Abstract
The remarkable high-frequency sensitivity and selectivity of the mammalian auditory system has been attributed to the evolution of mechanical amplification, in which sound waves are amplified by outer hair cells in the cochlea. This process is driven by the recently discovered protein prestin, encoded by the gene Prestin. Echolocating bats use ultrasound for orientation and hunting and possess the highest frequency hearing of all mammals. To test for the involvement of Prestin in the evolution of bat echolocation, we sequenced the coding region in echolocating and nonecholocating species. The resulting putative gene tree showed strong support for a monophyletic assemblage of echolocating species, conflicting with the species phylogeny in which echolocators are paraphyletic. We reject the possibilities that this conflict arises from either gene duplication and loss or relaxed selection in nonecholocating fruit bats. Instead, we hypothesize that the putative gene tree reflects convergence at stretches of functional importance. Convergence is supported by the recovery of the species tree from alignments of hydrophobic transmembrane domains, and the putative gene tree from the intra- and extracellular domains. We also found evidence that Prestin has undergone Darwinian selection associated with the evolution of specialized constant-frequency echolocation, which is characterized by sharp auditory tuning. Our study of a hearing gene in bats strongly implicates Prestin in the evolution of echolocation, and suggests independent evolution of high-frequency hearing in bats. These results highlight the potential problems of extracting phylogenetic signals from functional genes that may be prone to convergence.
Keywords: evolution, phylogenetics, convergence, cochlea, mammals
Acute and sensitive hearing is important in communication, prey detection, and predator avoidance (1). In mammals, remarkable high-frequency sensitivity and selectivity have been conferred by the evolution of a mechanical sound amplification system involving specialized outer hair cells (OHC) located in the organ of Corti in the cochlea (2, 3). Each OHC is characterized by a bundle of stereocilia, which when stimulated by incoming sound waves triggers a change in the cellular membrane potential that influences cell length by means of contraction and elongation (4–7). This so-called electromotility generates mechanical energy, and the resulting increase in the amplitude of the vibration patterns in the organ of Corti can enhance hearing sensitivity by >100-fold (by 40 dB) (refs 8 and 9, but see ref. 10). The membrane motor protein that drives the somatic amplification of OHCs was recently identified and named prestin (3, 11), a member of the SLC26 superfamily of anion transporters that is encoded by the gene Prestin. The prestin protein comprises 10–12 transmembrane domains linked by intra- and extracellular loops and flanked by cytoplasmic N and C termini (12). Studies of Prestin-knockout mice (8, 13) and humans with nonsyndromic deafness (ref. 14, but see ref. 15) have confirmed the importance of Prestin for cochlea function and hearing. Yet despite its pivotal role in mammalian auditory amplification, orthologues of Prestin have been sequenced in very few species. A phylogeny of SLC26 genes showed positive selection during the evolution of the Prestin gene on the branch leading to the mammals, but it suggested strong purifying selection among the four placental mammal species surveyed (16). This observation suggests that the origin of prestin was a key innovation during the evolution of auditory sensitivity in mammals, and that Prestin gene sequence has been largely conserved during the adaptive radiation of mammals.
Among all mammals, sensitivity to the highest frequencies occurs in echolocating cetaceans and bats (17), which use sound for orientation and often for the detection, localization, and classification of prey (18, 19). The processing of echolocation signals begins at the hair cells in the organ of Corti, continues along the auditory nerve, and terminates in the auditory cortex in the brain (1). Prestin seems to be of major importance for hearing high frequencies and for selective hearing, and both of these processes are vital for echolocation. Bats, in particular, show a tremendous diversity in signal design, with calls being shaped not only by phylogeny but also by perceptual constraints imposed by their habitat (20, 21). Bat echolocation calls range in dominant frequency from 11 kHz to 212 kHz (22), although most species emit ultrasonic calls dominated by frequencies between 20 and 60 kHz (23). Recent molecular phylogenies have placed important new perspectives on the evolution of bat echolocation (24, 25). Contrary to earlier views that grouped all bats that produce and transmit echolocation calls in the larynx (i.e., “laryngeal echolocators”), a wealth of recent molecular evidence has shown that laryngeal echolocators are paraphyletic. The resulting new arrangement, which we term the “species tree,” indicates that bats are classified as comprising two major clades, the Yangochiroptera and Yinpterochiroptera, the latter of which includes some laryngeal echolocators and the nonecholocating Old World fruit bats (25, 26). Thus laryngeal echolocation and associated high-frequency hearing has either evolved at least twice during bat evolution or has been lost in fruit bats. These competing scenarios remain contentious and unresolved; whereas a synthesis of molecular and fossil data supports a loss (24, 25), some workers favor multiple origins (27), and others still dispute the paraphyly of echolocating bats (28).
Echolocation calls show both similarities and differences in structure and function among bats in the two main clades. Most members of the Yangochiroptera use relatively brief (<20 ms) signals, the faint echoes of which are processed in the time window before the next call is emitted (e.g., see ref. 29). In contrast, horseshoe bats (Rhinolophidae) and leaf-nosed bats (Hipposideridae) within the Yinpterochiroptera emit relatively long calls dominated by a constant-frequency (CF) component, which are adapted to detect and classify the wing beats of insects (30). These bats are characterized by exceptional frequency selectivity, with enhanced sensitivity to the frequency they emit while resting and reduced sensitivity to frequencies around this (31, 32). This heightened tuning arises from an overrepresentation of the narrow frequency band (auditory fovea) in the cochlea (33) and specializations in the auditory centers in the brain (31). By lowering their call frequency in relation to flight speed, these bats compensate for Doppler shifts induced by their motion (34) and thus ensure that the echoes of their calls always return at the frequency of the acoustic fovea. The acoustic fovea appears to be more sharply tuned in horseshoe bats than in leaf-nosed bats (33).
