Abstract
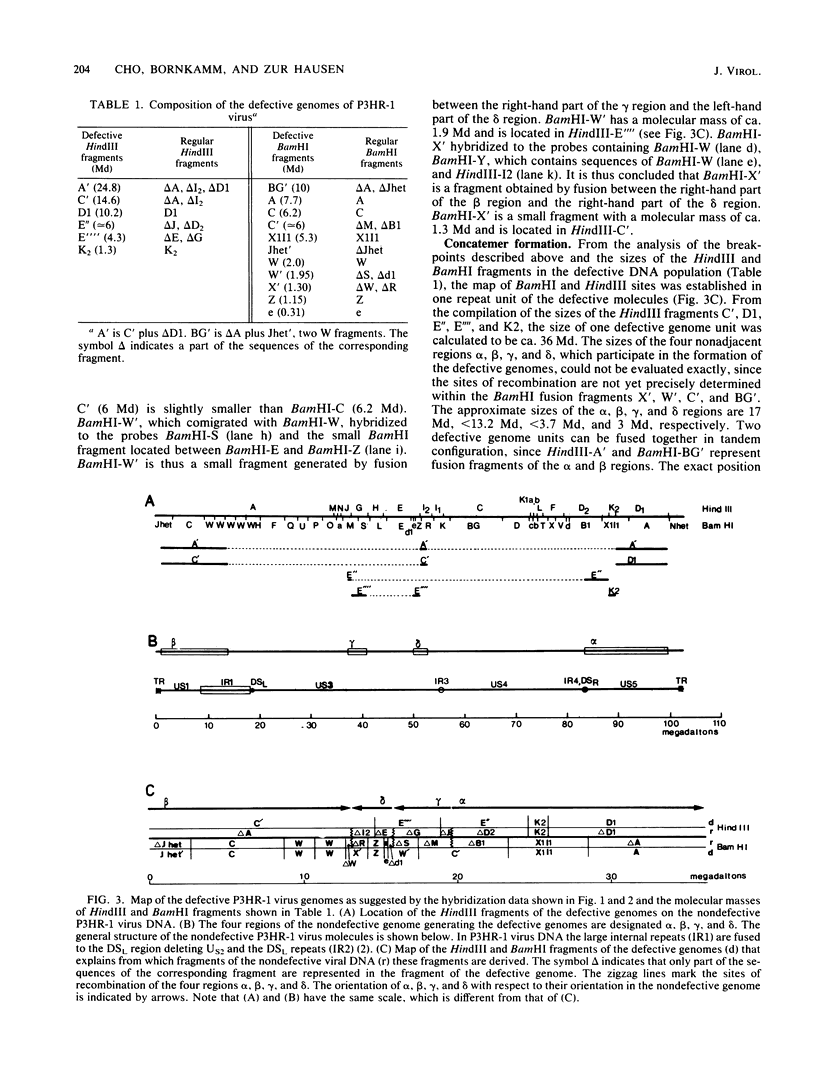
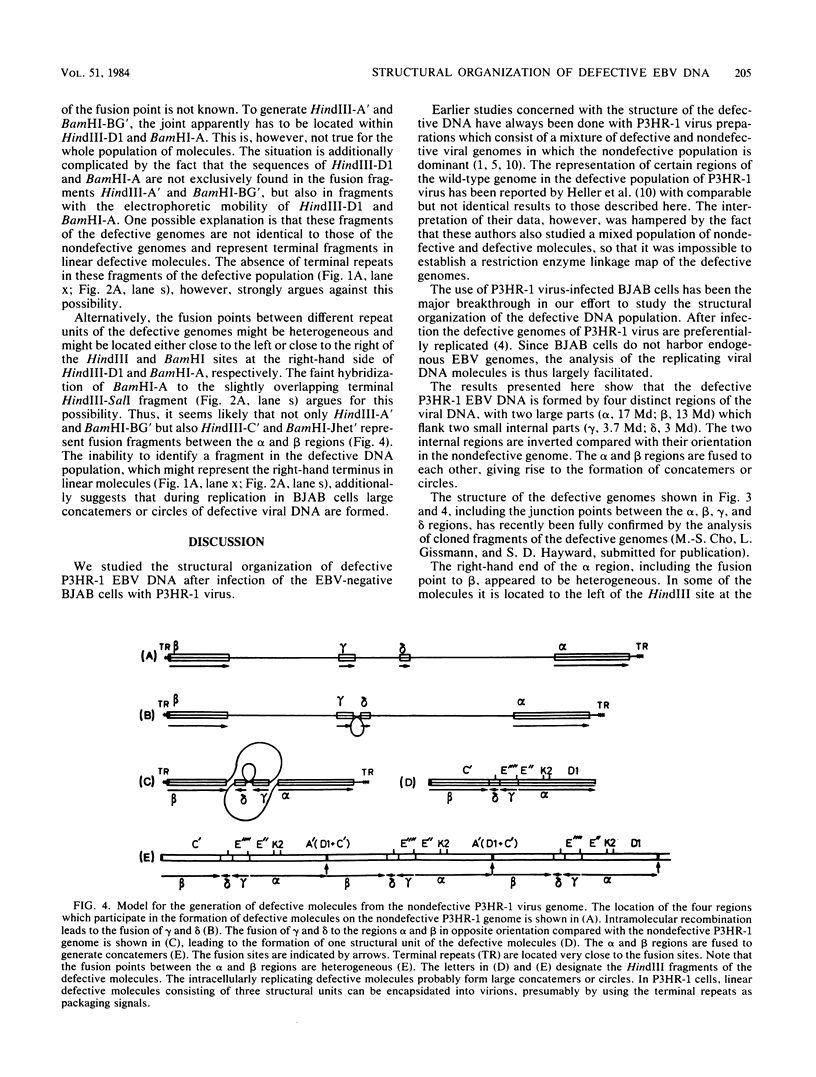
Epstein-Barr virus (EBV), isolated from P3HR-1 cells, induces early antigen and viral capsid antigen upon infection of human B-lymphoblasts. The strong early antigen- and viral capsid antigen-inducing activity is only observed in P3HR-1 virus preparations harboring particles with defective genomes, suggesting that this biological activity is directly associated with the defective DNA population. After infection of EBV genome-carrying Raji or EBV genome-negative BJAB cells, defective genomes of P3HR-1 EBV DNA are replicated in excess, depending on the multiplicity of infecting EBV particles. Hybridization of the DNA from such infected cells with 32P-labeled EBV DNA after HindIII cleavage reveals six hypermolar fragments. Mapping of these fragments shows that they form one defective genome unit containing four nonadjacent regions (alpha, beta, gamma, and delta) of the nondefective P3HR-1 EBV DNA. Two of the segments (alpha and beta) contain ca. 17 and 13 megadaltons, respectively, from the terminal regions of the P3HR-1 genome, whereas the two smaller segments (gamma and delta) contain ca. 3.7 and 3.0 megadaltons, respectively, originating from the central portion of the genome. In the defective molecule, the regions gamma and delta are present in the opposite orientation compared with nondefective P3HR-1 EBV DNA. Tandem concatemers are formed by fusion of the alpha and beta regions. Our model suggests that tandem concatemers of three defective genome units can be packaged into virions in P3HR-1 cells.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bornkamm G. W., Delius H., Zimber U., Hudewentz J., Epstein M. A. Comparison of Epstein-Barr virus strains of different origin by analysis of the viral DNAs. J Virol. 1980 Sep;35(3):603–618. doi: 10.1128/jvi.35.3.603-618.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm G. W., Hudewentz J., Freese U. K., Zimber U. Deletion of the nontransforming Epstein-Barr virus strain P3HR-1 causes fusion of the large internal repeat to the DSL region. J Virol. 1982 Sep;43(3):952–968. doi: 10.1128/jvi.43.3.952-968.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A., Kieff E. Long internal direct repeat in Epstein-Barr virus DNA. J Virol. 1982 Oct;44(1):286–294. doi: 10.1128/jvi.44.1.286-294.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M. S., Fresen K. O., zur Hausen H. Multiplicity-dependent biological and biochemical properties of Epstein-Barr virus (EBV) rescued from non-producer lines after superinfection with P3HR-1 EBV. Int J Cancer. 1980 Sep 15;26(3):357–363. doi: 10.1002/ijc.2910260316. [DOI] [PubMed] [Google Scholar]
