM, Carey KL, Macara IG. A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush MG, Drivas G, D’Eustachio P. The small nuclear GTPase Ran: how much does it run? Bioessays. 1996;18:103–112. doi: 10.1002/bies.950180206. [DOI] [PubMed] [Google Scholar]
- Saitoh H, Pu R, Cavenagh M, Dasso M. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc Natl Acad Sci USA. 1997;94:3736–3741. doi: 10.1073/pnas.94.8.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H, Sparrow DB, Shiomi T, Pu RT, Nishimoto T, Mohun TJ, Dasso M. Ubc9p and the conjugation of SUMO-1 to RanGAP1 and RanBP2. Curr Biol. 1998;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G, Wong DH, Koepp DM, Silver PA. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 1995;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki K, Yanagida M. Functional dissection of the phosphorylated termini of fission yeast DNA topoisomerase II. J Cell Biol. 1992;119:1023–1036. doi: 10.1083/jcb.119.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Eder PS, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Taura T, Krebber H, Silver PA. A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc Natl Acad Sci USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura T, Schlenstedt G, Silver PA. Yrb2p is a nuclear protein that interacts with Prp20p, a yeast Rcc1 homologue. J Biol Chem. 1997;272:31877–31884. doi: 10.1074/jbc.272.50.31877. [DOI] [PubMed] [Google Scholar]
- Turi TG, Mueller UW, Sazer S, Rose JK. Characterization of a nuclear protein conferring brefeldin A resistance in Schizosaccharomyces pombe. J Biol Chem. 1996;271:9166–9171. doi: 10.1074/jbc.271.15.9166. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghoffer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- Wilken N, Senecal JL, Scheer U, Dabauvalle MC. Localization of the Ran-GTP binding protein RanBP2 at the cytoplasmic side of the nuclear pore complex. Eur J Cell Biol. 1995;68:211–219. [PubMed] [Google Scholar]
- Wilson AC, Freiman RN, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Yokoyama N, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- Yoneda Y. How proteins are transported from cytoplasm to the nucleus. J Biochem. 1997;121:811–817. doi: 10.1093/oxfordjournals.jbchem.a021657. [DOI] [PubMed] [Google Scholar]
- Zolotukhin AS, Felber BK. Mutations in the nuclear export signal of human Ran-binding protein RanBP1 block the Rev-mediated posttranscriptional regulation of Human Immunodeficiency Virus Type 1. J Biol Chem. 1997;272:11356–11360. doi: 10.1074/jbc.272.17.11356. [DOI] [PubMed] [Google Scholar]