Abstract
Polα is the principal DNA polymerase for initiation of DNA replication and also functions in postinitiation DNA synthesis. In this study, we investigated the cell cycle responses induced by mutations in polα+. Germinating spores carrying either a deletion of polα+ (polαΔ) or a structurally intact but catalytically dead polα mutation proceed to inappropriate mitosis with no DNA synthesis. This suggests that the catalytic function, and not the physical presence of Polα, is required to generate the signal that prevents the cells from entering mitosis prematurely. Cells with a polαts allele arrest the cell cycle near the hydroxyurea arrest point, but, surprisingly, polαts in cdc20 (polε mutant) background arrested with a cdc phenoytpe, not a polαts-like phenotype. At 25°C, replication perturbation caused by polαts alleles induces Cds1 kinase activity and requires the checkpoint Rads, Cds1, and Rqh1, but not Chk1, to maintain cell viability. At 36°C, replication disruption caused by polαts alleles induces the phosphorylation of Chk1; however, mutant cells arrest with heterogeneous cell sizes with a population of the cells entering aberrant mitosis. Together, our results indicate that the initiation DNA structure synthesized by Polα is required to bring about the S phase to mitosis checkpoint, whereas replication defects of different severity caused by polαts mutations induce differential downstream kinase responses.
INTRODUCTION
Cells have a complex network of mechanisms to coordinate the completion of chromosome replication and repair of damaged DNA with mitotic entry. Early cell fusion experiments demonstrated that when an S phase cell is fused with a G2 cell, the G2 nucleus delays its mitotic entry until the S phase nucleus finishes DNA replication. This suggests that S phase cells have a mitotic inhibitor or an inhibitory signal that prevents premature mitosis. (Rao and Johnson, 1970). Subsequent genetic studies of Saccharomyces cerevisiae and Schizosaccharomyces pombe have substantially contributed to the understanding of how cells maintain the interdependency of S phase and mitosis. In S. pombe, deletion or mutation of genes involved in the initiation of S phase (cdc18+, cdt1+, cut5+, cdc30+, and polα+) allow the cells to enter inappropriate mitosis (Kelly et al., 1993a,b; Saka and Yanagida, 1993; Hofmann and Beach, 1994; Saka et al., 1994; D’Urso et al., 1995; Grallert and Nurse, 1996). In contrast, cells carrying deletion of genes such as polδ and pcn1 (proliferating cell nuclear antigen), which are involved in the elongation process of DNA replication, arrest with a cdc phenotype (Waseem et al., 1992; Francesconi et al., 1993). These findings suggest that it is the initiation of DNA replication that generates the signal, preventing cells from entering mitosis prematurely (Li and Deshaies, 1993; Nurse, 1994). However, it is not known whether it is the formation of the replication complex on the origin or the initiation DNA structure that is responsible for generating the S to M phase checkpoint.
Several proteins are essential for the initiation of DNA synthesis in S. pombe, including Orp1, Cdc18, and Polα. However, the roles played by each protein in this process are fundamentally distinct. A prerequisite for initiation of DNA replication is the assembly of a prereplication complex on the origin, which includes Orp1 and Cdc18 (Diffley, 1996; Aparicio et al., 1997; Donovan et al., 1997; Newlon, 1997; Tanaka et al., 1997), although neither Orp1 nor Cdc18 participates directly in the synthesis of the initiation DNA structure (Muzi and Kelly, 1995; Muzi et al., 1996; Stillman, 1996). In contrast, Polα is a component of the replication complex that directly participates in synthesis of the initiation DNA structure at the replication origin. Thus, the role of Polα in initiation is entirely different from that of Orp1 and Cdc18 (Stillman, 1996; Wang, 1996). In addition, Polα is also involved in postinitiation DNA synthesis (Wang, 1991, 1996; Campbell, 1993). Because Polα plays a dual role in both the formation of the replication complex and the synthesis of nascent DNA, Polα is the ideal replication enzyme to dissect the question of what generates the replication checkpoint signal during initiation.
Previous studies have shown that germinating spores carrying a disrupted polα+ gene entered mitosis when DNA synthesis was inhibited by hydroxyurea, thus implicating Polα as playing a role in the coordination of S phase with mitosis (D’Urso et al., 1995). However, this study did not resolve the question of whether the inappropriate mitotic entry was due to the physical absence of Polα, resulting in a failure to assemble the replication complex, or due to the absence of Polα catalytic activity and a subsequent inability to synthesize an initiation DNA structure. Thus the question remains as to why deletion of polα+ fails to bring about the appropriate replication surveillance responses in these cells.
Once DNA synthesis has initiated, cells have additional surveillance mechanisms to delay mitotic entry in the event of DNA damage or blocks to ongoing replication. Studies of S. cerevisiae and S. pombe have identified several genes involved in these mechanisms (Hartwell and Weinert, 1989; Enoch et al., 1993; Sheldrick and Carr, 1993; Nurse, 1994; Carr and Hoekstra, 1995; Carr, 1996; Elledge, 1996; Lydall and Weinert, 1996; Paulovich et al., 1997). In S. pombe, a group of six “checkpoint Rad” proteins (Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1) are thought to be involved in monitoring damaged DNA and S phase arrest caused by hydroxyurea or a cdc mutant (Al-Khodairy and Carr, 1992; Enoch et al., 1992; Rowley et al., 1992; Al-Khodairy et al., 1994). Downstream of the checkpoint Rad proteins are two effector proteins, Chk1 and Cds1. In response to DNA damage, the Chk1 protein is absolutely required for cell cycle arrest in G2 and undergoes a checkpoint Rad-dependent phosphorylation (Walworth and Bernards, 1996), which inhibits the activation of cdc2 kinase by regulating the phosphorylation of Tyr15 (O’Connell et al., 1997; Rhind et al., 1997). Interestingly, cells arrested by a cdc mutation in a chk1Δ background enter mitosis inappropriately (Francesconi et al., 1995; Uchiyama et al., 1997), whereas cells arrested by the S phase inhibitor hydroxyurea at 30°C do not activate Chk1 (Walworth and Bernards, 1996). A recent study has demonstrated that the primary effector responding to hydroxyurea block is not Chk1, but Cds1 (Lindsay et al., 1998). Cds1 was originally identified as a multicopy suppressor of a DNA polymerase α thermosensitive allele, swi7-H4 (Murakami and Okayama, 1995), and has recently been shown to be required for reversible S phase arrest. It is important for maintaining the viability of cells when S phase is arrested by hydroxyurea or DNA lesions (Lindsay et al., 1998). Another protein, Rqh1, is also required for reversible S phase arrest (Murray et al., 1997; Stewart et al., 1997). Therefore, in addition to the checkpoint Rad-Chk1 pathway, cells have a checkpoint Rad-Cds1-Rqh1 subpathway for recovery of cells during S phase perturbation. Because Polα is involved in both initiation and postinitiation DNA synthesis, studies with different mutant alleles of this enzyme will help further elucidate the different cell cycle surveillance responses during S phase progression.
