Abstract
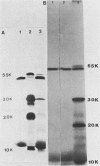
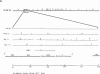
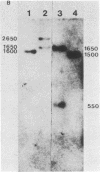
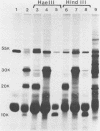
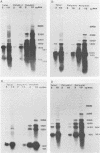
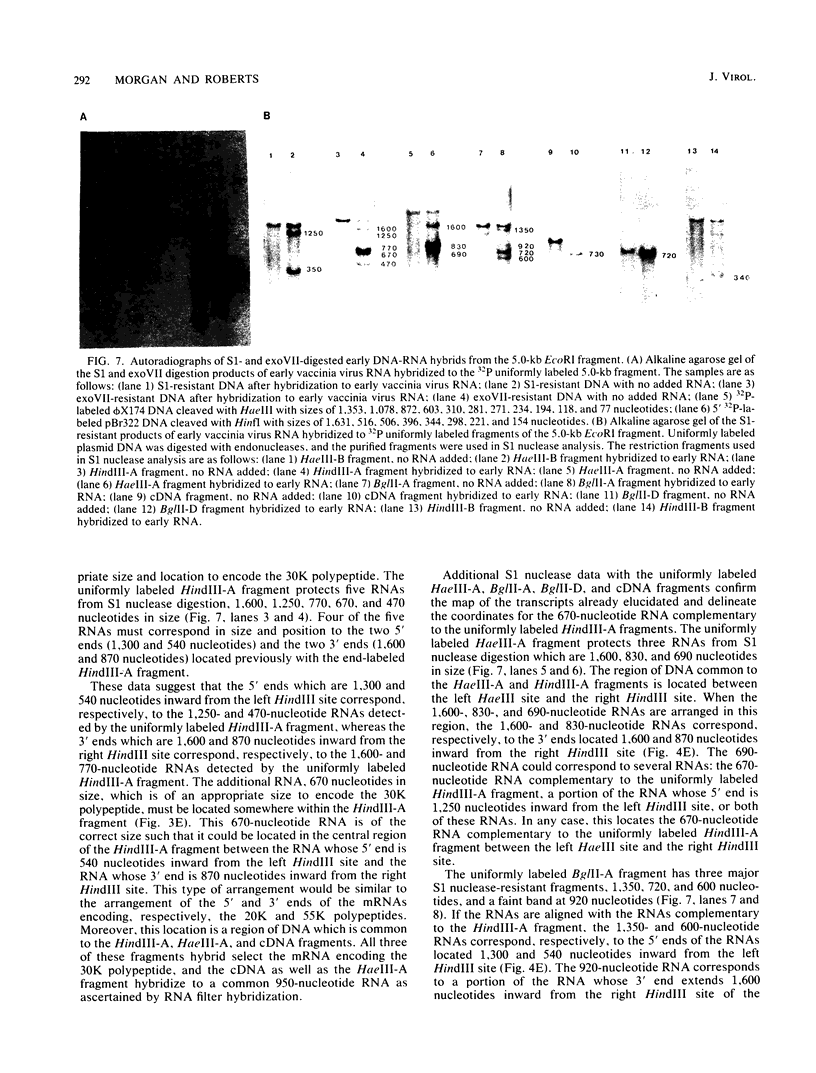
The detailed organization of the RNAs transcribed from an early gene cluster encoded by vaccinia virus has been determined from the information derived from several complementary techniques. These include hybrid selection coupled with cell-free translation to locate DNA sequences complementary to mRNAs encoding specific polypeptides; RNA filter hybridization to size and locate on the DNA mature RNAs as well as higher-molecular-weight RNAs; S1 nuclease mapping to precisely locate the 5' and 3' ends of the RNAs; S1 nuclease mapping to precisely locate the 5' and 3' ends of the RNAs; and fractionation of hybrid-selected mRNAs in an agarose gel containing methyl mercury hydroxide followed by the cell-free translation of these mRNAs to definitively ascertain the size of the mRNA encoding each polypeptide. The early gene cluster is located between 21 and 26 kilobases from the left end of the vaccinia virus genome and is encoded by a 5.0-kilobase EcoRI fragment which spans the HindIII-N, -M, and -K fragments. Transcribed towards the left terminus are four mature mRNAs, 1,450, 950, 780, and 400 nucleotides in size, encoding polypeptides of 55, 30, 20, and 10 kilodaltons, respectively. These mRNAs are colinear with the DNA template and are closely spaced such that the 5' terminus of one mRNA is within 50 