Abstract
Lipophilic yeasts of the genus Malassezia are part of the normal cutaneous microflora and are considered one of the factors that trigger atopic dermatitis (AD). We isolated two strains of Malassezia from a healthy Japanese female. Analysis of the D1/D2 26S ribosomal DNA and internal transcribed spacer region sequences of the isolates suggested that they are new members of the genus Malassezia. We propose the name Malassezia japonica sp. nov. for the isolates. M. japonica is easily distinguished from the seven known lipophilic species by its ability to assimilate Tween 40 and Tween 60 and its inability to assimilate Tween 20 and Tween 80 and to grow at 40°C. Furthermore, by applying transparent dressings to the skin lesions of 36 patients with AD and the skin of 22 healthy subjects, M. japonica DNA was detected by a non-culture-based method consisting of nested PCR with M. japonica species-specific primers. M. japonica DNA was detected from 12 of the 36 patients (33.3%) and 3 of the 22 healthy subjects (13.6%). Although it is not known whether M. japonica plays a role in AD, this species was part of the microflora in both patients with AD and healthy subjects.
Lipophilic yeasts of the genus Malassezia are members of the normal human cutaneous microflora and are also associated with several skin diseases. It is strongly suspected that Malassezia species are responsible for pityriasis versicolor, seborrheic dermatitis, Malassezia folliculitis, and atopic dermatitis (AD) (2, 4, 5, 19). The genus Malassezia includes eight species: Malassezia globosa, M. restricta, M. obtusa, M. slooffiae, M. furfur, M. sympodialis, M. dermatis, and M. pachydermatis (7, 22). M. pachydermatis is not a lipophilic species and is associated several animal skin diseases (1, 8). Recently, our research group found M. dermatis on Japanese patients with AD (22). Much research has examined the relationships between these eight species and their roles as causative agents of disease or factors that trigger disease. Most studies indicate that pityriasis versicolor and seborrheic dermatitis are likely affected by M. globosa and M. sympodialis (3, 9, 10, 15). The distribution of Malassezia species in the skin lesions of AD patients and healthy subjects was previously compared by a non-culture-based method (nested PCR) that is not affected by the isolation medium (21). Of the members of the genus Malassezia, M. globosa and M. restricta were associated with disease in more than 90% of AD patients, while the other species were detected in less than 50% of the patients. In our survey of the cutaneous Malassezia microflora, we isolated a new Malassezia species from a healthy subject. In the present study, we propose a new species, M. japonica, for the isolates from this subject and investigated the skin surfaces of patients with AD and healthy subjects for the presence of this species.
MATERIALS AND METHODS
Yeast isolate.
Malassezia strains were isolated from the left forearm of a healthy 22-year-old Japanese female. OpSite transparent dressings (3 by 7 cm; Smith and Nephew Medical Ltd., Hull, United Kingdom) were applied to the scalp, back, arm, and nape of the neck of the subject. The samples were then transferred onto modified Leeming and Notman agar (LNA; 20 g of glucose, 50 g of malt extract, 1 g of polypeptone, 20 g of bile salts [Oxoid, Basingstoke, United Kingdom], 1% Tween 40, 0.2% glycerol, and 50 μg of chloramphenicol [Sankyo, Tokyo, Japan]) and incubated at 32°C.
Direct DNA sequencing of rRNA genes.
Nuclear DNA was extracted by the method of Makimura et al. (14). The D1/D2 26S rRNA and internal transcribed spacer (ITS) regions of the rRNA gene were sequenced directly from the PCR products by using primer pair NL-1 (5′-GCATATCAATAAGCGGAGGAAAAG) and NL-4 (5′-GGTCCGTGTTTCAAGACGG) (13) and primer pair pITS-F (5′-GTCGTAACAAGGTTAACCTGCGG) and pITS-R (5′-TCCTCCGCTTATTGATATGC) (20), respectively. The PCR products were sequenced with an ABI 310 DNA sequencer and a BigDye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer Applied Biosystems, Foster, Calif.), according to the instructions of the manufacturer.
Molecular phylogenetic analysis.
The sequences were aligned by using Clustal W software (23). For the neighbor-joining analysis (18), the distances between sequences were calculated by using the two-parameter model of Kimura (12). A bootstrap analysis was conducted with 100 replications (6).
Morphological, physiological, and chemotaxonomic characteristics.
