Abstract
Expression of the Arabidopsis TRANSPARENT TESTA 2 (TT2) MYB family transcription factor leads to massive accumulation of proanthocyanidins (PAs) in hairy roots of Medicago truncatula. Microarray analysis showed that TT2 induces genes for flavonoid/PA biosynthesis, transcription factors, and a large number of genes of unknown function. A second microarray dataset identified genes that were preferentially expressed in the M. truncatula seed coat. Comparison of the two datasets defines target genes for steps that are yet unidentified in PA biosynthesis and accumulation. Of these genes, a glycosyltransferase, UGT72L1, was active specifically toward the PA precursor (−)−epicatechin, and its expression pattern in developing seeds correlated with the presence of epicatechin glucoside and accumulation of PAs. UGT72L1 may be involved in the production of epicatechin 3′-O-glucoside in the seed coat as a key step in PA biosynthesis or its regulation.
Keywords: glucosyltransferase, model legume, proanthocyanidin
Proanthocyanidins (PAs, also called condensed tannins), are oligomeric/polymeric flavonoid compounds that provide protective functions in the fruits, bark, leaves and seeds of many plants (1). PAs benefit human health through their antioxidant, anticancer (2, 3), anti-inflammatory (4), and cardioprotective activities (5), and PA products from a number of different plant species are widely used as dietary supplements.
The presence of PAs is a positive trait in forage crops. PAs bind to proteins and slow their fermentation in the rumen, reducing generation of methane and thereby protecting the animal from potentially lethal pasture bloat (1). Furthermore, the protein-protective effects of moderate levels of PAs in forages improve nitrogen nutrition, reduce urinary nitrogen excretion, and help counter intestinal parasites. Along with bloat protection, these traits have been major targets for genetic breeding of forage crops (6–8).
Analysis of mutants of Arabidopsis thaliana that exhibit a transparent testa (tt) as a result of genetic lesions in flavonoid/PA biosynthesis (9) has identified transcriptional regulators of PA biosynthesis and deposition (10–13), and transporters potentially involved in movement of PA precursors to the cell vacuole (14, 15) [supporting information (SI) Fig. S1]. However, the exact nature of the intermediates that are transported and subsequently polymerized to PAs is not known.
The model legume Medicago truncatula contains oligomeric PAs, comprised primarily of (−)−epicatechin units, in seed coats (16). Here, we report that ectopic expression of the Arabidopsis MYB family transcription factor TRANSPARENT TESTA 2 (TT2) (11) in hairy roots of M. truncatula results in massive accumulation of oligomeric PAs. Microarray analysis of transcripts induced by TT2 in the hairy roots and those preferentially expressed in the Medicago seed coat led to the identification of a glycosyltransferase with unique specificity for (−)−epicatechin and therefore likely associated with PA biosynthesis.
Results
TT2 Induces PA Accumulation in Medicago Hairy Roots.
A construct containing the ORF of Arabidopsis TT2 (11), driven by the double 35S promoter, was transformed into Agrobacterium rhizogenes to generate hairy roots of M. truncatula. TT2-expressing hairy roots were phenotypically identical to empty vector controls, exhibiting a strong, reddish purple pigmentation (Fig. 1 A and B). However, when stained with dimethylaminocinnamaldehyde (DMACA) reagent, the TT2-expressing lines, but not the vector controls, turned an intense blue-green color (Fig. 1 C and D), indicative of the presence of PA polymers, oligomers, or precursor flavan-3-ols (17) .
Fig. 1.
Phenotypic appearance of transgenic M. truncatula hairy roots. (A) Unstained TT2-expressing roots. (B) Unstained vector control roots. (C) DMACA-stained TT2-expressing roots. (D) DMACA-stained empty vector control roots.
Soluble PAs were analyzed by normal phase HPLC, coupled with postcolumn derivatization with DMACA (18). No signal was observed after separation of extracts from control roots (Fig. 2B). The soluble PA fraction from the TT2-expressing line 239-5 contained monomers, dimers, and a range of oligomers with an estimated degree of polymerization ≤ 10 (Fig. 2A), based on calibration of the HPLC column with PA size standards (18). Epicatechin monomer and a compound with the same retention time as procyanidin B2 (epicatechin-(4β → 8)-epicatechin) were among the major soluble components. The average soluble PA content in two independent TT2-expressing lines was > 10 times the level in the control lines (Fig. 2F).
Fig. 2.
PA content and composition in M. truncatula hairy roots. (A) The soluble PA fraction from TT2-expressing line 239-5 analyzed by normal phase HPLC with postcolumn derivatization. (B) As above, for control line 2300-11. Letters indicate the retention times of authentic standards of (−)−epicatechin (Epi), (+)−catechin (Cat), procyanidin B1 (B1), and procyanidin B2 (B2). (C) Dried residues from line 239-5 (1, 3) and 2300-11 (2, 4) before (Left) and after (Right) hydrolysis in acid-butanol. (D) HPLC chromatograph of acid-butanol hydrolyzed products from a TT2-expressing line. (E) As above, from a vector control line. Letters indicate retention times of authentic anthocyanidin standards. De, delphinidin; Cy, cyanidin; Pe, pelargonidin. (F) Levels of total soluble (filled bars) and insoluble (open bars) PAs in duplicate TT2-expressing and empty vector lines.