base pairs of the 3' terminus of the adjacent RNA. In addition to the mature size mRNAs, there are higher-molecular-weight RNAs, 5,000, 3,300, 2,350, 2,300, 1,800, 1,700, and 1,350 nucleotides in size. The 5' and 3' termini of the high-molecular-weight RNAs are coterminal with the 5' and 3' termini of the mature size mRNA. The implications of this arrangement and the biogenesis of these early mRNAs are discussed.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Parker B. A., Reiser J., Renart J., Stark G. R., Wahl G. M. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- Alwine J. C., Kemp D. J., Stark G. R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Bajszár G., Wittek R., Weir J. P., Moss B. Vaccinia virus thymidine kinase and neighboring genes: mRNAs and polypeptides of wild-type virus and putative nonsense mutants. J Virol. 1983 Jan;45(1):62–72. doi: 10.1128/jvi.45.1.62-72.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Moss B. In vitro translation of immediate early, early, and late classes of RNA from vaccinia virus-infected cells. Virology. 1979 Jul 30;96(2):368–380. doi: 10.1016/0042-6822(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Moss B. Transcription of vaccinia virus mRNA coupled to translation in vitro. Virology. 1978 Jul 1;88(1):149–165. doi: 10.1016/0042-6822(78)90118-6. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Wittek R., Moss B. Extension of the transcriptional and translational map of the left end of the vaccinia virus genome to 21 kilobase pairs. J Virol. 1981 Sep;39(3):733–745. doi: 10.1128/jvi.39.3.733-745.