- Delius H., Bornkamm G. W. Heterogeneity of Epstein-Barr virus. III. Comparison of a transforming and a nontransforming virus by partial denaturation mapping of their DNAs. J Virol. 1978 Jul;27(1):81–89. doi: 10.1128/jvi.27.1.81-89.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Fresen K. O., Cho M. S., zur Hausen H. Recovery of transforming EBV from non-producer cells after superinfection with non-transforming P3HR-1 EBV. Int J Cancer. 1978 Oct 15;22(4):378–383. doi: 10.1002/ijc.2910220403. [DOI] [PubMed] [Google Scholar]
- Fresen K. O., Hausen H. Establishment of EBNA-expressing cell lines by infection of Epstein-Barr virus (EBV)-genome-negative human lymphoma cells with different EBV strains. Int J Cancer. 1976 Feb 15;17(2):161–166. doi: 10.1002/ijc.2910170203. [DOI] [PubMed] [Google Scholar]
- Fresen K. O., Merkt B., Bornkamm G. W., Hausen H. Heterogeneity of Epstein-Barr virus originating from P3HR-1 cells. I. Studies on EBNA induction. Int J Cancer. 1977 Mar 15;19(3):317–323. doi: 10.1002/ijc.2910190306. [DOI] [PubMed] [Google Scholar]
- Heller M., Dambaugh T., Kieff E. Epstein-Barr virus DNA. IX. Variation among viral DNAs from producer and nonproducer infected cells. J Virol. 1981 May;38(2):632–648. doi: 10.1128/jvi.38.2.632-648.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle W., Henle G., Zajac B. A., Pearson G., Waubke R., Scriba M. Differential reactivity of human serums with early antigens induced by Epstein-Barr virus. Science. 1970 Jul 10;169(3941):188–190. doi: 10.1126/science.169.3941.188. [DOI] [PubMed] [Google Scholar]
- Heston L., Rabson M., Brown N., Miller G. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature. 1982 Jan 14;295(5845):160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- Hinuma Y., Konn M., Yamaguchi J., Wudarski D. J., Blakeslee J. R., Jr, Grace J. T., Jr Immunofluorescence and herpes-type virus particles in the P3HR-1 Burkitt lymphoma cell line. J Virol. 1967 Oct;1(5):1045–1051. doi: 10.1128/jvi.1.5.1045-1051.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudewentz J., Delius H., Freese U. K., Zimber U., Bornkamm G. W. Two distant regions of the Epstein-Barr virus genome with sequence homologies have the same orientation and involve small tandem repeats. EMBO J. 1982;1(1):21–26. doi: 10.1002/j.1460-2075.1982.tb01118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G., Lindahl T., Jondal M., Leibold W., Menézes J., Nilsson K., Sundström C. Continuous lymphoid cell lines with characteristics of B cells (bone-marrow-derived), lacking the Epstein-Barr virus genome and derived from three human lymphomas. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3283–3286. doi: 10.1073/pnas.71.8.3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller B., Delius H., Bünemann H., Müller W. The isolation of DNA from agarose gels by electrophoretic elution onto malachite green-polyacrylamide columns. Gene. 1978 Nov;4(3):227–239. doi: 10.1016/0378-1119(78)90020-3. [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Yajima Y., Nonoyama M. Mechanism of infection by Epstein-Barr virus. II. Comparison of viral DNA from HR-1 and superinfected Raji cells by restriction enzymes. Virology. 1977 Aug;81(1):17–24. doi: 10.1016/0042-6822(77)90054-x. [DOI] [PubMed] [Google Scholar]