The convergence of call types (and of echolocation itself) makes bats excellent subjects for studying genes associated with sensory performance. We previously showed that the FoxP2, a gene implicated in orofacial coordination, has undergone accelerated evolution in echolocating bats compared with all other vertebrates (35). Yet given the auditory specializations of bats, including their use of high frequencies, extreme auditory sensitivity, call diversity, and specialized audiograms, there are especially good a priori reasons to suspect that key genes underpinning the evolution and development of echolocation are expressed in the auditory system, and, in particular, the cochlea. To address this possibility, we studied the phylogenetic history and molecular evolution of Prestin in bats and other mammals, given that Prestin is fundamental to frequency sensitivity and selectivity in the mammalian auditory system.
Results
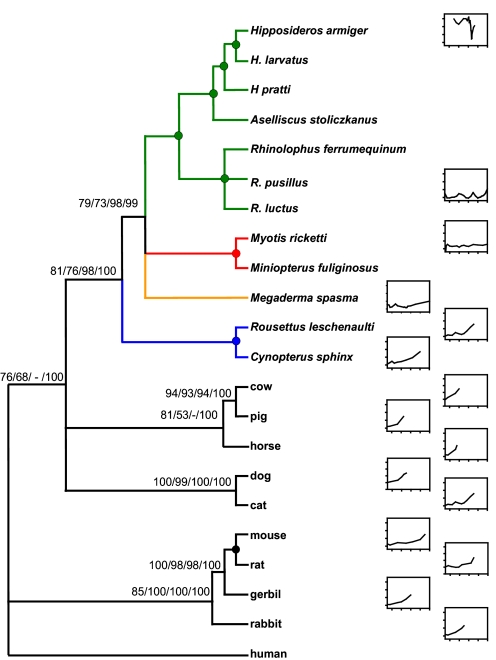
Our phylogenetic trees based on the Prestin coding sequence showed a clade that grouped together members of the superorder Euarchontoglires (rodents, rabbits, and humans) and a separate clade that grouped members of the superorder Laurasiatheria (carnivores, artiodactyls, perrisodactyls, and bats). However, arrangements within the bat clade did not conform to expectations on the basis of multigene phylogenies. Both the maximum-likelihood (ML) and Bayesian analyses recovered a tree (which we refer to as the putative gene tree) in which all laryngeal echolocators group together in a strongly supported clade [bootstrap values 73–98% and Bayesian posterior probability (BPP) of 99%], to the exclusion of nonlaryngeal echolocating fruit bats (Fig. 1). We assessed the relative confidence of the putative gene tree vs. the species tree by using the approximately unbiased test. We found that the putative gene tree, in which we forced monophyly of the laryngeal echolocators, was not significantly worse than the species tree with Yinpterochiroptera monophyly (P = 0.848 and P = 0.168, respectively) and was identical to the unconstrained tree (P = 0.848). Therefore, there was no greater support for the species tree over the observed putative Prestin tree, or vice versa. The results of both the split-network and spectral analyses also suggest a mixed phylogenetic signal, with considerable support for both the putative gene tree and species tree topologies [see supporting information (SI) Fig. S1]. Our reconciled tree analysis found that a single gene duplication in the ancestor of the Chiroptera, followed by independent losses in ancestral branches of the Yangochiroptera, fruit bats, and laryngeal echolocating Yinpterochiroptera would be needed to explain the evolution of the gene tree in terms of birth and death of gene copies. It is therefore unlikely that the observed Prestin tree (Fig. 1) arose as a consequence of gene duplication events.
Fig. 1.
Maximum-likelihood putative gene tree based on the complete Prestin coding sequence of 22 mammals. Values indicate statistical support (maximum likelihood, maximum parsimony, neighbor-joining, and posterior probability, respectively). A filled circle indicates 100% support across methods and a negative sign (−) indicates lack of support for a specific method. Audiograms are given on the basis of published data (for references see SI Materials and Methods). For each plot, the x-axis ranges from 10 to 180 kHz, with ticks at 50, 100, 150, and 200, and the y-axis ranges from −20 to 100 dB with increments of 20 dB. The Yinpterochiroptera comprise the CF bats (green), Megaderma (orange), and the Old World fruit bats (blue), and both Yangochiroptera species are red.
Repeated analyses of phylogenetic reconstruction based on nucleotides corresponding to variable amino acid sites only (414 bp), thus reflecting areas of nonsynonymous change, also recovered the putative gene tree topology with strong support for the clade of laryngeal echolocators (62% ML bootstrap and 97% BPP). Conversely, analyses of the remaining nucleotides (1,800 bp) recovered the species tree, albeit with reduced support for the Yinpterochiroptera clade (<50% ML bootstrap, 47% BPP). Therefore, the monophyly of laryngeal echolocators is supported when only those parts of the gene that lead to amino acid changes are analyzed, but this arrangement is lost and laryngeal echolocators become paraphyletic (as suggested by recent molecular phylogenies) when areas of the gene that do not result in amino acid changes are analyzed.