Butanol-HCl hydrolysis of the insoluble cell residue fraction from the TT2-expressing lines led to a massive release of colored anthocyanidins (Fig. 2C), shown by HPLC analysis to consist largely of cyanidin (Fig. 2D), which originates from epicatechin and/or catechin extension units in PAs. Very little anthocyanidin was released from the insoluble residue of empty vector control lines (Fig. 2 C and E). The average level of insoluble PAs in two independent TT2-expressing lines was >24-fold higher than in the empty vector control lines (Fig. 2F) and >50-fold higher than the level of soluble PAs produced in response to expression of TT2. The overall PA level of TT2-expressing roots was higher than that found naturally in the seed coat of M. truncatula (16).
TT2 Induces Anthocyanin and Flavonol Biosynthesis in Medicago.
TT2, in conjunction with two other transcription factors, TT8 and TRANSPARENT TESTA GLABRA 1 (TTG1), controls the PA-specific branch of the flavonoid pathway in the Arabidopsis seed coat (11, 13), whereas other transcription factors control anthocyanin and flavonol accumulation (9). Empty vector-transformed Medicago hairy roots contained a significant level of anthocyanins as determined by spectrophotometric analysis, but this amount was approximately double in lines expressing TT2 (Fig. S2A). HPLC analysis of line 239-5 revealed the presence of multiple anthocyanin peaks (Fig. S2B), all of which disappeared after acid hydrolysis and were converted predominantly to cyanidin (Fig. S2 C and D), the precursor for both anthocyanins and (−)−epicatechin units in PAs. HPLC analysis also revealed the presence of flavonols, particularly quercetin, in TT2-expressing but not in control roots (Fig. S3).
Genes Induced by Ectopic Expression of TT2 in Medicago Hairy Roots.
TT2 is necessary for transcriptional activation of anthocyanidin reductase (ANR) (Fig. S1) in Arabidopsis (13). A preliminary screen of transgenic hairy roots by RT-PCR indicated that lines positive for TT2 expression also exhibited high levels of ANR transcripts, but ANR transcripts were not detected in empty vector control lines (Fig. 3A).
Fig. 3.
Transcripts induced in M. truncatula hairy roots by expression of TT2, or expressed in the M. truncatula seed coat. (A) RT-PCR screen of individual hairy root lines for expression of the TT2 transgene and endogenous ANR transcripts. Actin was used as loading control. (B) Scatter plots of gene expression level differences between TT2-expressing and control lines from Affymetrix microarray analysis. (C) Venn diagram showing overlap between probe sets induced by TT2 in hairy roots and expressed preferentially in the seed coat. a, Number of probe sets up-regulated by TT2; b, Number of probe sets preferentially expressed in seed coat; c, Intersection of a and b.
Total RNA samples from duplicate biological replicates of TT2-expressing and empty vector controls were subjected to Affymetrix microarray analysis. Changes in expression level of all probe sets on the chip are shown in Fig. 3B. Four hundred and twenty two probe sets were up-regulated in the TT2-expressing lines, and 344 were down-regulated. The Gene Ontology classifications of the up-regulated probe sets are summarized in Fig. S4A.
Of the 30 probe sets up-regulated > 10-fold (Table 1), seven represented genes with unknown function. ANR was the most strikingly induced gene (473-times the expression level in the empty vector control line). A number of other flavonoid pathway genes required for PA biosynthesis were also up-regulated > 2-fold in the TT2-expressing lines (Table S1), including genes encoding anthocyanidin synthase and leucoanthocyanidin reductase, which converts leucocyanidin to (+)−catechin (Fig. S1). Consistent with the increase in flavonols in the hairy roots, flavonol synthase transcripts were induced 16.6-fold.
Table 1.