Morphology was examined on LNA after incubation at 32°C for 7 days. Tween 20, 40, 60, and 80 utilization, catalase reactions, and diazonium blue B reactions were performed as described by Guého et al. (7). Ubiquinone molecules were identified by the method of Nakase and Suzuki (16).
Direct detection of DNA in samples from patients with AD and healthy subjects. (i) Subjects.
The microfloras of 36 AD outpatients (24 men and 12 women; age range, 20 to 64 years; mean age, 33.3 ± 10.5 years) at Tokyo Medical University Hospital and 22 healthy students (7 men and 15 women; age range, 19 to 25 years; mean age, 20.7 ± 1.6 years old) at Meiji Pharmaceutical University were analyzed. AD was diagnosed according to the criteria of Hanifin and Rajka (11). Routine skin care, including intermittent applications of mild steroid ointment or petrolatum, was administered before sampling. Written informed consent was obtained from each subject.
(ii) Design of M. japonica species-specific primers for PCR.
The sequences of the intergenic spacer (IGS) 1 region, which is located between the 18S and 5.8S rRNA genes of M. japonica and the phylogenetically closely related species M. furfur and M. obtusa, were determined. The IGS 1 region was amplified with primer 26SBF (5′-AGCT GCTGCCAATGCTAGCTC), which hybridizes to a sequence located at the end of the 26S rRNA gene, and primer Mala-R (5′-TACTGCTGTGAATGCTCCAGC), which hybridizes to a sequence located in the 5.8S rRNA gene, and by use of the following program: 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min. The amplified PCR products were directly sequenced with the primer pair 26SBF and Mala-R. A species-specific primer pair was designed on the basis of IGS 1 sequence analysis: primer JP-IGS1F (5′-GACTGCTGATAATGCTCCAGT) and primer JP-IGS1R (5′-GTCTGCTG ATAAGTCTCACTG). To investigate the specificities of these primers, we used the other Malassezia and relevant species listed in Table 1.
TABLE 1.
Specificities of the primers for M. japonica and related species
| Species | Straina | Specificity of primer pair:
|
|
|---|---|---|---|
| 26SBF and Mala-R | JP-IGSIF and JP-IGSIR | ||
| M. japonica sp. nov. | M 9966 | + | + |
| M. japonica sp. nov. | M 9967 | + | + |
| M. dermatis | M 9927 | − | − |
| M. dermatis | M 9929 | − | − |
| M. furfur | CBS 4162 | + | − |
| M. furfur | CBS 6000 | + | − |
| M. furfur | CBS 7982 | + | − |
| M. globosa | CBS 7966 | + | − |
| M. globosa | M 9972 | + | − |
| M. obtusa | CBS 7876 | + | − |
| M. obtusa | Clinical isolate 2-17 | + | − |
| M. pachydermatis | CBS 1879 | + | − |
| M. restricta | CBS 7991 | + | − |
| M. restricta | M 9976 | + | − |
| M. slooffiae | CBS 7956 | + | − |
| M. slooffiae | M 9980 | + | − |
| M. sympodialis | CBS 7222 | − | − |
| M. sympodialis | M 9978 | − | − |
| Candida albicans | CBS 562 | − | − |
| Candida guilliermondii | CBS566 | − | − |
| Candida parapsilosis | CBS 604 | − | − |
| Rhodotorula mucilaginosa | CBS 17 | − | − |
| Staphylococcus aureus | Clinical isolate 4-2 | − | − |
CBS, Centraal Bureau voor Schimmelcultures, Baarn, The Netherlands; M, Meiji Pharmaceutical University, Tokyo, Japan.
(iii) Analysis of M. japonica microflora.
Samples of Malassezia were collected by applying OpSite transparent dressings (3 by 7 cm) to erythematous lesions on the faces and necks of patients with AD and the faces and necks of healthy subjects. Malassezia DNA was extracted from the OpSite dressing by a previously described method (21). The DNA extracted (3 μl) from each sample was added to 47 μl of the PCR master mixture, which consisted of 5 μl of 10× PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2; Takara Inc., Shiga, Japan), 4 μl of 200 μM deoxynucleoside triphosphates (an equimolar mixture of dATP, dCTP, dGTP, and dTTP; Takara), 10 pmol of each primer, and 2.5 U of Ex TaqDNA polymerase (Takara). PCR was performed with primers 26SBF and Mala-R, with an initial denaturation at 94°C for 3 min, followed by 30 cycles of 30 s at 94°C, 1 min at 57°C, and 30 s at 72°C, with a final extension at 72°C for 10 min. In the nested PCR step, 1 μl of the first amplification product was added to a new reaction mixture with the same composition as the first one. The PCR with primers JP-IGS1F and JP-IGS1R consisted of an initial denaturation at 94°C for 3 min, followed by 30 cycles of 30 s at 94°C, 1 min at 44°C, and 30 s at 72°C, with a final extension at 72°C for 10 min.