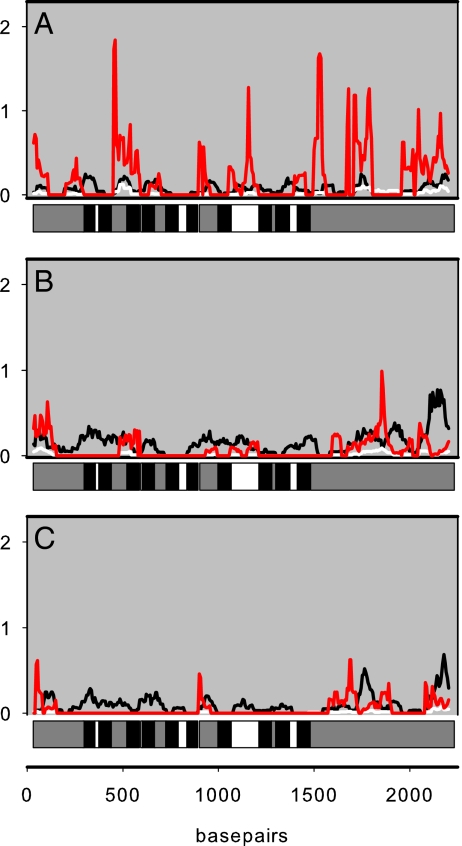
Sliding window analyses were used to highlight where rates of nonsynonymous substitution (dN) exceeded rates of synonymous substitution (dS) along specific branches, and hence to identity positive selection. Higher values of dN/dS were found along the branch ancestral to CF bats that use Doppler-shift compensation (DSC). Moreover, dN/dS values were relatively higher for exposed domains of the protein (extracellular loops and termini) than for transmembrane regions in all three branches (Fig. 2). Similar results were also obtained when this analysis was repeated using the putative gene tree topology (data not shown). After removal of the 10 putative transmembrane domains, we repeated the phylogenetic analysis on the basis of an alignment (1,539 bp) of concatenated loops and termini. This analysis also showed support for the clade of laryngeal echolocators (59% ML and 98% BPP), whereas the transmembrane domain alignment (681 bp) again recovered the species tree with limited support for the Yinpterochiroptera (<50% ML and 64% BPP). Site-wise likelihood values showed that sites supporting the species tree were concentrated in transmembrane and α-helix domains, whereas the putative gene tree topology was supported by the cytoplasmic and extracellular loops, and, more specifically, the coil domains and the sulfate transporters and antisigma factor antagonists (STAS) domain (see ref. 12) (Table 1 and Fig. S2). Again, these results suggest that differential support for the topology of the putative gene tree vs. the species tree arises from changes in key areas of the protein that are of functional importance. Our detailed comparisons of Prestin isoforms obtained from cochlea and brain tissue also revealed that such functional regions were preserved in nearly all cases.
Fig. 2.
Sliding windows of Nei–Gojobori estimates of dS (black), dN (white), and dN/dS (red) along the ancestral branches of CF bats (A), Yangochiroptera (B), and Old World fruit bats (C). Each plot is compared to a gene schematic showing the transmembrane (black), extracellular (white), and intracellular (gray) domains.
Table 1.
Relative support for gene tree and species tree topologies
| Domain | Length of domain | Sum of log-likelihood difference | Mean of log-likelihood difference |
|---|---|---|---|
| N terminus | 363 | 2.1285 | 0.0059 |
| Transmembrane | 750 | −0.5306 | −0.0007 |
| Extracellular and cytoplasmic loops | 474 | 1.5960 | 0.0034 |
| α-Helix | 186 | −1.6305 | −0.0088 |
| C terminus | 741 | 6.2428 | 0.0084 |
| STAS | 453 | 0.9662 | 0.0021 |
| Charge clusters | 33 | 0.7908 | 0.0247 |
| Coil regions | 120 | 3.1374 | 0.0261 |
| All nontransmembrane | 1578 | 9.9674 | 0.0063 |
Numbers are the sums and means of site-wise negative log-likelihood scores for the species tree minus the sum of site-wise negative log-likelihood scores for the gene tree, calculated for different domains of the Prestin gene. Positive values indicate greater support for the gene tree than the species tree.
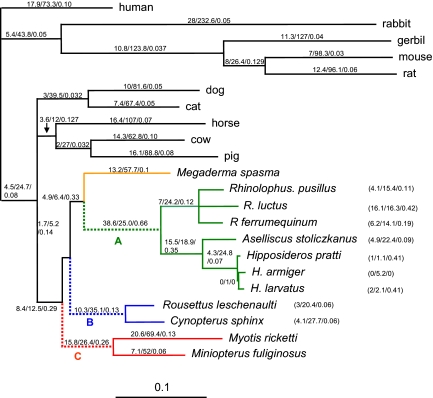
A free ratio test in which dN/dS was allowed to vary among branches showed a significantly better fit to the data than a model where dN/dS was fixed across the tree (one-ratio model) [log-likelihood ratio test (LRT) = 156.3, df = 39, P < 0.001]. The free ratio test also revealed that the values of dN/dS on the CF and Yangochiroptera ancestral branches (0.66 and 0.26, respectively) were an order of magnitude greater than the nonbat branches (see Fig. 3). This result, which is consistent with bursts of positive selection along these branches, was also obtained when the analysis was repeated with the putative gene tree topology.
Fig. 3.
Species tree topology based on refs. 24–27. Values given on the branches, or in parentheses, are the maximum-likelihood free-ratio estimates of the non-synonymous substitution rate (dN), the synonymous substitution rate (dS) and omega (dN/dS), respectively. Branch lengths are scaled by the number of nucleotide substitutions per codon. The Yinpterochiroptera comprise the CF bats (green), Megaderma (orange), and the Old World fruit bats (blue), and both Yangochiroptera species are red. The dashed lines A, B, and C show the ancestral branches of the three groups studied.
Branch-specific tests of positive selection undertaken separately for the ancestral branches of the Old World fruit bats, the Yangochiroptera, and CF bats (indicated in Fig. 3) found evidence of positive selection in the CF branch only (see Table S1). For this branch, the null hypothesis, in which ù was fixed at 1 (neutral evolution) gave a log-likelihood value of −11417.23, whereas the equivalent value for the alternative hypothesis in which the ù could exceed 1 (positive selection) was −11411.51. (LRT = 11.44, df = 1, P < 0.001). This finding supports the hypothesis that positive selection on the Prestin gene occurred around the time of origin of these CF bats. Bayes empirical-Bayes (BEB) analysis suggested that of 33 amino acid replacements in the CF ancestral branch, 23 sites were identified as positive selection sites at P > 0.5, 14 at P > 0.8, and 3 at P > 0.90. Repeated branch-site tests in which ancestral sequences were reconstructed from the species topology gave similar results, with positive selection detected only in the ancestral branch of the CF bats, and the same amino acid sites identified as under positive selection (data not shown).