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. A., Wittek R., Moss B. Hybridization selection and cell-free translation of mRNA's encoded within the inverted terminal repetition of the vaccinia virus genome. J Virol. 1981 Jan;37(1):284–294. doi: 10.1128/jvi.37.1.284-294.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban M., Metz D. H. Early virus protein synthesis in vaccinia virus-infected cells. J Gen Virol. 1973 May;19(2):201–206. doi: 10.1099/0022-1317-19-2-201. [DOI] [PubMed] [Google Scholar]
- Goldbach R. W., Evers R. F., Borst P. Electrophoretic strand separation of long DNAs with poly (U,G) in agarose gels. Nucleic Acids Res. 1978 Aug;5(8):2743–2754. doi: 10.1093/nar/5.8.2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golini F., Kates J. R. Transcriptional and translational analysis of a strongly expressed early region of the vaccinia virus genome. J Virol. 1984 Feb;49(2):459–470. doi: 10.1128/jvi.49.2.459-470.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mackett M., Archard L. C. Conservation and variation in Orthopoxvirus genome structure. J Gen Virol. 1979 Dec;45(3):683–701. doi: 10.1099/0022-1317-45-3-683. [DOI] [PubMed] [Google Scholar]
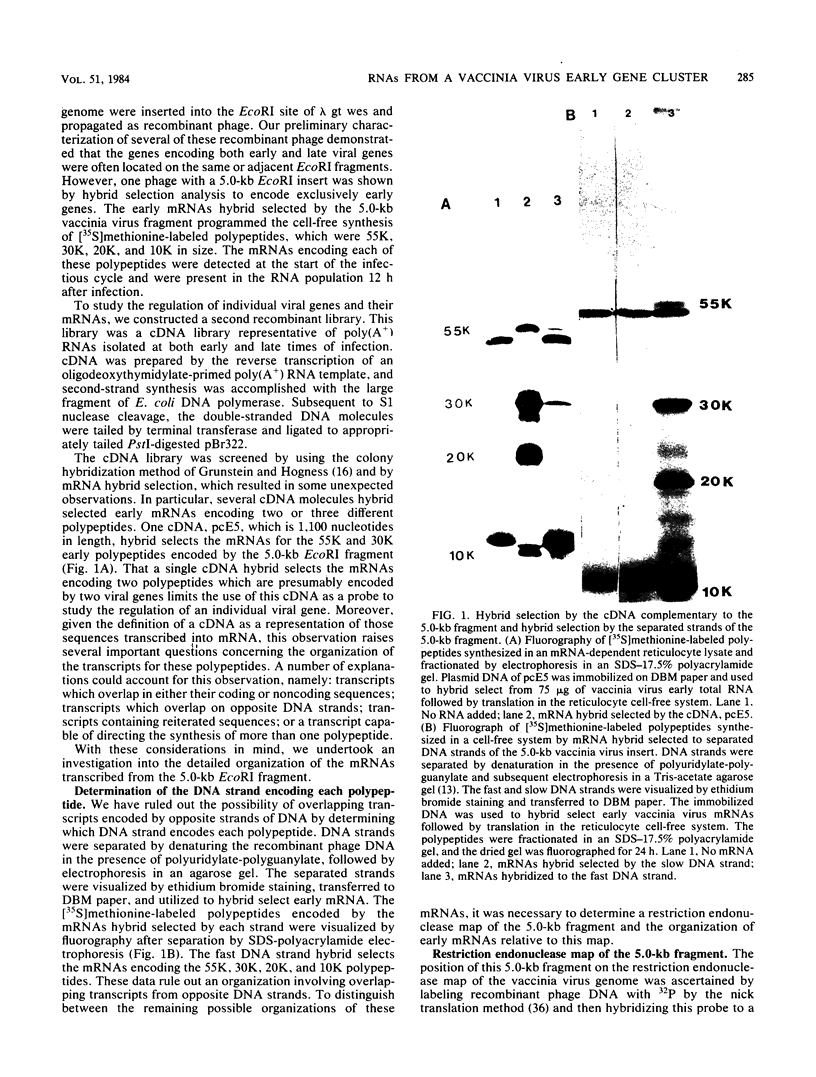
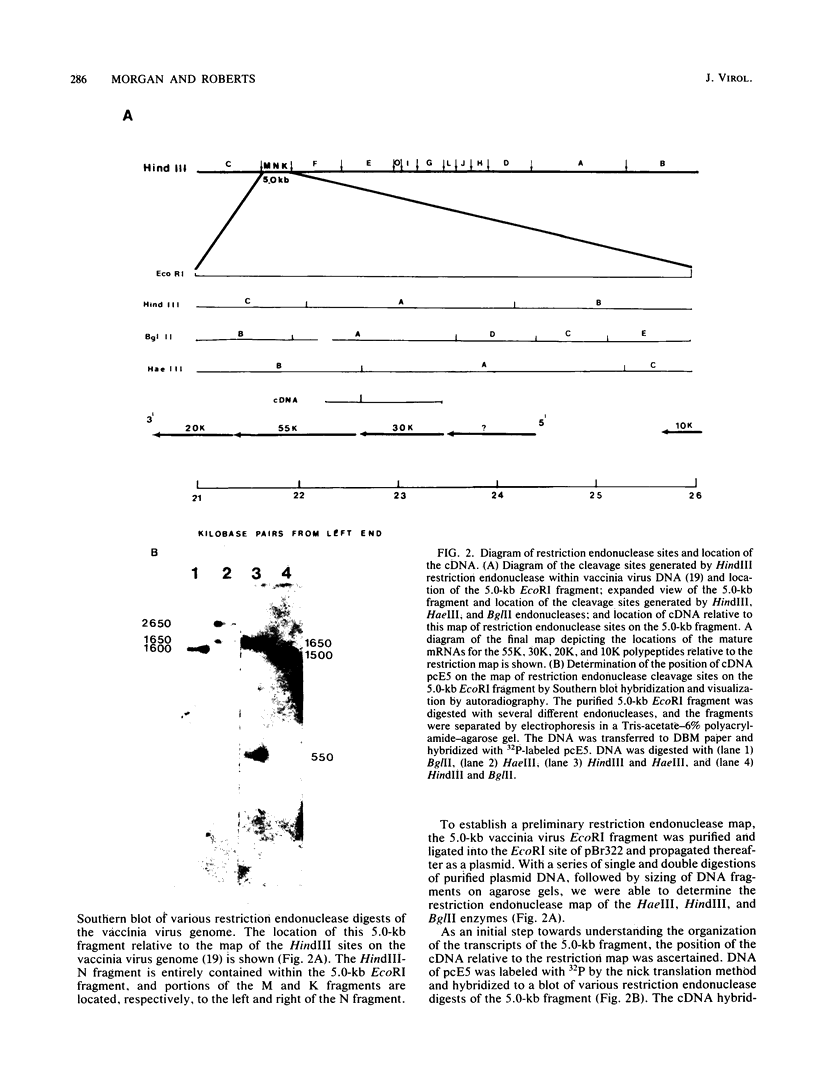
- Mahr A., Roberts B. E. Organization of six early transcripts synthesized from a vaccinia virus EcoRI DNA fragment. J Virol. 1984 Feb;49(2):497–509. doi: 10.1128/jvi.49.2.497-509.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonell M. W., Simon M. N., Studier F. W. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J Mol Biol. 1977 Feb 15;110(1):119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- Miller J. S., Ricciardi R. P., Roberts B. E., Paterson B. M., Mathews M. B. Arrangement of messenger RNAs and protein coding sequences in the major late transcription unit of adenovirus 2. J Mol Biol. 1980 Oct 5;142(4):455–488. doi: 10.1016/0022-2836(80)90258-2. [DOI] [PubMed] [Google Scholar]
- Moss B. Inhibition of HeLa cell protein synthesis by the vaccinia virion. J Virol. 1968 Oct;2(10):1028–1037. doi: 10.1128/jvi.2.10.1028-1037.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Salzman N. P. Sequential protein synthesis following vaccinia virus infection. J Virol. 1968 Oct;2(10):1016–1027. doi: 10.1128/jvi.2.10.1016-1027.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Winters E., Cooper J. A. Deletion of a 9,000-base-pair segment of the vaccinia virus genome that encodes nonessential polypeptides. J Virol. 1981 Nov;40(2):387–395. doi: 10.1128/jvi.40.2.387-395.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicali D., Davis S. W., Mercer S. R., Paoletti E. Two major DNA variants present in serially propagated stocks of the WR strain of vaccinia virus. J Virol. 1981 Mar;37(3):1000–1010. doi: 10.1128/jvi.37.3.1000-1010.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
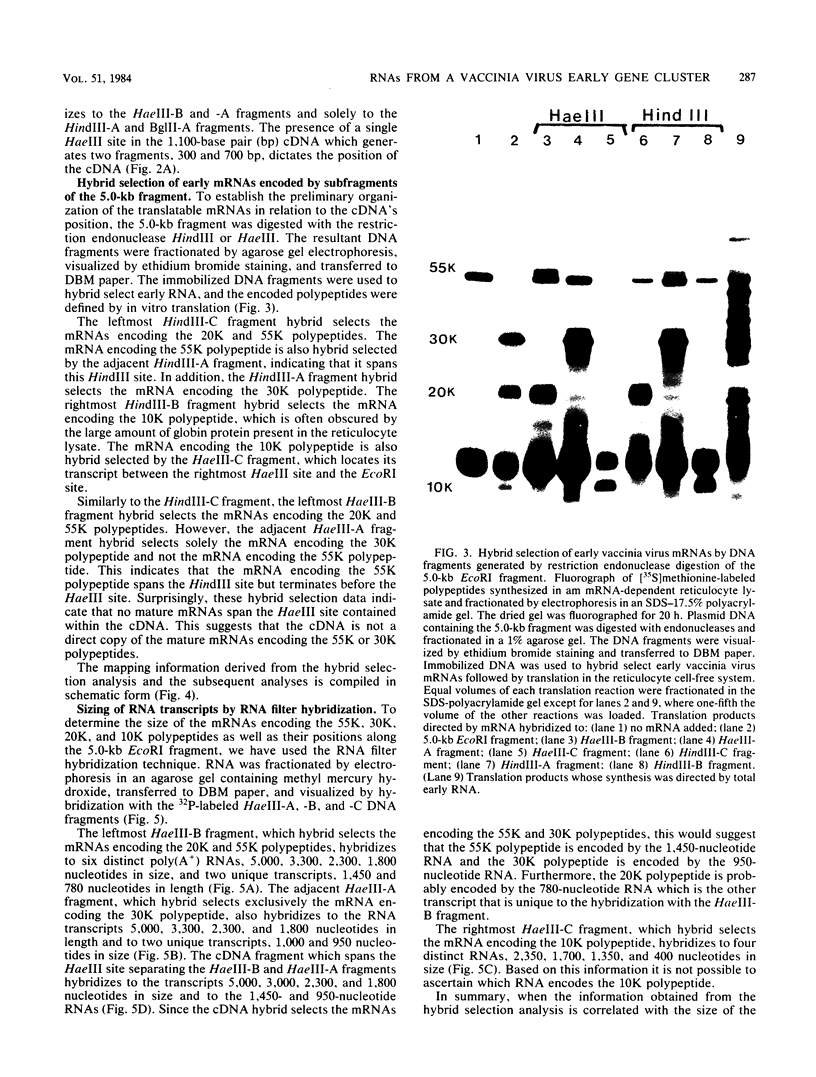