Nucleotide sequence accession numbers.
The nucleotide sequences of the D1/D2 26S rRNA, ITS, and IGS regions determined in this study have been deposited with DDBJ (DNA Data Bank of Japan) and are listed in Table 2.
TABLE 2.
Accession numbers of the IGS, ITS, and D1/D2 26S rRNA sequences
| Species | Strain | Accession no.
|
||
|---|---|---|---|---|
| D1/D2 26S rRNA gene | ITS region | IGS 1 region | ||
| M. japonica sp. nov. | M 9966Ta | AB100599 | AB100599 | AB105063 |
| M 9967 | AB105199 | AB105199 | AB105064 | |
| M. furfura | CBS 4162 | AB111460 | ||
| CBS 6000 | AB111459 | |||
| CBS 7982T | AB105198 | AB105150 | ||
| Clinical isolate 2-9 | AB105151 | |||
| Clinical isolate 2-4 | AB105152 | |||
| Clinical isolate 2-521 | AB105153 | |||
| Clinical isolate 2-52 | AB105154 | |||
| M. obtusa | CBS 7876T | AB105197 | AB105158 | AB111461 |
| Clinical isolate 2-17 | AB105155 | |||
| Clinical isolate 2-20 | AB105156 | |||
| Clinical isolate 2-35 | AB105157 | |||
T, type strain.
RESULTS AND DISCUSSION
Molecular phylogenetic analysis and taxonomic characteristics.
The isolates formed a cluster with M. furfur and M. obtusa, with 100 and 99% bootstrap support on trees constructed by using the D1/D2 26S rRNA gene and ITS 1 sequences, respectively (Fig. 1A and B). The dissimilarities between the D1/D2 regions of the 26S rRNA genes of the isolates and those of the M. furfur and M. obtusa strains were 4.6% (27 of 582 bp) and 6.9% (40 of 580 bp), respectively. The ITS 1 regions of the isolates had 12.5 to 24.2% and 15.3 to 20.3% dissimilarities to those of the M. furfur and M. obtusa strains, respectively; and the ITS 2 regions of the isolates had 15.6 to 17.9% and 20.2 to 21.3% dissimilarities to those of the the M. furfur and M. obtusa strains, respectively. Since the divergence between the M. furfur and M. obtusa strains and our isolates is sufficient to resolve them as individual species, we propose the name M. japonica for the isolates (17, 20). The species epithet used here refers to the country where the species was discovered. The characteristics differentiating the new species, M. japonica, and the known Malassezia species are summarized in Table 3. M. japonica is easily distinguished from the other species by its ability to assimilate Tween 40 and Tween 60 and its inability to assimilate Tween 20 and Tween 80 and to grow at 40°C.
FIG. 1.
Phylogenetic trees constructed by using the D1/D2 26S rRNA sequences of M. japonica sp. nov. and related species of the class Ustilaginomycetes (A) and the ITS1 region of M. japonica sp. nov. and other members of the genus Malassezia (B). The DDBJ and GenBank accession numbers are indicated in parentheses. The numerals represent the confidence levels from 100 replicate bootstrap samplings (frequencies less than 50% are not indicated). Knuc, Kimura's parameter (12).
TABLE 3.
Physiological characteristics of M. japonica sp. nov. and other Malassezia speciesa
| Species | Growth on SAb at 32°C | Growth on mDixonc at
|
Catalase reaction | Utilization of:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| 32°C | 37°C | 40°C | 10% Tween 20 | 0.5% Tween 40 | 0.5% Tween 60 | 0.1% Tween 80 | |||
| M. japonica sp. nov. | − | + | + | − | + | − | ± | + | − |
| M. slooffiaed | − | + | + | + | + | ± or + | + | + | − |
| M. sympodialisd | − | + | + | + | + | − | + | + | + |
| M. furfurd | − | + | + | + | + | + | + | + | + |
| M. dermatis | − | + | + | + | + | + | + | + | + |
| M. globosad | − | + | ± or − | − | + | − | − | − | − |
| M. obtusad | − | + | ± or + | − | + | − | − | − | − |
| M. restrictad | − | + | + | − | − | − | − | − | − |
| M. pachydermatisd | + | + | + | + | ± or + | − | + | + | + |
+, positive; −, negative; ±, weakly positive.