In this study using a polαΔ strain as well as a strain carrying a structurally intact but catalytically dead polα mutant, we demonstrate that the initiation DNA structure is required to generate the S phase to mitosis checkpoint signal. In addition, using polαts mutants, we clearly demonstrate that the different extents of perturbation and disruption of DNA replication caused by these mutations induce differential downstream cell cycle kinase responses.
MATERIALS AND METHODS
Strains, Media, and Genetic and Molecular Methods
S. pombe strains used in this study are listed in Table 1. Rich medium (yeast extract) and Edinburgh minimal medium (EMM) were as described by Moreno et al. (1991). All standard genetic methods were as described by Gutz et al. (1974). Standard molecular biology techniques were carried out as described by Maniatis et al. (1982). The plasmid pDblet (Brun et al., 1995) was modified by replacing the ura4+ marker with Leu2+, and the modified plasmid is named pDblet(leu). Transformation of fission yeast was performed by using the lithium acetate method described by Griffiths et al. (1995). For growth analysis of mutant strains, cells were first grown at 25°C to exponential phase and then shifted to 36°C. At the indicated time, cell number was determined by hemocytometer count. Cell viability measured at the restrictive temperature was performed by removal of a fixed number of cells at defined time intervals after shift to 36°C. Cells were diluted and plated onto yeast extract plates and incubated at 25°C for 3 d. Colonies were scored, and viability was expressed as a percentage of the colonies formed on cell samples plated immediately before shifting to 36°C.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| KG2 | h− ade6-M216 leu1-32 ura4-D18 his3-D1 | K. Gould |
| KG3 | h+ ade6-M210 leu1-32 ura4-D18 his3-D1 | K. Gould |
| KG23 | h−/h+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura-4D18/ura4-D18 his3-D1/his3-D1 | This study |
| DB23 | h−/h+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1 polα+/polαΔ::his3+ | This study |
| DB24 | h−/h+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1 polαts13/polαΔ::his3+ | This study |
| DB25 | h−/h+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1 polα+/polαΔ::his3+[pDblet(leu)polα+] | This study |
| DB26 | h−/h+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1 polα+/polαΔ::his3+[pDblet(leu)polαts13] | This study |
| DB27 | h−/h+ ade6-M216/ade6-M210 leu1-32/leu1-32 ura4-D18/ura4-D18 his3-D1/his3-D1 polα+/polαΔ::his3+[pDblet(leu)polαD984N] | This study |
| DB2 | h+ polαΔ::his3+ ade6-M216 leu1-32 ura4-D18 his3-D1(pRep81-polα+) | This study |
| DB3 | h+ polαΔ::his3+ ade6-M216 leu1-32 ura4-D18 his3-D1 pRep82-polα+) | This study |
| DB10 | h+ polαΔ::polα+ leu1+ his3+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| DBts11 | h+ polαΔ::polαts11 leu1+his3+ ade6-M216 leu1-32 ura4-D18 his3-D1 | This study |
| DBts13 | h+ polαΔ::polαts13 leu1+his3+ ade6-M210 leu1-32 ura4-D18 his3-D1 | This study |
| DBts131 | h+ polαΔ::polαts13 ade6-M210 leu1-32 ura4-D18 | This study |
| NW222 | h− chk1:ep ade6-M216 leu1-32 | N. Walworth |
| TE767 | h− rqh1::ura4+ ura4-D18 | T. Enoch |
| DB232 | h− chk1:ep polαΔ::polats13 leu1+ his3+ ade6-M210 leu1-32 ura4-D18 | This study |
| DB242 | h+ polαΔ::polαts13 leu1+ his3+ chk1::ura4+ ade6-M210 leu1-32 ura4-D18 | This study |
| DB252 | h+ polαΔ::polαts13 leu1+ his3+ cds1::ura4+ ade6-M216 leu1-32 ura4-D18 | This study |
| DB272 | h+ polαΔ::polαts11 leu1+ his3+ chk1::ura4+ ade6-M210 leu1-32 ura4-D18 | This study |
| polαts13 cdc10 | h+ polαΔ::polαts13 leu1+ his3+ cdc10-129 ade6-M210 leu1-32 ura4-D18 | This study |
| polαts13 cdc17 | h− polαΔ::polαts13 leu1+ his3+ cdc17-K42 ade6-M216 leu1-32 ura4-D18 | This study |
| polαts13 cdc18 | h− polαΔ::polαts13 leu1+ his3+ cdc18-K46 ade6-M210 leu1-32 ura4-D18 | This study |
| polαts13 cdc19 | h− polαΔ::polαts13 leu1+ his3+ cdc19-P1 ade6-M210 leu1-32 ura4-D18 | This study |
| polαts13 cdc20 | h− polαΔ::polαts13 leu1+ his3+ cdc20-M10 ade6-M210 leu1-32 ura4-D18 | This study |
| polαts13 cdc21 | h+ polαΔ::polαts13 leu1+ his3+ cdc21-M68 ade6-M210 leu1-32 ura4-D18 | This study |
| polαts13 cdc22 | h− polαΔ::polαts13 leu1+ his3+ cdc22-M45 ade6-M210 leu1-32 ura4-D18 | This study |
| polαts13 cdc25 | h− polαΔ::polαts13 leu1+ his3+ cdc25-22 ade6-M210 leu1-32 ura4-D18 | This study |
| polαts13 cdc2-3w | h− polαΔ::polαts13 leu1+ his3+ cdc2-3w ade6-M216 leu1-32 ura4-D18 | This study |
| polαts13 poldts03 | h− polαΔ::polαts13 leu1+ his3+ poldts03 ade6-M216 leu1-32 ura4-D18 | This study |
Construction of polαΔ Strains
Heterozygous diploid strain DB23 (Table 1) carrying a full deletion of the polα gene was constructed by a one-step gene replacement method. A 1.2-kb his3+ gene flanked by the polα+ genomic sequences (486-bp upstream sequence and 700-bp downstream sequence) was transformed into the diploid strain KG23 (Burke and Gould, 1994). Histidine prototrophic transformants were selected. The replacement of polα+ coding sequence by his3+ was confirmed by two methods: 1) genomic Southern analysis of the stable his3+ prototrophs, and 2) sporulation followed by tetrad dissection, which yielded two viable, histidine auxotrophic spores. For analysis of cells containing polαΔ, histidine prototrophic spores derived from DB23 (polα+/polαΔ) were selected to germinate at 30°C.
A haploid strain was constructed by transforming the heterozygous diploid DB23 (polα+/polαΔ) with pREP82-polα+ containing the ura4+-selectable marker (Maundrell, 1993). Histidine and uracil prototrophic transformants were selected, followed by sporulation and tetrad dissection. Haploid cells derived from the histidine and uracil prototrophic spores were designated DB3, which contains polαΔ::his3+[pREP82-polα+].