The probe sets with expression more than 10 fold up-regulated by TT2 in M. truncatula hairy roots
| Probe sets | Annotation | Ratio (TT2/CK) | P-value* | Q-value† |
|---|---|---|---|---|
| Mtr.44985.1.S1_at | Anthocyanidin reductase, complete | 473.3 | 0.00003 | 0.05024 |
| Mtr.21996.1.S1_x_at | Weakly similar to glucosyltransferase-13 (fragment) | 64.8 | 0.00029 | 0.06699 |
| Mtr.41147.1.S1_at | Unknown | 63.5 | 0.00068 | 0.08141 |
| Mtr.47691.1.S1_at | Unknown | 29.5 | 0.00081 | 0.08448 |
| Mtr.10917.1.S1_at | Cytochrome P450 77A3, partial (95%) | 25.6 | 0.00015 | 0.06019 |
| Mtr.4369.1.S1_at | Similar to At2 g41420, partial (90%) | 25.2 | 0.00112 | 0.08818 |
| Mtr.47777.1.S1_at | Weakly similar to UP O81190 (O81190) putative transposase | 23.9 | 0.00693 | 0.11234 |
| Mtr.47631.1.S1_s_at | Weakly similar to UP Q5UDR1 (Q5UDR1) transposase, partial (37%) | 23.5 | 0.00123 | 0.08818 |
| Mtr.52009.1.S1_s_at | Putative BED finger; HAT dimerisation; immunoglobulin major histocompatibility complex | 20.4 | 0.00219 | 0.09470 |
| Mtr.50650.1.S1_s_at | Plant MUDR transposase; SWIM Zn-finger; Zn-finger, CCHC Type | 19.9 | 0.02058 | 0.13148 |
| Mtr.23138.1.S1_s_at | Weakly similar to MUDR family transposase protein, partial (61%) | 18.6 | 0.00029 | 0.06699 |
| Mtr.9658.1.S1_at | Unknown | 18.1 | 0.00013 | 0.05831 |
| Mtr.11000.1.S1_at | Unknown | 17.2 | 0.00533 | 0.10722 |
| Mtr.14017.1.S1_at | Similar to flavonol synthase (FLS), partial (19%) | 16.6 | 0.00539 | 0.10735 |
| Mtr.39235.1.S1_at | Similar to AT4 g28740 F16A16_150, partial (18%) | 16.6 | 0.00595 | 0.10923 |
| Mtr.38712.1.S1_at | Similar to AT4 g28740 F16A16_150, partial (23%) | 16.4 | 0.00730 | 0.11356 |
| Mtr.7974.1.S1_at | Unknown | 16.1 | 0.00032 | 0.06896 |
| Mtr.17084.1.S1_at | LQGC hypothetical protein | 16.0 | 0.00252 | 0.09579 |
| Mtr.18767.1.S1_at | Hypothetical protein | 15.9 | 0.00061 | 0.08114 |
| Mtr.45980.1.S1_at | LQGC hypothetical protein | 15.4 | 0.00011 | 0.05609 |
| Mtr.36851.1.S1_at | Unknown | 14.9 | 0.00353 | 0.10103 |
| Mtr.32890.1.S1_at | Similar to UP Q6NV39 (Q6NV39) Zgc: 85612, partial (2%) | 14.2 | 0.00178 | 0.09131 |
| Mtr.16495.1.S1_at | Cyclin-like F-box | 12.1 | 0.00009 | 0.05481 |
| Mtr.17982.1.S1_s_at | Hypothetical protein | 11.9 | 0.01932 | 0.13060 |
| Mtr.25016.1.S1_at | Unknown | 11.7 | 0.01440 | 0.12507 |
| Mtr.6531.1.S1_at | Similar to UP PGS1_XENLA (Q9IB75) biglycan precursor, partial (3%) | 11.5 | 0.01231 | 0.12194 |
| Mtr.51818.1.S1_at | Predicted protein | 11.4 | 0.00003 | 0.05024 |
| Mtr.28306.1.S1_at | Weakly similar to (GPI-anchored protein) (At5 g63500), complete | 10.5 | 0.03016 | 0.14094 |
| Mtr.33218.1.S1_at | Similar to F14N23.12 (At1 g10240 F14N23_12), partial (4%) | 10.4 | 0.01317 | 0.12292 |
| Mtr.18503.1.S1_s_at | LQGC hypothetical protein | 10.0 | 0.00778 | 0.11485 |
Two putative homologs of TT8, which encodes a bHLH protein involved in PA biosynthesis (12) were up-regulated by 2.0 and 2.3-fold, and a homolog of Arabidopsis TTG1, a WD40 repeat protein that regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis (19), was also induced by 2.3-fold (Table S1). Several probe sets with weak sequence similarity to the Arabidopsis transporters TT12 and TT19 (14, 15), and the proton translocating ATPase AHA10 necessary for PA biosynthesis (20), were weakly up-regulated by expression of TT2 (Table S2).
Genes Preferentially Expressed in the Medicago Seed Coat.
Ectopic, high level expression of transcription factors can result in artifactual pleiotropic effects (21). Therefore, we further interrogated TT2-induced genes for preferential expression in the seed coat, the natural site of PA biosynthesis in Medicago (16). Coats were dissected from developing seeds [from 16–24 days after pollination (dap)] and total RNA from pooled material analyzed by hybridization to Affymetrix arrays. A total of 1,546 gene probe sets were expressed in the seed coat at a level at least twice that in any other organ, and their Gene Ontology classifications are summarized in Fig. S4B. The gene with the highest seed coat specificity was a putative legumin J precursor (Table S3). Among the seed coat preferentially expressed genes, 45 probe sets were also up-regulated >2-fold by TT2 expression (Fig. 3C).