- Menezes J., Patel P., Dussault H., Bourkas A. E. Comparative studies on the induction of virus-associated nuclear antigen and early antigen by lymphocyte-transforming (B95-8) and nontransforming (P3HR-1) strains of Epstein-Barr virus. Intervirology. 1978;9(2):86–94. doi: 10.1159/000148926. [DOI] [PubMed] [Google Scholar]
- Miller G., Robinson J., Heston L., Lipman M. Differences between laboratory strains of Epstein-Barr virus based on immortalization, abortive infection, and interference. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4006–4010. doi: 10.1073/pnas.71.10.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PULVERTAFT J. V. A STUDY OF MALIGNANT TUMOURS IN NIGERIA BY SHORT-TERM TISSUE CULTURE. J Clin Pathol. 1965 May;18:261–273. doi: 10.1136/jcp.18.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab-Traub N., Dambaugh T., Kieff E. DNA of Epstein-Barr virus VIII: B95-8, the previous prototype, is an unusual deletion derivative. Cell. 1980 Nov;22(1 Pt 1):257–267. doi: 10.1016/0092-8674(80)90173-7. [DOI] [PubMed] [Google Scholar]
- Rabson M., Heston L., Miller G. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc Natl Acad Sci U S A. 1983 May;80(9):2762–2766. doi: 10.1073/pnas.80.9.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Shaw J. E., Seebeck T., Li J. L., Pagano J. S. Epstein-Barr virus DNA synthesized in superinfected Raji cells. Virology. 1977 Apr;77(2):762–771. doi: 10.1016/0042-6822(77)90497-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima Y., Marczynska B., Nonoyama M. Transforming activity of Epstein-Barr virus obtained by superinfection of Raji cells. Proc Natl Acad Sci U S A. 1978 Apr;75(4):2008–2010. doi: 10.1073/pnas.75.4.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima Y., Nonoyama M. Mechanisms of infection with Epstein-Barr virus. I. Viral DNA replication and formation of noninfectious virus particles in superinfected Raji cells. J Virol. 1976 Jul;19(1):187–194. doi: 10.1128/jvi.19.1.187-194.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H., O'Neill F. J., Freese U. K., Hecker E. Persisting oncogenic herpesvirus induced by the tumour promotor TPA. Nature. 1978 Mar 23;272(5651):373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]
- zur Hausen H., Schulte-Holthausen H., Klein G., Henle W., Henle G., Clifford P., Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970 Dec 12;228(5276):1056–1058. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]