Another diploid strain, DB24, heterogeneous for polαΔ, was constructed by crossing DB3 with the thermosensitive haploid strain DBts13 (polαts13). After 5-fluoro-orotic acid (FOA) selection, the diploid was sporulated in EMM and inoculated in media minus leucine for selective germination of spores carrying polαts13 at 36°C. Diploid strains DB25, DB26, and DB27 were constructed by transforming the diploid strain DB23 (polα+/polαΔ) with pDblet(leu)polα+, pDblet(leu)polαts13, and pDblet(leu)polα(D984N), respectively. The diploids were sporulated and germinated at 25°C in EMM containing adenine and uracil for selective germination of spores containing polαΔ::his3+ [pDblet(leu)polα].
Isolation of Temperature-sensitive polα Mutants
The polα+ gene on plasmid pREP81 (Maundrell, 1993) was mutagenized using hydroxylamine as described (Rose et al., 1990). After mutagenesis, the DNA was transformed into Escherichia coli strain CJ236 (ung−) (Kunkel et al., 1987). Mutagenized plasmid DNAs were prepared from 1 × 105 ampicillin-resistant colonies.
Thermosensitive polα mutants were isolated by two different approaches. 1) Mutagenized plasmid pREP81-polα DNAs were transformed into the haploid strain DB3 containing polαΔ::his3+ [pREP82-polα+] followed by plasmid shuffling (Boeke et al., 1987). Transformants were replica plated onto EMM plates lacking histidine and leucine but containing FOA and incubated at 25°C for 4 d. Colonies that survived FOA selection were then replica plated onto selective medium containing phloxin B at 36°C for 24 h. Red colonies were selected as putative polα thermosensitive mutants and confirmed by several rounds of temperature selection. 2) Mutagenized pREP81-polα plasmid DNAs were transformed into the heterozygous diploid strain DB23 (polαΔ/polα+). Transformants were pooled, sporulated, and germinated in selective EMM medium. The haploid cells derived from histidine and leucine prototrophic spores were replica plated at 36°C onto selective EMM medium containing phloxin B. Red colonies were selected as potential thermosensitive mutant clones and confirmed as described above. After screening ∼5 × 104 colonies, 18 thermosensitive polα mutants were isolated. Four representative thermosensitive mutant alleles were identified by sequence analysis (Table 2).
Table 2.
Representative thermosensitive mutant alleles of polα+
| Allele | Mutation
|
Amino acid change | |
|---|---|---|---|
| Nucleotide | Amino acid | ||
| polαts11 | ACT → ATT | 840 | Thr-Ile |
| polαts13 | Deletion | 470–472 | Leu, Ser, arg, (deleted) |
| polαts16 | ACA → CCA | 759 | Thr-Pro |
| polαts17 | GCC → GTC | 463 | Ala-Val |
| CAT → TAT | 624 | His-Tyr | |
| GAC → AAC | 1183 | Asp-Asn | |
Integration of Wild-Type and Mutant polα
Wild-type polα+ and the mutant polαts13 gene under its endogenous chromosomal promoter and terminator sequences in tandem with the S. pombe leu1 sequence was cloned into the plasmid pJK148 (Keeney and Boeke, 1994). Plasmid pJK148 containing the polα sequence was linearized at an unique PstI site in the polα+ upstream region to facilitate recombination at the polα chromosomal locus. Linearized plasmid DNA was transformed into the heterozygous diploid strain DB23 containing polαΔ::his3+ followed by sporulation and germination. Haploid leucine and histidine prototrophs were selected. Stable integrants DB10 (polα+), DBts11 (polαts11), and DBts13 (polαts13) were identified by several rounds of selection on nonselective media and further confirmed by genomic Southern analysis. DBts13 (polαts13) was further crossed with wild-type SP808 to remove the leu1+ marker, and the resulting strain was named DBts131 (polαts13/leu−). Strains DBts13 and DBts131 yielded identical results in all studies. Thus, DBts13 (polαts13) was used as the representative thermosensitive mutant for most of the studies in this paper.
Generation and Purification of Cds1 Antibody
Cds1 protein expressed in S. pombe as a GST fusion protein was affinity purified on a glutathione-agarose column followed by a Hitrap Q column (Pharmacia). The purified GST-Cds1 protein (300 μg) was used as antigen to immunize rabbits. The crude sera was affinity purified on a tandem GST column and GST Cds1 column. The affinity-purified antibody was used to test cross-reactivity against the purified protein and crude extracts from S. pombe wild-type cells and cds1 null mutant cells. The antibody recognized a single Cds1-specific band in the crude extract from wild-type cells, and this band was not present in extracts derived from the cds1Δ strain.
Cds1 Kinase Assay
Cds1 kinase assay was performed as described by Lindsay et al. (1998) with modification. Cells were grown to midlog phase, washed in PBS, and then washed in lysis buffer (150 mM HEPES, pH 7.9, 250 mM KCl, 50 mM NaF, 60 mM β-glycerol phosphate, 15 mM p-nitrophenyl phosphate, 1 mM DTT, 1 mM EDTA, supplemented with a mixture of protease inhibitors). Cells suspended in lysis buffer were disrupted by vortexing with glass beads. The protein extracts were spun at 15,000 rpm for 15 min at 4°C to remove the glass beads and cell debris. Protein concentrations of the supernatant were determined, and 300 μg of the protein extract in 500 μl of lysis buffer were incubated with a 1:400 dilution of the affinity-purified Cds1 antibody at 4°C for 2 h. Immunocomplexes were further incubated with 30 μl of protein A beads (50% slurry) at 4°C for an additional 1 h. The protein A beads were precipitated and washed three times with lysis buffer and three times with kinase buffer (10 mM HEPES, pH 7.5, 75 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT). The immunocomplex-protein A pellet was incubated in a 20-μl reaction containing 100 μM ATP, 5 μg of myelin basic protein (MBP), 5 μCi of [γ-32P]ATP at 30°C for 10 min. The reaction was terminated by the addition of 5 μl of 5× SDS sample buffer. After boiling for 3 min, the samples were run on 15% gels, fixed in 40% methanol and 10% acetic acid, and dried before exposure to films. Equal amounts of Cds1-immunoprecipitate used in the kinase assay were quantitated by gel analysis. The extent of phosphorylated MBP was quantitated by using an IS-1000 digital imaging system (Alpha Innotech, San Leandro, CA).