The genes encoding enzymes of PA biosynthesis have a clearly defined expression pattern in developing seed, with maximal transcript level at 10–12 dap followed by a decline to very low levels by 36 dap, paralleling the deposition pattern of PAs in the seed coat (16). Of the TT2-induced seed coat preferentially expressed genes, many exhibited the same expression pattern as flavonoid/PA biosynthetic genes such as ANR and chalcone synthase (CHS) (for example the TTG1 ortholog) (Fig. 4 A–C), as shown by mining the Medicago Gene Expression Atlas (22). Others, however, were expressed later in seed development, and likely reflect transcripts present in contaminating seed tissue that do not play a role in PA biosynthesis.
Fig. 4.
Transcript levels of selected genes during M. truncatula seed development and in different organs as determined by microarray analysis. (A-G) Normalized relative transcript levels of indicated genes during seed development. Numbers on the x axes represent dap. (H) Relative transcript level of UGT72L1 in different organs. a, fold up-regulated by TT2 versus control; b, fold preferentially expressed in seed coat versus non-seed organs.
Characterization of UGT72L1.
Two TT2-induced seed coat preferentially expressed genes were annotated as encoding uridine diphosphate glycosyltransferases (UGTs). One, UGT72L1, exhibited a > 10-fold higher expression in the seed coat than in any other organ (Fig. 4H), and a 64.8-fold higher expression in roots expressing TT2 as compared with controls. Furthermore, its expression kinetics in developing seeds were similar to those of ANR, CHS, and the TTG1 ortholog (Fig. 4D).
The genomic sequence of UGT72L1 present in Medicago BAC clone AC124966 (www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nuccore&id=90659913) contains no introns. Its coding sequence was obtained by RT-PCR from total RNA isolated from TT2-expressing hairy roots. It encodes a protein of 482 aa, with a putative isoelectric point of 5.16 and molecular mass of 53 kDa, and shows 52% amino acid identity to arbutin synthase (AS) from Rauvolfia serpentina and ≈30% identity to UGT71G1 and other flavonoid UGTs from M. truncatula (Fig. S5). The most related sequence in soybean showed 50% amino acid identity. Phylogenetic analysis indicated that UGT72L1 clustered in an outlying clade with AS but separate from the (iso)flavonoid-specific UGTs from M. truncatula (23) (Fig. S6). DNA gel blot analysis indicated that UGT72L1 is likely represented by three copies in the M. truncatula genome (data not shown).
The opening reading frame of UGT72L1 was expressed in E. coli as a maltose-binding protein (MBP) fusion (Fig. S7A). With UDP-glucose as a sugar donor, recombinant UGT72L1-MBP showed high activity for glucosylation of (−)−epicatechin (Fig. 5A), significant activity (27%) with (−)−epigallocatechin, and weak activity with (+)−catechin and cyanidin (<15% of the activity with epicatechin). It was not active with procyanidin B1, procyanidin B2, dihydroquercetin, kaempferol, quercetin, apigenin, luteolin, isoliquiritigenin, daidzein, or genistein. The pH optimum for glycosylation of epicatechin was 7.5–8.5 (Fig. S7B). After removal of the MBP tag by proteolytic cleavage, the native enzyme exhibited the same overall activity and substrate specificity as the fusion protein but was less stable on storage (data not shown).
Fig. 5.
Characterization of the product of recombinant MBP-UGT72L1 fusion protein. (A) HPLC analysis of products from 1-h incubation of MBP-UGT72L1 fusion protein with UDP-glucose and epicatechin (epi). (B) As above, but with boiled enzyme. (C) Mass fragment patterns and (D) UV absorption spectrum of epicatechin glucoside (epi-glc).
The product of the UGT72L1 reaction exhibited the mass fragmentation pattern of an epicatechin glycoside and a UV absorption spectrum similar to that of epicatechin (Fig. 5 C and D), and was converted to (−)−epicatechin on incubation with almond β-glucosidase (data not shown). NMR analysis (Table S4) showed a cross peak between H-1 of β-glucose and C-3′ of epicatechin in the HMBC spectrum, indicating linkage of glucose to O-3′ of the aglycone (Fig. S8A). This structure was confirmed by a cross peak in the NOESY spectrum between H-1 of glucose and H-2′ of epicatechin (Fig. S8B).
Kinetic analysis of recombinant MBP-UGT72L1 fusion protein revealed Km values for epicatechin and UDP glucose of 11.5 and 140 μM, respectively, and a Kcat value of 9.89 × 10−3·s−1.
Eight Medicago UGTs (GenBank accession nos. ABI94020, ABI94021, ABI94022, AAW56092, ABI94023, ABI94024, ABI94025 and ABI94020) are active with a range of flavonoid and isoflavonoid acceptor molecules (23), including cyanidin and quercetin. However, none of these enzymes could glycosylate (−)−epicatechin (data not shown).