SA, Sabouraud dextrose agar.
mDixon, modified Dixon agar.
Data are from Guého et al. (7).
Direct detection of M. japonica DNA by a non-culture-based method.
The sensitivity of the nested PCR assay was examined by using M. japonica DNA purified from the culture. The limit of detection for purified DNA was approximately 10 fg by the nested PCR assay. The specificities of the M. japonica species-specific primers are shown in Table 1. Our PCR primers amplified only the targeted M. japonica DNA and did not amplify the DNA of any other Malassezia species. We confirmed the absence of false-positive reactions by determining the DNA sequences of the PCR products after they were cloned in the pCR2.1 vector (Invitrogen), since various species, including bacteria and filamentous fungi, colonize the skin surface. We collected and analyzed 142 samples from 36 patients with AD and 66 samples from 22 healthy subjects. M. japonica DNA was detected in 12 patients (33.3%) and 3 healthy subjects (13.6%). A non-culture-based method (PCR) was previously developed to analyze the Malassezia microflora on the skin surface, since the isolation media and technique influence the growth of Malassezia species and the growth rates of each species differ. As a result, the detection rate is higher by the PCR-based method than by traditional culture methods (21). The previous study suggests that M. globosa and M. restricta make up the major part of the microflora in patients with AD. The frequency of detection of the new species, M. japonica, is the same as that of M. sympodialis and M. furfur.
In conclusion, we have described a novel species, M. japonica, isolated from the skin surface of a healthy Japanese subject. It is not known whether this microorganism plays a significant role in AD or other skin diseases. M. japonica was part of the microfloras in both patients with AD and healthy subjects.
Latin description of Malassezia japonica Sugita, Takashima, Kodama, Tsuboi, et Nishikawa.
In LNA, post dies 6 ad 32°C, cellulae vegetativae sphaericae, ovoideae vel ellipsoideae 2-5 × 2-7 μm; sympodiales gemmantes proferentes. Cultura xanthoalba, semi-nitida aut hebetata, rugosa, et butyracea et margo glabra aut lobulata. In agaro glucoso-peptonico Tween 40 et Tween 60 (0.5%) addito crescit. H2O2 hydrolysatur. Commutatio colori per diazonium caeruleum B positiva. GTC acid: deoxyribonucleati 60.4 mol%. Ubiquinonum majus Q-9 est. Teleomorphis ignota. Typus: JCM 11963T, ex cute, feminae sani, Tokyo, Japonia, 2002, T. Kodama (originaliter ut M 9966), conservatur in collectionibus culturarum quas “Japan Collection of Microorganisms,” Saitama, Japan, sustentat.
Description of Malassezia japonica Sugita, Takashima, Kodama, Tsuboi, et Nishikawa sp. nov.
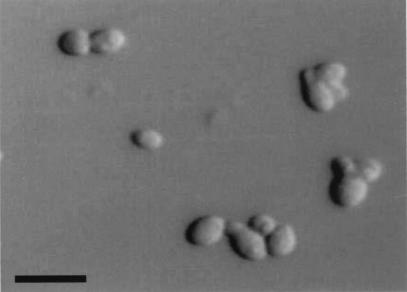
On LNA, after 6 days at 32°C, the vegetative cells are spherical, oval, or ellipsoidal and 2 to 5 by 2 to 7 μm, and sympodial budding is observed (Fig. 2). The colony is pale yellowish, semishining to dull, wrinkled, and butyrous and has an entire to lobed margin. The organism grows on glucose-peptone agar with 0.5% Tween 40 and Tween 60 as the sole source of lipid. The catalase reaction is positive. The diazonium blue B reaction is positive. The G+C content of nuclear DNA is 60.4 mol%, and the major ubiquinone is Q-9. The teleomorph is unknown.
FIG. 2.
Vegetative cells of M. japonica M 9966 grown in LNA for 7 days at 32°C. Bar, 10 μm.