Reciprocal Shift Experiments Using Hydroxyurea Block and Release
Reciprocal shift experiments using a hydroxyurea block and release were performed with either single mutant DBts13 (polαts13) or double mutants harboring polαts13 and cdc20 or cdc25 mutant alleles as described by Nasmyth and Nurse (1981). Hydroxyurea was added to a final concentration of 12 mM to each cell culture at 25°C and incubated in YES. After 4 h in hydroxyurea, cells were washed extensively with prewarmed (36°C) YES and then resuspended and grown in YES at 36°C. Cell samples were removed at indicated time intervals for analysis of growth rate, viability, DNA content, and nuclear and cell morphology.
Cytological Analysis
Cells were fixed in 70% ethanol and stained by addition of DAPI followed by calcofluor, processed, examined, and photographed as described (Uchiyama et al., 1997).
Flow Cytometry Analysis
Cells were collected, washed in water, and fixed in 70% ethanol before staining with propidium iodide as described by Paulovich and Hartwell (1995). DNA contents was measured by a Coulter Electronics (Hialeah, FL) fluorescence-activated cell sorter.
RESULTS
Cells with polαΔ Enter Mitosis with a 1C DNA Content
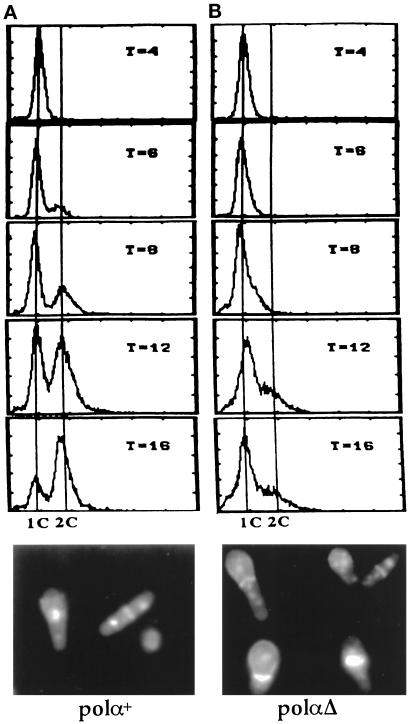
We have previously shown that cells carrying a disrupted polα gene display heterogeneous cell morphology (Francesconi et al., 1993). D’Urso et al. (1995) have shown that cells harboring a disrupted polα gene arrested with variable amounts of DNA and entered aberrant mitosis. The variable amounts of DNA synthesis observed in spores carrying a disrupted polα gene were thought to be due to residual Polα being carried over from the original diploid after sporulation (D’Urso et al., 1995). To definitively discern the DNA content of cells in the absence of polα+, we constructed a diploid strain (DB23) that is heterozygous for a complete deletion of the polα+ coding sequence and polα+. polαΔ spores, derived from the diploid DB23 (polαΔ/polα+), were selected for germination. Sixteen hours after inoculation, no DNA synthesis was observed in spores deleted of polα+. After 12 h, ∼60% of the cells were either anucleated or had missegregated nuclear material across the septum (Figure 1). The phenotype of the polαΔ spores is similar to that shown by Francesconi et al. (1993) and D’Urso et al. (1995) and identical to that of cdc18Δ and cdc30Δ germinating spores (Kelly et al., 1993a; Grallert and Nurse, 1996). To further substantiate this observation, we constructed a heterozygous diploid DB24 (polαΔ/polαts13) carrying a complete deletion of polα+ and a copy of the polα gene containing a thermosensitive polαts allele in tandem with the leu+ gene (see description of polαts alleles below and Table 1 for strain description). Spores derived from DB24 (polαΔ/polαts13) were germinated in a leucine-minus medium at 36°C for selective germination of spores carrying polαts13. After 10 h at 36°C, these spores displayed aberrant nuclear phenotypes identical to those of polαΔ spores. These results demonstrate that Polα plays a critical role in coordinating S phase with mitosis.
Figure 1.
polαΔ germinating spores undergo mitosis with 1C DNA content. FACS profile and phenotype of polα+ (A) and polαΔ (B) germinating spores at 30°C. Shown here are germinating spores 12 h after inoculation into selective medium.
The Catalytic Function, Not the Physical Presence of Polα, Is Required to Generate the S Phase to Mitosis Checkpoint
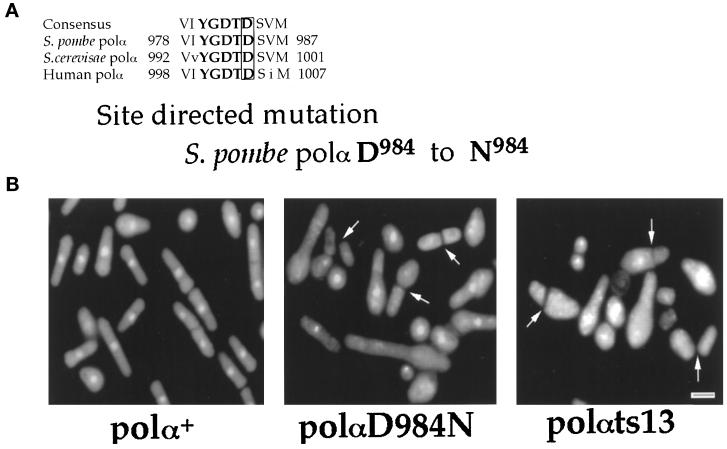
To distinguish whether it is only the physical presence of Polα in the replication complex or whether the catalytic function of Polα for synthesis of an initiation DNA structure is necessary for bringing about the replication checkpoint, we constructed a catalytically dead but structurally intact Polα mutant. Asp984 of S. pombe Polα is a critical residue in region I, the most conserved region of the α-like DNA polymerases (Figure 2A) (Delarue et al., 1990; Ito and Braithwaite, 1991; Wang, 1991, 1996). Previous mutational studies have shown that conservative mutation of the second Asp residue of human Polα Asp1004 to Asn completely abolishes the catalytic activity of Polα. This mutation, however, does not alter either the protein structure of Polα or the ability of the mutant Polα protein to assemble into the Polα–primase complex (Copeland and Wang, 1993a,b). We therefore introduced an identical mutation into the S. pombe Polα by changing Asp984 to Asn and investigated the effect of the physically intact but catalytically dead Polα mutant, polα(D984N), on the S phase to mitosis checkpoint.
Figure 2.