Multivariate analysis of protein polymorphism (MAPP) scores of physicochemical impact were estimated for 33 amino acid variants among CF bats and predicted that 20 of these would have had a large functional effect [mean MAPP score = 17.7 ± 2.1 (SE), range 9.2–37.6] and 13 a small effect (4.4 ± 0.4, range 2.0–7.00). Ranks of MAPP scores and BEB values were significantly correlated (Pearson coefficient = 0.43, P = 0.042) with two of the three sites with highest BEB values (A585I and A720E) also assigned the highest MAPP scores (37.2 and 37.6).
Discussion
Our phylogenetic reconstruction based on the Prestin coding sequence was congruent with the superordinal arrangement of mammals of recent molecular phylogenies, recovering both the Euarchontoglires and Laurasiatheria clades (36, 37). However, arrangements within the bat clade did not conform to expectations on the basis of multigene phylogenies. Instead, phylogenies of bats based on the Prestin gene revealed consistent conflict with the established species tree derived from large-scale analyses of both mitochondrial and nuclear genes (24–26). All three methods of phylogenetic reconstruction that we used recovered trees in which the species with laryngeal echolocation formed a monophyletic clade to the exclusion of the nonecholocating Old World fruit bats. This arrangement is particularly intriguing because it resembles the earlier classification of two suborders, Microchiroptera and Megachiroptera, a view that has been largely rejected in recent years in light of overwhelming molecular evidence (25–27, 38).
Conflicts between species and gene trees can arise for several reasons. The possibility that the Prestin gene has undergone a past duplication with secondary loss, leading to shared paralogous sequences among echolocators, seems extremely unlikely given the requisite number of independent gene losses. Moreover, a gene duplication event would be expected to result in a single dominant phylogenetic signal in the data with a bimodal distribution of pairwise sequence distances, neither of which were supported by our results. We also found no evidence of gene duplicates from searches of published mammal genome databases, or from amplification using universal primers. Instead, spectral analysis revealed that the data contained two phylogenetic signals, corresponding to the putative gene and species trees, and we were unable to reject either of these as the most likely hypothesis.
An alternative explanation is that the clustering of the echolocating bats is a consequence of accelerated evolution along the fruit bat lineage, resulting from either relaxed or positive selection. Although relaxation in Prestin would resonate with the theory that fruit bats have lost the ability to echolocate, the branch lengths of the fruit bat clade were not unusually long. Indeed, we found evidence for strong purifying selection in the Prestin gene along this branch.
Instead, we suggest that the most likely explanation is that the Prestin gene tree reflects convergent evolution at the molecular level, associated with high-frequency hearing among echolocating species. Indeed, removal of synonymous changes resulted in a reversion to the species topology; whereas a tree based solely on amino acid differences gave a well-supported putative gene tree. These findings thus appear to add credence to the view that the high-frequency hearing associated with echolocation has evolved more than once. However, it is important to emphasize that echolocation is likely to involve many hundreds of genes, and thus additional evidence is needed to confirm whether echolocation itself has evolved on more than one occasion.
The adaptive evolution of the Prestin gene in bats is also suggested by the results of our selection tests. We found evidence of significant positive selection along the ancestral branch of horseshoe and leaf-nosed bats. Therefore, contrary to a previous study that suggested that Prestin is under purifying selection within mammals (16), we show that this gene has continued to undergo adaptive evolution since mammals diversified, and, in particular, that this appears to be related to the origin of the specialized form of constant-frequency (CF) echolocation that characterizes these bats.
The distribution of codons under selection and site-wise likelihood values are also highly informative. Over 80% of sites under positive selection occurred in regions thought to be functionally important, including the extracellular loops (4 sites), the transmembrane domains (3 sites), and the N and C termini (17 sites), both of which are known to be important for normal voltage sensing (12, 39). The N terminus is also thought to mediate prestin–prestin interactions (12), while the C terminus controls prestin's cellular localization and function (40). It is interesting that 9 sites found to be under selection are located in the C-terminal region known as the sulfate transporters and STAS domain, which is common to all members of the SLC26A family and was previously reported to be a hotspot of adaptive change in Prestin associated with the origin of electromotility in mammals (16). Our own results suggest that changes in the STAS domain, mutations in which are known to compromise both protein function and targeting (41), might also be key to the hearing of some or all echolocating bats. An examination of the site-wise likelihood values confirm that specific sites in the STAS domain and neighboring coil region contribute much of the support for the putative gene tree topology, suggesting that Prestin's role in high-frequency hearing is associated with convergence in these regions in both lineages of echolocating bats.
Detection of positive selection in the CF bats, but not along the branch ancestral to the other clade of echolocating bats (Yangochiroptera), either could point to a specific role of prestin in CF echolocation or could reflect the fact that CF bats often emit calls of higher frequencies than other echolocating bats of equivalent body mass (42), in part because most of the signal energy is focused into the second harmonic. Indeed the recovery of the putative gene tree from a nucleotide alignment is based on concatenated intra-/extracellular loops and termini, yet the species tree from the concatenated transmembrane sections does suggest that areas under positive selection in CF bats are also of adaptive significance in the other echolocating bat species. This suggestion is also reflected in the broadly concordant distribution of sites with high BEB posterior probabilities in the echolocating bats from both major clades.