- Panicali D., Davis S. W., Weinberg R. L., Paoletti E. Construction of live vaccines by using genetically engineered poxviruses: biological activity of recombinant vaccinia virus expressing influenza virus hemagglutinin. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5364–5368. doi: 10.1073/pnas.80.17.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti E. High molecular weight virion-associated RNA of vaccinia. A possible precursor to 8 to 12 S mRNA. J Biol Chem. 1977 Feb 10;252(3):872–877. [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Sykes J. M., Hunt T. Characteristics of a coupled cell-free transcription and translation system directed by vaccinia cores. Eur J Biochem. 1978 Jan 2;82(1):199–209. doi: 10.1111/j.1432-1033.1978.tb12012.x. [DOI] [PubMed] [Google Scholar]
- Pelham H. R. Use of coupled transcription and translation to study mRNA production by vaccinia cores. Nature. 1977 Oct 6;269(5628):532–534. doi: 10.1038/269532a0. [DOI] [PubMed] [Google Scholar]
- Pennington T. H. Vaccinia virus polypeptide synthesis: sequential appearance and stability of pre- and post-replicative polypeptides. J Gen Virol. 1974 Dec;25(3):433–444. doi: 10.1099/0022-1317-25-3-433. [DOI] [PubMed] [Google Scholar]
- Ricciardi R. P., Miller J. S., Roberts B. E. Purification and mapping of specific mRNAs by hybridization-selection and cell-free translation. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4927–4931. doi: 10.1073/pnas.76.10.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice A. P., Roberts B. E. Vaccinia virus induces cellular mRNA degradation. J Virol. 1983 Sep;47(3):529–539. doi: 10.1128/jvi.47.3.529-539.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Berk A. J., Berget S. M. Transcription maps of adenovirus. Methods Enzymol. 1980;65(1):750–768. doi: 10.1016/s0076-6879(80)65071-x. [DOI] [PubMed] [Google Scholar]