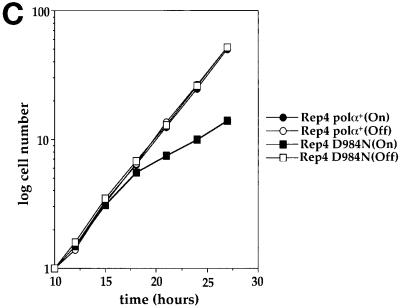
Germinating spores lacking Polα catalytic activity undergo inappropriate mitosis. (A) Primary sequence conservation of the region I of α-like DNA polymerases (Delarue et al., 1990; Ito and Braithwaite, 1991; Wang, 1991, 1996). Asp984 of S.pombe DNA polymerase α was mutagenized to Asn. (B) Phenotype of germinating spores containing a chromosomal polαΔ and plasmids pDblet(leu)polα+, pDblet(leu)polα(D984N), and pDblet(leu)polαts13. Diploid DB23 with one copy of the chromosomal polα+ deleted was transformed with pDblet(leu)polα+ or pDblet(leu)polα(D984N) and inoculated into selective media for germination of spores containing polαΔ/pDblet(leu)polα+ and polαΔ/pDblet(leu)polα+ at 30°C. The phenotype of the cells shown here is 14 h after inoculation. Diploid DB23 cells transformed with pDblet(leu)polαts13 were inoculated into selective media for germination of spores containing polαΔ/pDblet(leu)polαts13 at 36°C, and the phenotype shown is 10 h after inoculation. Bar, 4 μm. (C) Dominant negative effect of overexpressing Polα(D984N) mutant. Cell number increase after induction by removal of thiamine or repression by addition of thiamine was measured by counting cells starting from 10 h using a hemocytometer. After 16-h removal of thiamine from the media (Maundrell, 1993), the overexpression of catalytically dead Polα(D984N) caused a significant slowdown of cell growth.
Mutant polα(D984N) was cloned into the vector pDblet(leu) and transformed into the diploid DB23 (polαΔ/polα+). As controls, plasmids pDblet(leu)polα+ and pDblet(leu)polαts13 (see description of polαts mutations below) were also constructed and transformed into the diploid DB23 (polαΔ/polα+). The diploid cells carrying each of the three individual pDblet(leu)polα constructs were sporulated and selectively germinated for the polαΔ/pDblet(leu)polα. Fourteen hours after inoculation of the spores at 30°C, the spores sustained with plasmid pDblet(leu)polα+ had germinated into normal cells (Figure 2B). In contrast, spores containing the plasmid pDblet(leu)polα(D984N) entered mitosis in the absence of DNA synthesis. Approximately 50% of these germinating spores displayed an aberrant mitotic nuclear phenotype, with either anucleated cells or cells with missegregated nuclear material across the septum (Figure 2B). Furthermore, none of these cells arrested with a cdc phenotype. An identical phenotype was observed when polαΔ spores harboring the plasmid pDblet(leu)polαts13 were germinated at the restrictive temperature. To further ensure that the observed aberrant mitotic phenotype is caused by the catalytically dead mutant, polα(D984N) was constructed into an inducible vector (Maundrell, 1990). Cells harboring the pRep4 polα(D984N) plasmid under uninduced conditions displayed a similar growth rate as the cells harboring the wild-type polα+ plasmid, with a doubling time of 3 h. In contrast, induced cells with overexpressed Polα(D984N) had a doubling time of 6 h, showing that expression of the catalytically dead Polα(D984N) mutant has a dominant negative effect on cell growth (Figure 2C). Furthermore, 24 h after induction, ∼20% of the cells had an elongated phenotype. Dominant negative effects are usually attributed to assembly of the defective protein into complexes with other cellular components, rendering a population of nonfunctional complex. Thus, our results indicate that the Polα(D984N) is competent to assemble into the replication complex, disabling the replication complex, and causing the observed slower cell growth rate. This result strongly supports the notion that the aberrant nuclear morphology observed in cells containing the Polα(D984N) (Figure 2B) is caused by the presence of a catalytically nonfunctional mutant Polα in the replication complexes. Our results thus indicate that it is the catalytic function of Polα, essential for the synthesis of an initiation DNA structure, and not the physical presence of Polα in the replication complex, that is required for generating the signal that prevents cells from entering inappropriate mitosis.
Thermosensitive Mutant Alleles of polα
To further investigate how mutations of Polα affect cell cycle events during S phase progression, we isolated 18 thermosensitive polα mutants by two approaches described in MATERIALS AND METHODS. Four mutants carrying polαts11, polαts13, polαts16, and polαts17 alleles display aberrant mitotic nuclear morphology at the restrictive temperature of 36°C. We identified and sequenced these four mutant alleles (Table 2). Because polαts13 contains a deletion of three contiguous amino acid residues, we further tested whether mutation of each of the individual amino acid residues of polαts13 would cause temperature-sensitive cell growth. Ser470, Leu471, and Arg472 were individually mutagenized to Ala and found to have no effect on cell growth at 36°C. This indicates that the observed thermosensitivity of DBts13 (polαts13) is caused by the deletion of more than one amino acid residue. In this study, we characterized two mutants, DBts11 (polαts11) and DBts13 (polαts13), and investigated the effects of these two polαts alleles on different cell cycle events.
Characterization of polα Thermosensitive Mutants
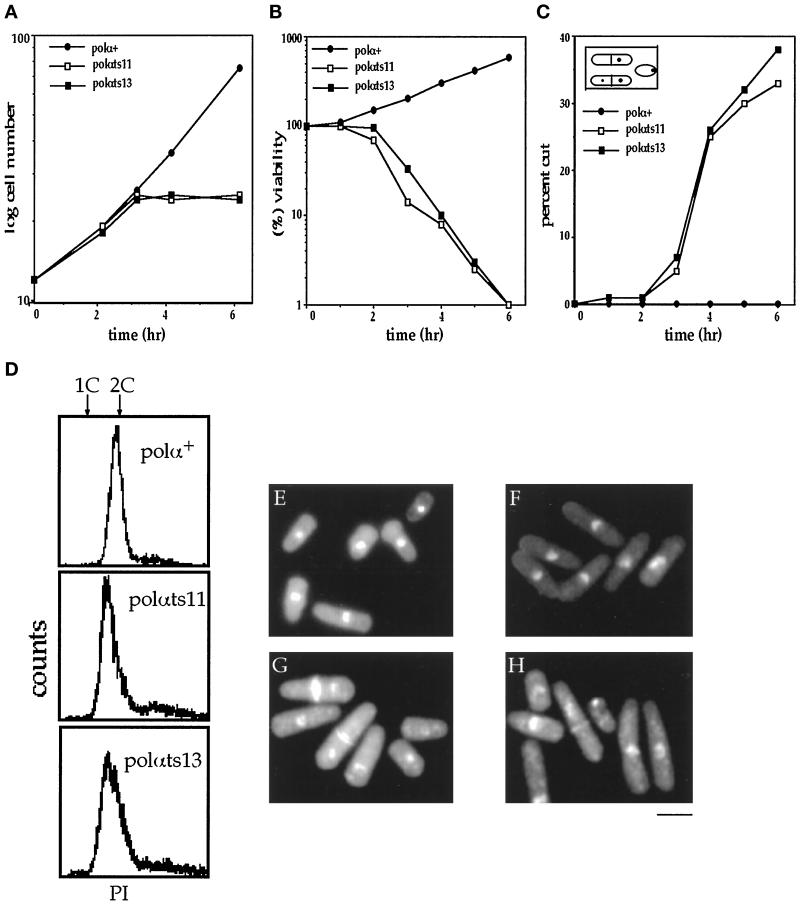