Although the extent to which the cochlear amplifier can tune to high frequencies has been questioned (43, 44), studies on guinea pigs have shown that isolated outer hair cells (OHCs) can react to frequencies of up to 100 kHz at 20°C (45), and that these frequencies induce correspondingly rapid basilar membrane oscillations in vivo (46). There is also mounting evidence to show that, in addition to somatic electromotility, cochlear amplification is augmented by rapid force generation by the stereocilia bundles themselves (47), a process that also appears to involve prestin (48). Moreover, the OHCs of echolocating bats from both the Yinpterochiroptera (49, 50) and Yangochiroptera (51) show structural characteristics that might be adaptations to high-frequency reception, including cell shape, angle, and stiffness. In particular, OHCs and stereocilia are shorter in bats than in all other mammals (52, 53). Thus while there is no evidence to date that bats use a specialized form of cochlear amplifier (54), our results do support a link between Prestin and ultrasonic stimuli. Studies are now needed to determine whether the residue changes in Prestin reported in this study are responsible for the morphological characteristics of bat outer hair cells and so modify their electromotility.
Alternatively, positive selection in Prestin in the CF bat lineage might relate not to the use of high frequencies per se but to the unique cochlear adaptations associated with this form of biosonar that enhance selectivity. The auditory fovea that characterizes CF bats contains nerve cells that are extremely sharply tuned to the dominant frequency emitted at rest (33, 55). Cochlea threshold curves based on distortion-product otoacoustic emissions (DPOAEs) show that Q10dB values in CF bats that use DSC can reach 200–600 at the resting frequency, compared with values of ∼20 in other mammals (56). This level of tuning at the resting frequency exceeds the capabilities of a normal cochlear amplifier, and it remains unclear how bats that use Doppler-sensitive echolocation are capable of possessing such narrow frequency filters without showing marked deterioration in temporal acuity through the resonance involved (54). Distortion measurements in the cochlea imply that specializations in cochlear mechanics are responsible for the steep variations in threshold and frequency tuning (56). Therefore, in light of our results, and the observation that Prestin-knockout mice lose frequency selectivity (57), it is tempting to propose that the adaptive changes in Prestin seen in the ancestral CF branch might relate to the evolution of their exquisite frequency selectivity.
To our knowledge, previous work on prestin in bat hearing has been limited to a single preliminary published study in which Prestin-like expression was confirmed in the OHCs of adult mustached bats (Pteronotus parnelli) (58). Our study, which sequences a hearing gene in bats, strongly implicates a role for importance and associated modification of prestin in the evolution of echolocation. Moreover, our findings support the scenario that high-frequency hearing, which is intimately linked to echolocation, has evolved more than once in bats. Finally, these results from Prestin highlight the dangers of using phylogenies based on sequences of putative functional genes for reconstructing the evolutionary history of taxa, where, like many morphological traits, such genes are likely to be subject to convergent evolution.
Materials and Methods
Taxonomic Coverage, Nucleic Acid Amplification, and Sequencing.
We sequenced the Prestin coding region in 12 bat species with divergent auditory characteristics. From the Yinpterochiroptera, Cynopterus sphinx and Rousettus leschenaulti (family Pteropodidae) are Old World fruit bats without laryngeal echolocation or associated specialized high-frequency auditory sensitivity and selectivity, although Rousettus does echolocate in caves by tongue clicking (59). Megaderma spasma (Megadermatidae) produces brief multiharmonic broadband signals. The horseshoe bats Rhinolophus ferrumequinum, R. luctus, and R. pusillus (family Rhinolophidae) and leaf-nosed bats Hipposideros armiger, H. pratti, H. larvatus, and Aselliscus stoliczkanus (Hipposideridae) all possess an auditory fovea and emit constant-frequency calls. From the Yangochiroptera, Myotis ricketti (Vespertilionidae) emits brief broadband signals (60) and Miniopterus fuliginosus (Miniopteridae) emits brief broadband calls ending in a narrowband tail (unpublished data). Accession numbers of all taxa, including outgroups obtained from direct sequencing and from sequence databases, are given in the SI Materials and Methods.
We used RT-PCR to amplify genes from total RNA isolated from brain tissue, as previously described (35). See SI Materials and Methods for details of primers and protocols used. All PCR products were isolated from a 1% agarose gel and cloned using the pGEM-T-easy vector (Promega). Positive clones were cycle sequenced in both directions by using Big Dye Terminator kits (Applied Biosystems) on an ABI 3730 automated DNA sequencer. To check for the existence of cochlea-specific Prestin isoforms, we also repeated this method for cochlea tissue obtained from single individuals of Hipposideros armiger, Rousettus leschenaulti, Miniopterus fuliginosus, and Myotis ricketti. Our results based on 220 sequenced clones revealed the existence of several cochlea splice variants; however, the most common transcripts were identical to those obtained from brain tissue (see Table S2).
Phylogenetic Reconstruction.
Nucleotide sequences (2,232 bp) of 22 species were aligned in the software ClustalX (61). After alignments of indels by eye with reference to amino acids translated in MEGA 3.1 (62), the alignment comprised 744 amino acid sites, of which 141 amino acids (∼19%) were variable in eutherian mammals (Fig. S3). Maximum-likelihood and Bayesian phylogenies were constructed and bootstrap support for particular relationships was estimated. We also estimated phylogenies constraining monophyly of the Yinpterochiroptera (the species tree) and laryngeal echolocating bats (the putative gene tree) and compared support for these topologies by using the approximately unbiased test (63). See SI Materials and Methods for details of these analyses. To explore conflicts in the phylogenetic signal, tree reconstruction was repeated separately for synonymous and nonsynonymous sites only and also for different parts of the protein (external loops and termini vs. transmembrane domains), which are considered to differ in functional importance. We determined the number of duplications and losses that would be required to reconcile gene and species topologies by using GENETREE (64) and also assessed whether topological ambiguities arose from multiple phylogenetic signals by using two additional methods. First we undertook a neighbor-net analysis in SPLITSTREE4 (65, 66) to generate a distance-based (uncorrected-p) split network. Second, we converted the sequence alignment to a spectrum of splits and visualized the support and conflict for the major splits by means of a Lento-Plot in SPECTRONET (67). We also identified the relative support for the two topologies for individual sites and domains along the gene by using site-wise likelihood scores.