Identification of Epicatechin Glucoside (epi-glc) in M. truncatula's Seed.
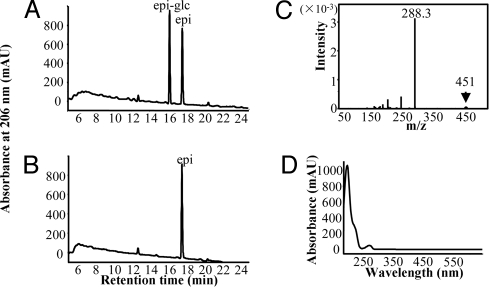
Flavonoid profiles of various organs and developing seeds were analyzed by liquid chromatography-mass spectrometry. Conjugates of apigenin, luteolin, and quercetin (quercetin-3-O-glucoside) were found in all organs examined (data not shown), as previously shown in alfalfa (24). In contrast, a compound with the same HPLC retention time, and UV- and mass-spectral characteristics as epi-glc, was found only in developing seeds (Fig. 6A, C, and D). This compound disappeared, with a corresponding increase in free epicatechin, when extracts were treated with β-glucosidase (Fig. 6B). More than 75% of the epicatechin in seed coats at 12 dap was present as a hydrolysable glucoside (Fig. S9). Epi-glc declined during seed development and was not detected in mature seeds (Fig. 6E). It was also detected in soluble extracts from TT2-expressing hairy roots (data not shown).
Fig. 6.
Identification of epicatechin glucoside in developing Medicago seed. (A) HPLC analysis of flavonoids from seeds at 12 dap. epi-glc, glucosylated epicatechin; epi, free epicatechin. Puerarin was internal standard. (B) As above, but after overnight hydrolysis with almond β-glucosidase. (C) UV absorption spectrum and (D) mass spectrum of epi-glc from M. truncatula seed. (E) Levels of epi-glc at different dap, based on analysis of 100-mg samples of pooled seed at each developmental stage.
Discussion
A Model System for the Study of PA Biosynthesis in Legumes.
Despite progress in the understanding of genetic lesions in Arabidopsis tt mutants (10–13, 25), there are still gaps in the understanding of PA biosynthesis, transport, and assembly (9, 26, 27). There are two rationales for developing M. truncatula as a model for understanding PA biosynthesis. First, it is a model with extensive genomic and genetic resources, including transposon insertion lines (28). Second, its very close relative, alfalfa, is a target for the introduction of the PA pathway through genetic engineering (16, 29).
The bulk of the PAs that accumulate in TT2-expressing Medicago hairy roots are insoluble polymers. Thus, TT2 activates genes for precursor synthesis, transport, oligomerization, and ultimate accumulation as high molecular weight polymers, unless some of these are already expressed in control roots. Medicago genes similar to the multidrug and toxic compound extrusion transporter TT12, the GST TT19, and the proton pumping ATPase AHA10, all of which are implicated in PA transport and/or accumulation (14, 15, 20), were only weakly induced by TT2 in the hairy roots. These genes are regulated by TT2 in Arabidopsis (9, 30). It is possible that their true Medicago orthologs were absent from the microarray chip, or that their induction is not necessary to support PA accumulation in Medicago roots.
Many of the probe sets up-regulated by TT2 in Medicago hairy roots may be induced as a result of off-target transactivation by ectopic expression of a transcription factor (21), or may represent responses to the abnormal metabolic load placed upon the cells. Combining the TT2 induction profile with a microarray dataset for seed coat preferentially expressed genes, directed us to the potential importance of UGT72L1 in PA biosynthesis. However, because of difficulties in obtaining uncontaminated seed coats, it was necessary to include the expression kinetics during seed development as an additional filter for exclusion of false positives.
UGT72L1 Is an Epicatechin Glucosyltransferase.
UGT72L1 appears specific for 2,3-cis-flavan-3-ol. Neither the 2,3-trans isomer, (−)−catechin, nor the achiral cyanidin, are good substrates, indicating a preference for a specific stereochemistry at C2-C3. In contrast, eight Medicago UGTs active with flavonoids all function with more than one class of compound; for example, GT83F (UGT78G1) is active with anthocyanidins, flavonols, flavones, coumestans, pterocarpans, and isoflavones (23). All these enzymes preferentially glycosylate at the C-ring 3-position if a hydroxyl group is present there, in contrast to UGT72L1, which glycosylates the B-ring at the 3′-position. However, none of the Medicago enzymes is active with (−)−epicatechin. The low Km of UGT72L1 for epicatechin, and the high Kcat/Km value when compared with other flavonoid UGTs, suggests that epicatechin is likely the natural substrate for the enzyme in vivo.
UGT72B1 from Arabidopsis and AS from Rauvolfia serpentina are the most closely related enzymes to UGT72L1. Both are multifunctional. UGT72B1 catalyzed both N- and O-glycosylation of xenobiotics (31), and AS glucosylated 45 out of 74 potential phenolic acceptors (32), in remarkable contrast to the apparently strict substrate specificity of UGT72L1.