At the permissive temperature, the mutants DBts11 (polαts11) and DBts13 (polαts13) exhibit a slightly elongated cell morphology with normal nuclear morphology (Figure 3F). The growth rate is comparable to the wild-type DB10 (polα+) cells (our unpublished observations). When midlog phase cultures of DBts11 (polαts11) and DBts13 (polαts13) were shifted to 36°C, they doubled their cell number once and then arrested cell growth after 3 h. In contrast, wild-type DB10 (polα+) cells continued to double every 2 h (Figure 3A). Viability analysis showed that the mutant cells could be recovered 2 h after shift to 36°C, but there was an overt decrease in their ability to recover after 3 h (Figure 2B). Both DBts11 (polαts11) and DBts13 (polαts13) began to display aberrant nuclear morphology 3 h after shift to 36°C. After 6–8 h, ∼40% of the mutant cells exhibited heterogeneous cell sizes and aberrant nuclear morphology with a mixed population of anucleated cells, cells with unevenly distributed nuclear material, or small cells with condensed nucleus localized at one end of the cell (Figure 3, C and inset, G, and H). We further investigated the aberrant phenotypes of these two mutants in cells synchronized in a lactose gradient. The kinetics of the appearance of the aberrant phenotypes at 36°C of the synchronized mutant cells were found to be identical to that of the asynchronous culture (our unpublished observations). Flow cytometry analysis of mutant cells 4 h after shift to 36°C indicated that both mutants arrested in early to mid S phase (Figure 3D). As a comparison, the polαts mutant pol1-1, isolated by D’Urso et al. (1995), was analyzed in parallel. After 4 h at the restrictive temperature, pol1-1 displayed a cdc phenotype and 2C DNA flow cytometry profile as described by D’Urso et al. (1995). This indicates that the polαts alleles isolated in this study induce different cell cycle responses than the pol1-1 allele previously isolated by D’Urso et al. (1995). Furthermore, the four polαts mutant alleles shown in Table 2 were not substantially sensitive to either UV irradiation or hydroxyurea at the permissive temperature (our unpublished results).
Figure 3.
Characterization of temperature-sensitive mutants of polα+. Cell number increase and viability were determined as described in MATERIALS AND METHODS. (A) Cell number increase of wild-type DB10 (polα+) and mutants upon shift to 36°C. DBts11 (polαts11) and DBts13 (polαts13) arrested at 36°C after one cell division. (B) Viability of wild type and thermosensitive mutants. (C) Percentage of cells displaying aberrant nuclear phenotype. Aberrant phenotype described as cut was scored by microscopic examination of DAPI- and calcofluor-stained cells. The inset shows the three types of aberrant phenotypes observed. (D) Flow cytometry analysis of DB10 (polα+), DBts11 (polαts11), and DBts13 (polαts13) 4 h after shift to the restrictive temperature. 1C and 2C standards are arrested cdc10 cells and exponentially growing haploid wild-type cells, respectively. (E–H). Photomicrographs of DB10 (polα+) at 36°C; DBts13 (polαts13) 4 and 6 h after shift to 36°C. Bar, 5.8 μm.
Genetic Interactions of polαts Mutant with Other Cell Cycle Mutants
Studies of budding yeast have shown that S phase mutants have an extensive network of synthetic interactions with other cell cycle genes (Hennessy et al., 1991; Yan et al., 1991a,b; Li and Herskowitz, 1993). We thus explored potential genetic interactions of polαts mutants with cdc mutants. We used polαts13 as a representative for this study and constructed double mutants of polαts13 and several cdc mutants (Table 3). As expected, polαts13 cdc10 and polαts13 cdc25 arrested with the elongated cdc10 and cdc25 phenotype, respectively, not the polαts13-like phenotype. polαts13 in cdc18, cdc19 (MCM protein), or cdc21 (MCM protein) backgrounds arrested with a mid S-phase flow cytometry profile and polαts13-like phenotype. Although cdc18+ and the MCM proteins are involved in initiation of S phase, the alleles used in this study, cdc18-K46, cdc19-P1, and cdc21-M68, all arrest the cell cycle with a G2 DNA content (Kelly et al., 1993a; Forsburg and Nurse, 1994; Forsburg, 1996; Maiorano et al., 1996). Thus double mutants of these genes with polαts13 arrest with a polαts13 phenotype. cdc2-3w is a semidominant mutant (Enoch and Nurse, 1990). The double mutant polαts13 cdc2-3w arrested with a cdc2-3w-like phenotype. Double mutant polαts13 cdc22 (cdc22 encodes the large subunit of ribonucleotide reductase) arrested with a cdc22-like phenotype with a very low percent of abnormal nuclear morphology. In agreement with the known biochemical functions of Polα, Polδ, and DNA ligase, polαts13 arrested the cell cycle in either polδts03 or cdc17 (DNA ligase) background with a polαts-like phenotype. The recovery of both double mutants polαts13 polδts03 and polαts13 cdc17 was lower than that of the single mutant DBts13 (polαts13), indicating that cells with two essential replication enzymes impaired have lower viability. The double mutant polαts13 cdc20 (cdc20+ is Polε in S. pombe) arrested with a G1-S flow cytometry profile, cdc20-like elongated phenotype, and a very low percent of abnormal nuclear morphology. After 4 h at the restrictive temperature, in contrast to the single mutant polαts13, the double mutant polαts13 cdc20 recovered with full viability. This was surprising, because Polα is thought to be the first DNA polymerase that functions at the replication fork; polαts13 cdc20 is expected to arrest with a polαts-like phenotype, not a cdc20 phenotype.
Table 3.
Analysis of polαts13 in cdc mutant backgrounds
| Strain | Flow cytometry profilea | Phenotype at 36°C | Abnormal nuclear morphology (%)b
|
Viability at 4 h (%) | |
|---|---|---|---|---|---|
| 4 h | 6 h | ||||
| polαts13 | Mid S | polαts13-like | 26 | 35 | 14 |
| polαts13 cdc10 | G1 | cdc-like | 0 | 7 | 55 |
| polαts13 cdc20 | G1-S | cdc-like | 0 | 2 | 100 |
| polαts13 cdc22 | G1-S | cdc-like | 0 | 2 | 50 |
| polαts13 cdc2-3w | Early S | small, cut | 25 | 33 | 5 |
| polαts13 cdc21 | Early S | polαts13-like | 25 | 35 | 6 |
| polαts13 cdc18 | Mid S | polαts13-like | 25 | 33 | 7 |
| polαts13 cdc19 | Mid S | polαts13-like | 16 | 17 | 8 |
| polαts13 polδts03 | Mid S | polαts13-like | 28 | 35 | 10 |
| polαts13 cdc17 | Mid S | polαts13-like | 17 | 31 | 12 |
| polαts13 cdc25 | G2 | cdc-like | 0 | 0 | 114 |
Flow cytometry profile was analyzed 4 h after shift to the restrictive temperature.