Molecular Evolution.
To characterize variation in the rate of molecular evolution over the Prestin gene, we performed sliding-window analyses (window = 20 codons, step = 2 codons) of Nei–Gojobori estimates of synonymous and nonsynonymous substitutions per site (dS and dN, respectively) and the dN/dS (omega, ù) in SWAAP1.0.2. These analyses were performed on the branches from the common ancestor of all bats to the ancestral nodes of the Old World fruit bats, Yangochiroptera, and CF bats, using ancestral sequences inferred by using the empirical Bayes (EB) method implemented in PAML 3.15 (68). Sliding-window analyses were repeated for both species and putative gene tree topologies. We also tested for positive selection by using PAML. Briefly, we obtained maximum-likelihood estimates of dS and dN under a free ratio model in which the dN/dS ratio (ù) can vary among branches (69). In addition, given our a priori hypothesis that Prestin has undergone accelerated evolution associated with the evolution of high-frequency sensitivity and selectivity, we tested for positive selection along the same three ancestral branches as in the sliding-window analysis (labeled in Fig. 3). In each case, we applied Test 2 of the branch-site model (70), which compares the likelihood when the focal branch is fixed at ù = 1 (neutral evolution) to when it can exceed one (ù > 1) and is thus under positive selection. (See ref. 35 for a full description of this approach.) Given ambiguities in the phylogenetic signal, we also repeated the selection tests based on both the species and putative gene tree topologies, and, to confirm the robustness of our results, we repeated the analyses multiple times with different initial ù values. Where tests indicated positive selection, we recorded sites under selection according to high posterior probabilities following Bayes empirical-Bayes (BEB) prediction. To estimate the possible physicochemical impact of amino acid replacements along branches identified as under selection, we used the multivariate analysis of protein polymorphism implemented in MAPP (71). Physicochemical constraints for each site were estimated from an alignment of Prestin orthologues of all other mammals and corrected for phylogenetic similarity (see ref. 35).
Supplementary Material
Acknowledgments.
We thank Bei-Bei He for assistance in the lab and two anonymous referees for helpful comments on the manuscript. This work was funded by grants awarded to S.Z. under the Key Construction Program of the National “985” Project and the “211” Project, a Royal Society Research Fellowship to S.J.R., a Darwin Initiative Grant (14–008) to G.J., and a Research Councils UK Academic Fellowship to J.A.C.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU914923–EU914937).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802097105/DCSupplemental.
References
- 1.Fay RR, Popper AN. Evolution of hearing in vertebrates: The inner ears and processing. Hear Res. 2000;149:1–10. doi: 10.1016/s0378-5955(00)00168-4. [DOI] [PubMed] [Google Scholar]
- 2.Dallos P, Fakler B. Prestin, a new type of motor protein. Nat Rev Mol Cell Biol. 2002;3:104–111. doi: 10.1038/nrm730. [DOI] [PubMed] [Google Scholar]
- 3.Zheng J, et al. Prestin is the motor protein of cochlear outer hair cells. Nature. 2000;405:149–155. doi: 10.1038/35012009. [DOI] [PubMed] [Google Scholar]
- 4.Ashmore JF. A fast motile response in guinea-pig outer hair-cells: The cellular basis of the cochlear amplifier. J Physiol. 1987;388:323–347. doi: 10.1113/jphysiol.1987.sp016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos-Sacchi J. Asymmetry in voltage-dependent movements of isolated outer hair-cells from the organ of Corti. J Neurosci. 1989;9:2954–2962. doi: 10.1523/JNEUROSCI.09-08-02954.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownell WE, Bader CR, Bertrand D, Deribaupierre Y. Evoked mechanical responses of isolated cochlear outer hair-cells. Science. 1985;227:194–196. doi: 10.1126/science.3966153. [DOI] [PubMed] [Google Scholar]
- 7.Dallos P, Evans BN, Hallworth R. Nature of the motor element in electrokinetic shape changes of cochlear outer hair-cells. Nature. 1991;350:155–157. doi: 10.1038/350155a0. [DOI] [PubMed] [Google Scholar]
- 8.Liberman, et al. Prestin is required for electromotility of the outer hair cell and for the cochlear amplifier. Nature. 2002;419:300–304. doi: 10.1038/nature01059. [DOI] [PubMed] [Google Scholar]
- 9.Santos-Sacchi J. New tunes from Corti's organ: The outer hair cell boogie rules. Curr Opin Neurobiol. 2003;13:459–468. doi: 10.1016/s0959-4388(03)00100-4. [DOI] [PubMed] [Google Scholar]