Involvement of UGT72L1 in PA Biosynthesis.
UGT72L1's specificity for (−)−epicatechin, regulation by TT2, preferential expression in the seed coat, and expression kinetics that parallel those of PA biosynthetic genes, along with the inverse correlation between epi-glc and insoluble PA levels in the seed coat, together suggest a role for UGT72L1 in PA biosynthesis.
The tonoplastic multidrug and toxic compound extrusion family transporter TT12 can facilitate transport of cyandin 3-O-glucoside into yeast vesicles but does not transport catechin-3-O-glucoside (which inhibits transport of cyanidin glucoside), free cyanidin, or epicatechin (33) (Fig. S1). Epi-glc was not tested in this study due to lack of availability, but was suggested as the likely substrate for TT12. TT12 could recognize glycosides of epicatechin and the achiral cyanidin, but not catechin. After transport to the vacuole, free epicatechin could be released by the activity of a β-glucosidase. A glucosidase was induced 3.2-fold by TT2 in the Medicago hairy roots, and its transcripts followed the same developmental pattern as ANR and UGT72L1 during seed development (data not shown). Alternative hypotheses are: (a) that UGT72L1 regulates the levels of free epicatechin, either as a potential detoxification mechanism, or to control the relative levels of starter and extension units for PA biosynthesis; or (b) that glycosylation of epicatechin at the 3′-position prevents formation of an o-diquinone on the B-ring during oxidative polymerization, and thus directs epicatechin units into the correct 4–8 linkage pattern; the glucose units may then be removed subsequently. Preliminary genetic analysis indicates that seeds of M. truncatula harboring transposon insertions in UGT72L1 have unaltered or even increased levels of PAs, more consistent with the two latter hypotheses. Further analysis of PA size and linkage type will be necessary to determine whether these knock-outs have a measurable PA phenotype.
Materials and Methods
Generation of TT2-expressing M. truncatula Hairy Roots.
pSB239, containing the ORF of Arabidopsis TT2 driven by the double 35S CaMV promoter (30), and empty pCAMBIA2300 vector for controls, were transformed into Agrobacterium rhizogenes strain ARqual1 (34) by using the freezing-thaw method (35). Transformed colonies were grown on LB-agar medium with selection at 28°C for 2 d, then used to inoculate radicles of M. truncatula (cv. Jemalong A17) seedlings as described previously in ref. 36. The resulting hairy roots were maintained on B5 agar media in Petri dishes supplied with 50 mg/l kanamycin under fluorescent light (140 μ E/m2·s1) with a 16-h photoperiod, and were subcultured every month onto fresh media. Screening of hairy root clones by RT-PCR, and by staining with DMACA reagent for the presence of PAs, are described in SI Materials and Methods.
Microarray Analysis.
TT2-expressing line 239-5 and vector control line 2300-11 were inoculated onto fresh B5 media supplied with 50 mg/l kanamycin one month before sampling. Total RNA was isolated with Tri-reagent according to the manufacturer's protocol (Gibco-BRL). Forty μg of total RNA from each sample was cleaned and concentrated by using the RNeasy MinElute Cleanup Kit (Qiagen, WN), and 10 μg of purified RNA used for microarray analysis of two biological replicates (with analytical duplicates).
For each sample, the .CEL file was exported from Genechip Operating System (GCOS) program (Affymetrix). All four .CEL files were imported into robust multichip average (RMA) and normalized as described in ref. 37. The presence/absence call for each probe set was obtained from dCHIP (38). Differentially expressed genes between vector control and TT2-expressing line were selected using Associative Analysis (39). Type I family-wise error rate was reduced by using a Bonferroni corrected P value threshold of 0.05/N, where N represents the number of genes present on the chip. The false discovery rate was monitored and controlled by calculating the Q-value (false discovery rate) by using extraction of differential gene expression (EDGE, www.biostat.washington.edu/software/jstorey/edge) (40, 41).
To identify genes preferentially expressed in the seed coat, coats were isolated from seeds collected at 16 to 24 dap. RNA isolation and microarray analysis were performed as described above, and data for three biological replicates were obtained and compared with gene expression levels in multiple organs extracted from the Medicago gene atlas (22). Seed and pod were removed from the analysis because these tissues also contained seed coats. Data from all replicates and samples were normalized together with RMA to reduce technical variation. The maximum expression level of each probe set was calculated for all of the organs, and only those genes showing at least 2-fold higher expression in seed coat were considered to have seed coat preferential expression.
Analysis of PAs, Anthocyanins, and Total Flavonoids in Hairy Roots.
Extraction and subsequent analysis of the various classes of flavonoids from hairy roots and other organs of M. truncatula by UV spectroscopy; DMACA plate assay; and reverse phase or normal phase HPLC coupled to postcolumn derivatization, UV diode array detection, or mass spectrometry are described in SI Materials and Methods.