Percent of abnormal nuclear morphology was determined 4 and 6 h after shift to the restrictive temperature.
polαts13 cdc20 Double Mutant Arrests Early in S Phase with a cdc Phenotype
To confirm the cell cycle arrest point of polαts13 relative to cdc20, we carried out reciprocal shift experiments using hydroxyurea (see MATERIALS AND METHODS). The single mutant DBts13 (polαts13) and the double mutant polαts13 cdc20 were used for the experiment, and the double mutant polαts13 cdc25 was used as a control. Mutants were first arrested in S phase by hydroxyurea at 25°C for 4 h. The hydroxyurea was then removed, and cells were shifted to the restrictive temperature. As the cells proceed through the cell cycle, they are expected to arrest with either a cdc phenotype or a polαts13 phenotype, depending on their point of execution in the cell cycle with respect to hydroxyurea. Mutants that arrest the cell cycle after the hydroxyurea block will not increase their cell number at the restrictive temperature, whereas mutants that arrest before the hydroxyurea block will double their cell number once, before arrest in the cell cycle.
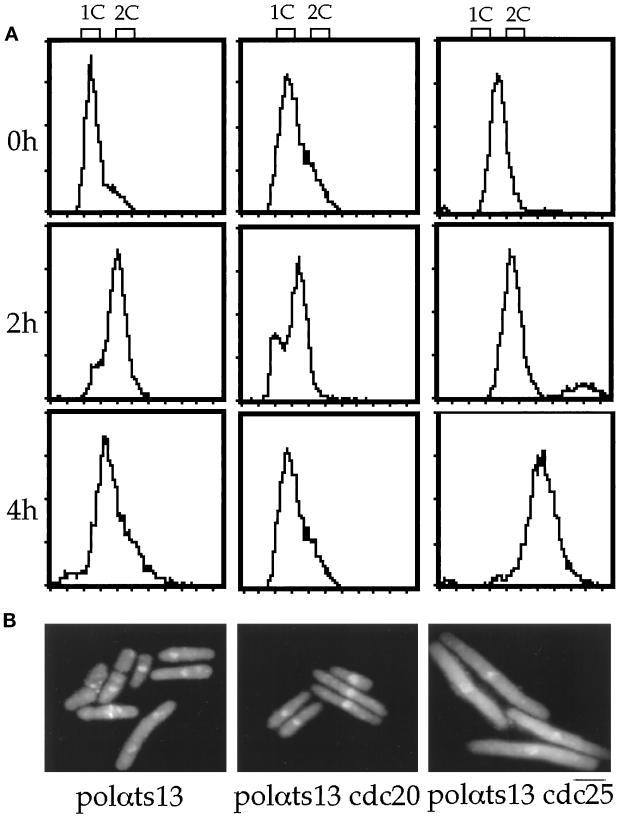
After a 4-h block in hydroxyurea at the permissive temperature, both DBts13 (polαts13) and the double mutant polαts13 cdc20 had a 1C DNA profile (Figure 4A). Four hours after shifting to the restrictive temperature, the single mutant DBts13 (polαts13) arrested with 1.5 C DNA (Figure 4A), and 40% of the cells displayed aberrant nuclear phenotypes (Figure 4B). However, the cell number of DBts13 (polαts13) only increased 1.5-fold (our unpublished observation), suggesting that polαts13 arrests the cell cycle very near the hydroxyurea block point. It has been reported that cdc20 arrests the cell cycle before the hydroxyurea block point (Nasmyth and Nurse, 1981) and with 1C DNA content (D’Urso and Nurse, 1997). Double mutant polαts13 cdc20 arrested the cell cycle with a DNA content slightly greater than 1C (Figure 4A), doubled in cell number, and displayed a cdc phenotype with no abnormal nuclear morphology, similar to the cdc20 single mutant (Figure 4B). As expected, double mutant polαts13 cdc25 had no increase in cell number after 4 h at 36°C and arrested with a phenotype and DNA content identical to the single mutant cdc25 (Figure 4, A and B).
Figure 4.
polαts13 cdc20 double mutant arrested with cdc phenotype. (A) FACS analysis of single mutant DBts13 (polαts13) and double mutants polαts13 cdc20 and polαts13 cdc25 in a hydroxyurea reciprocal shift experiment at 0, 2, and 4 h after shift to the restrictive temperature. The hydroxyurea reciprocal shift experiments were performed as described in MATERIALS AND METHODS. Double mutant polαts13 cdc25 at 25°C is moderately elongated, resulting in a >1C profile at 0 h. (B) Phenotypes of single and double mutants of polαts13 strains. Cells were stained with DAPI and calcofluor after shift to the restrictive temperature for 4 h. Bar, 5.8 μm.
Previous study has shown that cdc20 arrests the cell cycle in late G1 or early S phase with 1C DNA content (D’Urso and Nurse, 1997). In addition, p25rum1, a specific inhibitor of the p34cdc2/p56cdc13 mitotic kinase, accumulates only in preSTART cells, not in postSTART cells. It has been reported that p25rum1 is not present in cdc20-arrested cells (Correa-Bordes and Nurse, 1995). This indicates that cdc20 arrests the cell cycle postSTART. In addition, our reciprocal shift experiments clearly showed that polαts13 cdc20 doubles its cell numbers and arrests with a slightly greater than 1C DNA content (Figure 4A). Together, this indicates that the double mutant polαts13 cdc20 arrests postSTART and the cdc phenotype of the double mutant is not caused by cells arresting at preSTART.
Replication Perturbation Caused by polαts Alleles Activates Cds1 Kinase and Requires the Checkpoint Rads, Cds1, and Rqh1, but Not Chk1, for Maintenance of Cell Viability
Our observation that polαts mutants have a slightly elongated cell morphology at the permissive temperature compared with the wild-type cells (Figure 3, E and F) suggests that polαts11 and polαts13 cause mild replication perturbations even at the permissive temperature. We thus investigated the cell cycle surveillance responses that could be induced by polαts alleles at 25°C. We found that cells carrying either of the polαts11 or polαts13 mutant alleles are synthetic lethal in all checkpoint rad gene deletion backgrounds (Table 4). Thus, the replication perturbation caused by these two polαts alleles requires the function of checkpoint Rads for viability of the cells at 25°C.
Table 4.