- 10.Ren T, Gillespie PG. A mechanism for active hearing. Curr Opin Neurobiol. 2007;17:498–503. doi: 10.1016/j.conb.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belyantseva IA, Adler HJ, Curi R, Frolenkov GI, Kachar B. Expression and localization of prestin and the sugar transporter GLUT-5 during development of electromotility in cochlear outer hair cells. J Neurosci. 2000;20:RC116, 1–5. doi: 10.1523/JNEUROSCI.20-24-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navaratnam D, Bai JP, Samaranayake H, Santos-Sacchi J. N-terminal-mediated homomultimerization of prestin, the outer hair cell motor protein. Biophys J. 2005;89:3345–3352. doi: 10.1529/biophysj.105.068759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu XD, Gao JG, Guo YK, Zuo J. Hearing threshold elevation precedes hair-cell loss in prestin knockout mice. Mol Brain Res. 2004;126:30–37. doi: 10.1016/j.molbrainres.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 14.Liu XZ, et al. Prestin, a cochlear motor protein, is defective in nonsyndromic recessive deafness. Hum Mol Genet. 2003;12:1155–1162. doi: 10.1093/hmg/ddg127. [DOI] [PubMed] [Google Scholar]
- 15.Tang HY, Xia AP, Oghalai JS, Pereira FA, Alford RL. High frequency of the IVS2–2A>G DNA sequence variation in SLC26A5, encoding the cochlear motor protein prestin, precludes its involvement in hereditary hearing loss. BMC Med Genet. 2005;6:30. doi: 10.1186/1471-2350-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franchini LF, Elgoyhen AB. Adaptive evolution in mammalian proteins involved in cochlear outer hair cell electromotility. Mol Phylogenet Evol. 2006;41:622–635. doi: 10.1016/j.ympev.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 17.Heffner HE, Heffner RS. In: Handbook of the Senses: Audition. Dallos P, Oertel D, Hoy R, editors. New York: Elsevier; 2007. pp. 55–60. [Google Scholar]
- 18.Griffin D. Listening in the Dark. New Haven, CT: Yale Univ Press; 1958. [Google Scholar]
- 19.Thomas JT, Moss CF, Vater M, editors. Advances in the Study of Echolocation in Bats and Dolphins. Chicago: Univ of Chicago Press; 2004. [Google Scholar]
- 20.Schnitzler HU, Moss CF, Denzinger A. From spatial orientation to food acquisition in echolocating bats. Trends Ecol Evol. 2003;18:386–394. [Google Scholar]
- 21.Jones G, Teeling EC. The evolution of echolocation in bats. Trends Ecol Evol. 2006;21:149–156. doi: 10.1016/j.tree.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Fenton MB, Bell GP. Recognition of species of insectivorous bats by their echolocation calls. J Mammal. 1981;62:233–243. [Google Scholar]
- 23.Fenton MB, Portfors CV, Rautenbach IL, Waterman JM. Compromises: Sound frequencies used in echolocation by aerial-feeding bats. Can J Zool. 1998;76:1174–1182. [Google Scholar]
- 24.Springer MS, Teeling EC, Madsen O, Stanhope MJ, de Jong WW. Integrated fossil and molecular data reconstruct bat echolocation. Proc Natl Acad Sci USA. 2001;98:6241–6246. doi: 10.1073/pnas.111551998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teeling EC, et al. Molecular evidence regarding the origin of echolocation and flight in bats. Nature. 2000;403:188–192. doi: 10.1038/35003188. [DOI] [PubMed] [Google Scholar]
- 26.Teeling EC, et al. Microbat paraphyly and the convergent evolution of a key innovation in Old World rhinolophoid microbats. Proc Natl Acad Sci USA. 2002;99:1431–1436. doi: 10.1073/pnas.022477199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eick GN, Jacobs DS, Matthee CA. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera) Mol Biol Evol. 2005;22:1869–1886. doi: 10.1093/molbev/msi180. [DOI] [PubMed] [Google Scholar]
- 28.Hutcheon JM, Kirsch JAW, Pettigrew JD. Base-compositional biases and the bat problem. III. The question of microchiropteran monophyly. Philos Trans R Soc London B Biol Sci. 1998;353:607–617. doi: 10.1098/rstb.1998.0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holderied MW, von Helversen O. Echolocation range and wingbeat period match in aerial-hawking bats. Proc R Soc London B Biol Sci. 2003;270:2293–2299. doi: 10.1098/rspb.2003.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnitzler H-U. In: Recent Advances in the Study of Bats. Fenton MB, Racey P, Rayner JMV, editors. Cambridge, UK: Cambridge Univ Press; 1987. pp. 226–243. [Google Scholar]
- 31.Schuller G, Pollak G. Disproportionate frequency representation in the inferior colliculus of Doppler-compensating greater horseshoe bats: Evidence for an acoustic fovea. J Comp Physiol A Sens Neural Behav Physiol. 1979;132:47–54. [Google Scholar]
- 32.Long GR, Schnitzler HU. Behavioral audiograms from the bat, Rhinolophus ferrumequinum. J Comp Physiol A Sens Neural Behav Physiol. 1975;100:211–219. [Google Scholar]
- 33.Kössl M, Foeller E, Faulstich M. In: Echolocation in Bats and Dolphins. Thomas JA, Moss CF, Vater M, editors. Chicago: Univ of Chicago Press; 2004. pp. 104–109. [Google Scholar]
- 34.Trappe M, Schnitzler HU. Doppler-shift compensation in insect-catching horseshoe bats. Naturwissenschaften. 1982;69:193–194. [Google Scholar]
- 35.Li G, Wang J, Rossiter SJ, Jones G, Zhang S. Accelerated FoxP2 evolution in echolocating bats. PLoS ONE. 2007;2:e900. doi: 10.1371/journal.pone.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy WJ, et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- 37.Reyes A, et al. Congruent mammalian trees from mitochondrial and nuclear genes using Bayesian methods. Mol Biol Evol. 2004;21:397–403. doi: 10.1093/molbev/msh033. [DOI] [PubMed] [Google Scholar]
- 38.Teeling EC, et al. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- 39.Bai JP, Navaratnam D, Samaranayake H, Santos-Sacchi J. En block C-terminal charge cluster reversals in prestin (SLC26A5): Effects on voltage-dependent electromechanical activity. Neurosci Lett. 2006;404:270–275. doi: 10.1016/j.neulet.2006.05.062. [DOI] [PubMed] [Google Scholar]