Cloning and Expression of UGT72L1.
The genomic sequence of UGT72L1 was retrieved from the Medicago BAC clone AC124966. The physical sequence was cloned from M. truncatula A17 wild-type genomic DNA with the primers MtGT1365CF and MtGT1365R (see SI Materials and Methods). Expression vector construction and expression of UGT72L1 as a MBP-fusion protein are described in SI Materials and Methods.
Recombinant UGT72L1-MBP was purified by affinity chromatography on an amylase resin (New England Biolabs), and UGT72L1 released from MBP by cleavage with Factor Xa protease (New England Biolabs) according to the manufacturer's instructions. Proteins were analyzed by electrophoresis on a 10–20% SDS polyacrylamide gel stained with Coomassie brilliant blue.
UGT72L1 was assayed in a reaction of 50 μl containing 100 mM Tris·HCl pH7.5, 10-μl protein (≈1.29 μg/μl) with 0.1 mM potential acceptor substrates, and 0.25 mM 14C-UDP-Glucose (8.8nCi/nmol). All assays were performed in triplicate for 1 h at 30°C along with boiled enzyme controls.
For studying pH optima, the buffers were 179 mM Mes pH 5.0–7.0, and 179 mM Tris·HCl pH 7.0–9.0. Potential acceptor substrates were (−)−epicatechin, (−)−epigallocatechin, (+)−catechin, (+)−gallocatechin, procyanidins B1 and B2, cyanidin, dihydroquercetin, quercetin, kaemferol, apigenin, luteolin, liquiritigenin, daidzein, and genistein (Sigma–Aldrich).
Phylogeny Analysis.
A multiple alignment of the deduced amino acid sequences of UGT72L1 and other UGTs was constructed by using MAFFT (42) and edited manually by using MacClade 4.0 (Sinauer Associates). Node support was estimated by using neighbor-joining bootstrap analysis (1000 bootstrap replicates) and unweighted parsimony bootstrap analysis (100 bootstrap replicates, 5 RAS per bootstrap replicate, limiting the search to 500 trees per RAS) by using PAUP* 4.0b10 (Sinauer Associates).
Supplementary Material
Acknowledgments.
We thank Dr. Tracy S. Feldman for assistance with phylogenetic analyses, David Huhman for assistance with liquid chromatography-mass spectrometry analysis, Dr. Luzia Modolo for advice on UGT assays, and Drs. Jiangqi Wen and Luzia Modolo for critical reading of the manuscript. This work was supported by the Samuel Roberts Noble Foundation and Forage Genetics International.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. EU434684). Microarray data are deposited at ArrayExpress (accession nos. E-MEXP-1115 and -1116).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805954105/DCSupplemental.
References
- 1.Dixon RA, Sharma SB., Xie D. Proanthocyanidins- a final frontier in flavonoid research? New Phytologist. 2005;165:9–28. doi: 10.1111/j.1469-8137.2004.01217.x. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad N, Gupta S, Mukhtar H. Green tea polyphenol epigallocatechin-3-gallate differentially modulates nuclear factor κB in cancer cells versus normal cells. Arch Biochem Biophys. 2000;376:338–346. doi: 10.1006/abbi.2000.1742. [DOI] [PubMed] [Google Scholar]
- 3.Kandil FE, et al. Composition of a chemopreventive proanthocyanidin-rich fraction from cranberry fruits responsible for the inhibition of 12-O-tetradecanoyl phorbol-13-acetate (TPA)-induced ornithine decarboxylase (ODC) activity. J Agric Food Chem. 2002;50:1063–1069. doi: 10.1021/jf011136z. [DOI] [PubMed] [Google Scholar]
- 4.Noreen Y, Serrano G, Perera P, Bohlin L. Flavan-3-ols isolated from some medicinal plants inhibiting COX-1 and COX-2 catalyzed prostaglandin biosynthesis. Planta-Med. 1998;64:520–524. doi: 10.1055/s-2006-957506. [DOI] [PubMed] [Google Scholar]
- 5.Serafini M, et al. Plasma antioxidants from chocolate. Nature. 2003;424:1013. doi: 10.1038/4241013a. [DOI] [PubMed] [Google Scholar]
- 6.Reed JD. Nutritional toxicology of tannins and related polyphenols in forage legumes. J Animal Sci. 1995;73:1516–1528. doi: 10.2527/1995.7351516x. [DOI] [PubMed] [Google Scholar]
- 7.Aerts RJ, Barry TN, McNabb WC. Polyphenols and agriculture: Beneficial effects of proanthocyanidins in forages. Agric, Ecosystems and Environ. 1999;75:1–12. [Google Scholar]
- 8.Douglas GB, Stienezen M, Waghorn GC, Foote AG, Purchas RW. Effect of condensed tannins in birdsfoot trefoil (Lotus corniculatus) and sulla (Hedysarum coronarium) on body weight, carcass fat depth, and wool growth of lambs in New Zealand. New Zealand J Agric Res. 1999;42:55–64. [Google Scholar]