- Smith G. L., Mackett M., Moss B. Infectious vaccinia virus recombinants that express hepatitis B virus surface antigen. Nature. 1983 Apr 7;302(5908):490–495. doi: 10.1038/302490a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spencer E., Loring D., Hurwitz J., Monroy G. Enzymatic conversion of 5'-phosphate-terminated RNA to 5'-di- and triphosphate-terminated RNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4793–4797. doi: 10.1073/pnas.75.10.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan S., Moss B. In vitro transcription of the inverted terminal repetition of the vaccinia virus genome: correspondence of initiation and cap sites. J Virol. 1981 Feb;37(2):738–747. doi: 10.1128/jvi.37.2.738-747.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
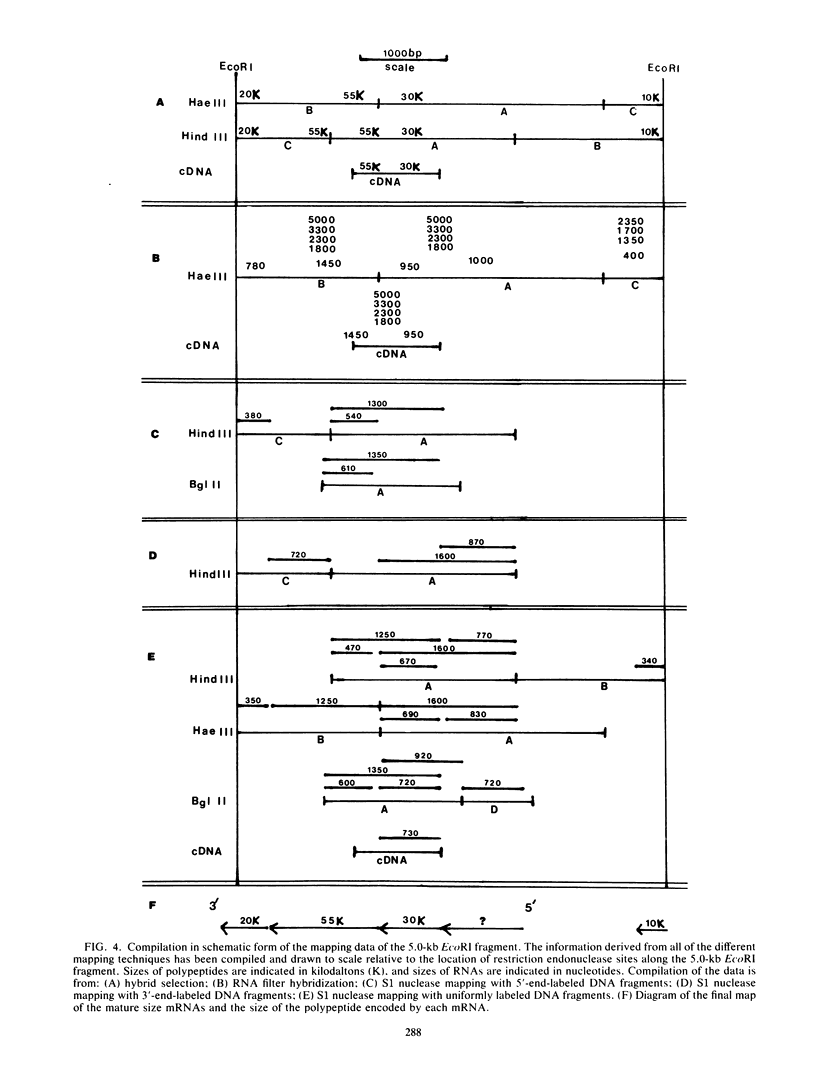
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Cooper J. A., Barbosa E., Moss B. Expression of the vaccinia virus genome: analysis and mapping of mRNAs encoded within the inverted terminal repetition. Cell. 1980 Sep;21(2):487–493. doi: 10.1016/0092-8674(80)90485-7. [DOI] [PubMed] [Google Scholar]
- Wittek R., Cooper J. A., Moss B. Transcriptional and translational mapping of a 6.6-kilobase-pair DNA fragment containing the junction of the terminal repetition and unique sequence at the left end of the vaccinia virus genome. J Virol. 1981 Sep;39(3):722–732. doi: 10.1128/jvi.39.3.722-732.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]