JCM 11963T (CBS 9431T; originally strain M 9966) was isolated from the skin of a healthy Japanese subject in Tokyo, Japan, by M. Kodama in November 2002 and is maintained in the Japan Collection of Microorganisms (JCM), Saitama, Japan. The other strain, M 9967, has also been deposited in the JCM and CBS collections as strain 11964 and strain 9432, respectively.
Acknowledgments
This study was supported in part by a Grant for the Promotion of the Advancement of Education and Research in Graduate Schools from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by a grant from Iatron Laboratries Inc. (to A.N.).
REFERENCES
- 1.Akerstedt, J., and I. Vollset. 1996. Malassezia pachydermatis with special reference to canine skin disease. Br. Vet. J. 152:269-281. [DOI] [PubMed] [Google Scholar]
- 2.Ashbee, H. R., and E. G. Evans. 2002. Immunology of diseases associated with Malassezia species. Clin. Microbiol. Rev. 15:21-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aspiroz, C., M. Ara, M. Varea, A. Rezusta, and C. Rubio. 2002. Isolation of Malassezia globosa and M. sympodialis from patients with pityriasis versicolor in Spain. Mycopathologia 154:111-117. [DOI] [PubMed] [Google Scholar]
- 4.Faergemann, J. 1998. The role of Malassezia species in the ecology of human skin and as pathogens. Med. Mycol. 36(Suppl. 1):220-229. [PubMed] [Google Scholar]
- 5.Faergemann, J. 2002. Atopic dermatitis and fungi. Clin. Microbiol. Rev. 15:545-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 7.Guého, E., G. Midgley, and J. Guillot. 1996. The genus Malassezia with description of four new species. Antonie Leeuwenhoek 69:337-355. [DOI] [PubMed] [Google Scholar]
- 8.Guillot, J., and R. Bond. 1999. Malassezia pachydermatis: a review. Med. Mycol. 37:295-306. [DOI] [PubMed] [Google Scholar]
- 9.Gupta, A. K., Y. Kohli, J. Faergemann, and R. C. Summerbell. 2001. Epidemiology of Malassezia yeasts associated with pityriasis versicolor in Ontario, Canada. Med. Mycol. 39:199-206. [DOI] [PubMed] [Google Scholar]
- 10.Gupta, A. K., Y. Kohli, R. C. Summerbell, and J. Faergemann. 2001. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med. Mycol. 39:243-251. [DOI] [PubMed] [Google Scholar]
- 11.Hanifin, J. M., and G. Rajka. 1980. Diagnostic features of atopic dermatitis. Acta Dermatol. Venereol. 92:4-47. [Google Scholar]
- 12.Kimura, M. 1980. A simple method for estimation evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 13.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makimura, K., Y. S. Murayama, and H. Yamaguchi. 1994. Detection of a wide range of medically important fungal species by polymerase chain reaction (PCR). J. Med. Microbiol. 40:358-364. [DOI] [PubMed] [Google Scholar]
- 15.Nakabayashi, A., Y. Sei, and J. Guillot. 2000. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med. Mycol. 38:337-341. [DOI] [PubMed] [Google Scholar]
- 16.Nakase, T., and M. Suzuki. 1986. Bullera megalospora, a new species of yeast forming large ballistospores isolated from dead leaves of Oryza sativa, Miscanthus sinensis, and Sasa sp. in Japan. J. Gen. Appl. Microbiol. 32:225-240. [Google Scholar]
- 17.Peterson, S. W., and C. P. Kurtzman. 1991. Ribosomal RNA sequence divergence among sibling species of yeasts. Syst. Appl. Microbiol. 14:124-129. [Google Scholar]
- 18.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 19.Scheynius, A., C. Johansson, E. Buentke, A. Zargari, and M. T. Linder. 2002. Atopic eczema/dermatitis syndrome and Malassezia. Int. Arch. Allergy Immunol. 127:161-169. [DOI] [PubMed] [Google Scholar]
- 20.Sugita, T., A. Nishikawa, R. Ikeda, and T. Shinoda. 1999. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J. Clin. Microbiol. 37:1985-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugita, T., H. Suto, T. Unno, R. Tsuboi, H. Ogawa, T. Shinoda, and A. Nishikawa. 2001. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. J. Clin. Microbiol. 39:3486-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sugita, T., M. Takashima, T. Shinoda, H. Suto, T. Unn, R. Tsuboi, H. Ogawa, and A. Nishikawa. 2002. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J. Clin. Microbiol. 40:1363-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]