- 40.Zheng J, et al. The C-terminus of prestin influences nonlinear capacitance and plasma membrane targeting. J Cell Sci. 2005;118:2987–2996. doi: 10.1242/jcs.02431. [DOI] [PubMed] [Google Scholar]
- 41.Shibagaki N, Grossman AR. The role of the STAS domain in the function and biogenesis of a sulfate transporter as probed by random mutagenesis. J Biol Chem. 2006;281:22964–22973. doi: 10.1074/jbc.M603462200. [DOI] [PubMed] [Google Scholar]
- 42.Jones G. Scaling of echolocation call parameters in bats. J Exp Biol. 1999;202:3359–3367. doi: 10.1242/jeb.202.23.3359. [DOI] [PubMed] [Google Scholar]
- 43.Ashmore J, Geleoc GSG. Cochlear function: Hearing in the fast lane. Curr Biol. 1999;9:R572–R574. doi: 10.1016/s0960-9822(99)80358-3. [DOI] [PubMed] [Google Scholar]
- 44.Gale JE, Ashmore JF. An intrinsic frequency limit to the cochlear amplifier. Nature. 1997;389:63–66. doi: 10.1038/37968. [DOI] [PubMed] [Google Scholar]
- 45.Frank G, Hemmert W, Gummer AW. Limiting dynamics of high-frequency electromechanical transduction of outer hair cells. Proc Natl Acad Sci USA. 1999;96:4420–4425. doi: 10.1073/pnas.96.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grosh K, Zheng JF, Zou Y, de Boer E, Nuttall AL. High-frequency electromotile responses in the cochlea. J Acoust Soc Am. 2004;115:2178–2184. doi: 10.1121/1.1695431. [DOI] [PubMed] [Google Scholar]
- 47.Kennedy HJ, Crawford AC, Fettiplace R. Force generation by mammalian hair bundles supports a role in cochlear amplification. Nature. 2005;433:880–883. doi: 10.1038/nature03367. [DOI] [PubMed] [Google Scholar]
- 48.Kennedy HJ, Evans MG, Crawford AC, Fettiplace R. Depolarization of cochlear outer hair cells evokes active hair bundle motion by two mechanisms. J Neurosci. 2006;26:2757–2766. doi: 10.1523/JNEUROSCI.3808-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vater M, Lenoir M. Ultrastructure of the horseshoe bats organ of Corti. 1. Scanning electron-microscopy. J Comp Neurol. 1992;318:367–379. doi: 10.1002/cne.903180403. [DOI] [PubMed] [Google Scholar]
- 50.Vater M, Lenoir M, Pujol R. Ultrastructure of the horseshoe bats organ of Corti. 2. Transmission electron-microscopy. J Comp Neurol. 1992;318:380–391. doi: 10.1002/cne.903180404. [DOI] [PubMed] [Google Scholar]
- 51.Reuter G, et al. Electromotility of outer hair-cells from the cochlea of the echolocating bat, Carollia perspicillata. J Comp Physiol A Sens Neural Behav Physiol. 1994;175:449–455. doi: 10.1007/BF00199252. [DOI] [PubMed] [Google Scholar]
- 52.Pujol R, Lenoir M, Ladrech S, Tribillac F, Rebillard G. In: Auditory Physiology and Perception, Advances in Biosciences. Cazals Y, Demany L, Horner K, editors. New York: Pergamon; 1992. pp. 45–52. [Google Scholar]
- 53.Yao Q, et al. Characteristics of echolocating bats' auditory stereocilia length, compared with other mammals. Sci China Ser C Life Sci. 2007;50:492–496. doi: 10.1007/s11427-007-0055-8. [DOI] [PubMed] [Google Scholar]
- 54.Vater M, Kossl M. In: Advances in the Study of Echolocation in Bats and Dolphins. Thomas JT, Moss CF, Vater M, editors. Chicago: Univ of Chicago Press; 2004. pp. 89–99. [Google Scholar]
- 55.Vater M. In: Ontogeny, Functional Ecology, and Evolution of Bats. Adams RA, Pedersen SC, editors. Cambridge, UK: Cambridge Univ Press; 2000. pp. 137–173. [Google Scholar]
- 56.Kössl M, Vater M. Hearing by Bats. New York: Springer; 1995. pp. 191–234. [Google Scholar]
- 57.Cheatham MA, Huynh KH, Gao J, Zuo J, Dallos P. Cochlear function in Prestin knockout mice. J Physiol. 2004;560:821–830. doi: 10.1113/jphysiol.2004.069559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vater M, Weber T, Knipper M. Prestin-like immunoreactivity in outer hair cells of the mustached bat (Pteronotus parnellii) Acta Otolaryngol Belgica. 2002;56:295. [Google Scholar]
- 59.Grinnell AD, Hagiwara S. Studies of auditory neurophysiology in non-echolocating bats, and adaptations for echolocation in one genus, Rousettus. Z Vgl Physiol. 1972;76:82–96. [Google Scholar]
- 60.Ma J, et al. Dietary analysis confirms that Rickett's big-footed bat (Myotis ricketti) is a piscivore. J Zool. 2003;261:245–248. [Google Scholar]
- 61.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinf. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 63.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 64.Page RDM. GeneTree: Comparing gene and species phylogenies using reconciled trees. Bioinformatics. 1998;14:819–820. doi: 10.1093/bioinformatics/14.9.819. [DOI] [PubMed] [Google Scholar]
- 65.Huson DH. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 66.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 67.Huber KT, Langton M, Penny D, Moulton V, Hendy M. Spectronet: A package for computing spectra and median networks. Appl Bioinformatics. 2002;1:159–161. [PubMed] [Google Scholar]
- 68.Yang ZH. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 69.Yang ZH. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 70.Zhang JZ, Nielsen R, Yang ZH. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 71.Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.