- 9.Lepiniec L, et al. Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol. 2006;57:405–430. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- 10.Nesi N, et al. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell. 2002;14:2463–2479. doi: 10.1105/tpc.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniecm L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nesi N, et al. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–1878. doi: 10.1105/tpc.12.10.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baudry A, et al. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 14.Debeaujon I, Peeters AJM, Leon-Kloosterziel KM, Korneef M. The TRANSPARENT TESTA 12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell. 2001;13:853–871. doi: 10.1105/tpc.13.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitamura S, Shikazono N, Tanaka A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004;37:104–114. doi: 10.1046/j.1365-313x.2003.01943.x. [DOI] [PubMed] [Google Scholar]
- 16.Pang Y, Peel GJ, Wright E, Wang Z, Dixon RA. Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol. 2007;145:601–615. doi: 10.1104/pp.107.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treutter D. Chemical reaction detection of catechins and proanthocyanidins with p-dimethylaminocinnamaldehyde. J Chromatogr. 1989;467:185–193. [Google Scholar]
- 18.Peel GJ, Dixon RA. Detection and quantification of engineered proanthocyanidins in transgenic plants. Nat Prod Commun. 2007;2:1009–1014. [Google Scholar]
- 19.Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- 20.Baxter IR, et al. A plasma membrane H+−ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc Nal Acad Sci USA. 2005;102:2649–2654. doi: 10.1073/pnas.0406377102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broun P. Transcription factors as tools for metabolic engineering in plants. Curr Opin Plant Biol. 2004;7:202–209. doi: 10.1016/j.pbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Benedito VA, et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- 23.Modolo L, et al. A functional genomics approach to (iso)flavonoid glycosylation in the model legume Medicago truncatula. Plant Mol Biol. 2007;64:499–518. doi: 10.1007/s11103-007-9167-6. [DOI] [PubMed] [Google Scholar]
- 24.Deavours BE, Dixon RA. Metabolic engineering of isoflavonoid biosynthesis in alfalfa (Medicago sativa L. ) Plant Physiol. 2005;138:2245–2259. doi: 10.1104/pp.105.062539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pourcel L, et al. TRANSPARENT TESTA 10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell. 2005;17:2966–2980. doi: 10.1105/tpc.105.035154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie D-Y, Dixon RA. Proanthocyanidin biosynthesis- still more questions than answers? Phytochemistry. 2005;66:2126–2143. doi: 10.1016/j.phytochem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Grotewold E. The challenges of moving chemicals within and out of cells: Insights into the transport of plant natural products. Planta. 2004;219:906–909. doi: 10.1007/s00425-004-1336-0. [DOI] [PubMed] [Google Scholar]
- 28.Tadege M, et al. Large scale insertional mutagenesis using Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- 29.Lees GL. Condensed tannins in some forage legumes: Their role in the prevention of ruminant pasture bloat. Basic Life Sci. 1992;59:915–934. doi: 10.1007/978-1-4615-3476-1_55. [DOI] [PubMed] [Google Scholar]
- 30.Sharma SB, Dixon RA. Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. Plant J. 2005;44:62–75. doi: 10.1111/j.1365-313X.2005.02510.x. [DOI] [PubMed] [Google Scholar]
- 31.Brazier-Hicks M, et al. Characterization and engineering of the bifunctional. N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci USA. 2007;104:20238–20243. doi: 10.1073/pnas.0706421104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hefner T, Arend J, Warzecha H, Siems K, Stockigt J. Arbutin synthase, a novel member of the NRD1 beta glycosyltransferase family, is a unique multifunctional enzyme converting various natural products and xenobiotics. Bioorg Med Chem. 2002;10:1731–1741. doi: 10.1016/s0968-0896(02)00029-9. [DOI] [PubMed] [Google Scholar]
- 33.Marinova K, et al. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+−antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell. 2007;19:2023–2038. doi: 10.1105/tpc.106.046029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quandt HJ, Puhler A, Broer I. Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact. 1993;6:699–706. [Google Scholar]
- 35.Chen H, Nelson RS, Sherwood JL. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques. 1994;16:664–668. 670. [PubMed] [Google Scholar]
- 36.Limpens E, et al. RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot. 2004;55:983–992. doi: 10.1093/jxb/erh122. [DOI] [PubMed] [Google Scholar]
- 37.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dozmorov I, Centola M. An associative analysis of gene expression array data. Bioinformatics. 2003;19:204–211. doi: 10.1093/bioinformatics/19.2.204. [DOI] [PubMed] [Google Scholar]
- 40.Storey JD, Tibshirani R. Statistical significance for genome wide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leek JT, Monsen E, Dabney AR, Storey JD. EDGE: Extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- 42.Katoh K, Kuma K-i, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.