Genetic interactions of polαts mutants with cell cycle response genes
| Allele | Growth at 25°C | Phenotype at 36°C
|
|
|---|---|---|---|
| Cell | Nuclear | ||
| polαts | ++a | Heterogeneousb | Aberrantb |
| polαts rad1Δ | Synthetic lethalc | ||
| polαts rad3Δ | Synthetic lethalc | ||
| polαts rad9Δ | Synthetic lethalc | ||
| polαts rad17Δ | Synthetic lethalc | ||
| polαts rad26Δ | Synthetic lethalc | ||
| polαts11 cds1Δ | Synthetic lethalc | ||
| polαts13 cds1Δ | Forms microcolonies at 22°C | Heterogeneousb | Aberrantb |
| polαts rqh1Δ | —d | ||
| polαts chk1Δ | ++ | Small | Cut |
++ represents growth.
After 6 h at 36°C, cells exhibit a mixture of elongated and small cell morphology with 40% of cells displaying a mixture of aberrant nuclear morphology.
Spores fail to germinate.
Spores either fail to germinate or form microcolonies with reduced growth rate.
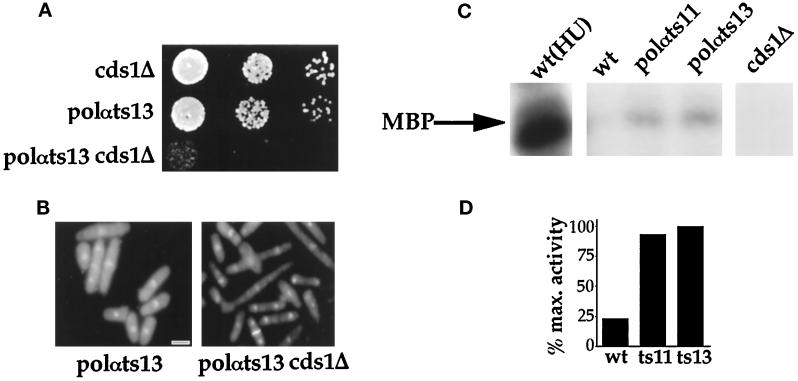
Because Cds1 is thought to be involved in a checkpoint Rad-dependent “S-phase recovery” subpathway to maintain cell viability in the event of S phase perturbation (Lindsay et al., 1998), attempts were made to construct double mutants of polαts11 and polαts13 in a cds1Δ background. The double mutant polαts11 cds1Δ was found to be synthetic lethal at either 22 or 25°C (Table 4). The double mutant polαts13 cds1Δ formed microcolonies at 22°C (Table 4). At 25°C, polαts13 cds1Δ had a severely reduced growth rate in comparison with either of the single mutants cds1Δ or polαts13 (Figure 5A) and displayed elongated cell morphology but normal nuclear morphology (Figure 5B).
Figure 5.
At 25°C, Cds1 is required to maintain normal growth of polαts13 mutant and is activated. (A) Double mutant polαts13 cds1Δ has reduced growth rate at 25°C compared with the respective single mutant cds1Δ and polαts13. Serial dilutions of exponentially growing cells at 25°C by 10-fold were spotted on YES plates. Plates were incubated at 25°C for 3 d. (B) Double mutant polαts13 cds1Δ displays an elongated phenotype compared with the respective single mutant. Shown are phenotypes of single mutant polαts13 and double mutant polαts13 cds1Δ at 25°C. Bar, 3.5 μm. (C) Cds1 kinase is activated in polαts mutants at 25°C. Cds1 protein was immunoprecipitated from logarithmically growing wild-type cells, (wt), wild-type cells treated with 20 mM hydroxyurea [wt(HU)], polαts11 and polαts13, and cds1Δ cells. The immunoprecipitated Cds1 proteins were used to assay for kinase activity using MBP as the substrate as described in MATERIALS AND METHODS. Shown here is the phosphorylation of MBP by Cds1 kinase derived from different strains. (D) The histogram shows that Cds1 kinase activity is fourfold higher in polαts11 and polαts13 as compared with the wild-type polα+ integrant DB10 cells. In DB10 cells treated with hydroxyurea, the Cds1 kinase activity is 25-fold higher than in untreated cells. The Cds1 kinase activity from polαts13 is defined as the 100% maximum activity.
Finding that Cds1 is required to maintain the viability of polαts mutants at the permissive temperature prompted us to assay the levels of Cds1 kinase activity in these polαts mutants. The Cds1 kinase activity in both polαts mutants was fourfold higher than that of the wild-type cells at 25°C (Figure 5, C and D). Cells treated with hydroxyurea was used as a control for the kinase assay, and cells containing a cds1Δ were used as a kinase-negative control. Similar to previous observations (Lindsay et al., 1998) the Cds1 kinase activity was activated ∼25-fold in wild-type cells treated with hydroxyurea, whereas no detectable MBP phosphorylation was observed in cds1Δ cell lysates (Figure 5C), similar to the Cds1 kinase dead mutant described by Lindsay et al. (1998).
In addition to Cds1, Rqh1 is also thought to be involved in the checkpoint Rad-dependent recovery subpathway to prevent inappropriate recombination or to bypass lesions during S phase arrest or DNA damage (Murray et al., 1997; Stewart et al., 1997). Attempts to generate double mutants of polαts11 or polαts13 in an rqh1Δ background indicated that spores carrying the double mutants either did not germinate or formed microcolonies with reduced growth rates (Table 4). Thus, the replication perturbation caused by polαts11 or polαts13 at the permissive temperature requires both Cds1 and Rqh1 for maintaining normal growth and cell viability.
Previous studies have shown that S phase arrest or S phase delay of cells caused by a polδts mutation requires the checkpoint Rad-Chk1 pathway to prevent inappropriate mitotic entry (Francesconi et al., 1995, 1997; Uchiyama et al., 1997). To test the requirement of Chk1 in polαts mutants at 25°C, double mutants of polαts in a chk1Δ background were analyzed. The double mutants polαts11 chk1Δ and polαts13 chk1Δ at 25°C had the same growth rate and identical cell size as those of the single polαts mutants (Table 4). This suggests that at the permissive temperature, Chk1, unlike Cds1 and Rqh1, does not play a role in maintaining the viability of the polαts mutants. Furthermore, at 25°C Chk1 was not phosphorylated in the polαts13 mutant (Figure 6B, lane 4).
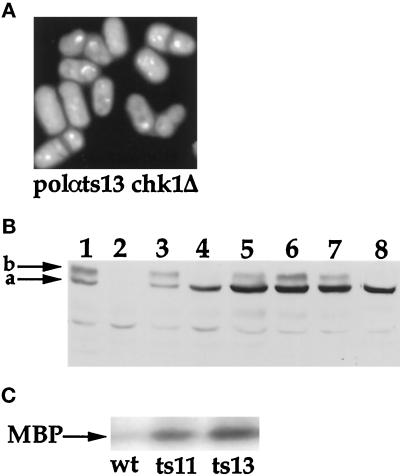
Figure 6.
Cds1 and Chk1 are both activated in polαts mutants at 36°C. (A) Phenotype of double mutant DB242 (polαts13 chk1Δ) at the restrictive temperature. Midlog phase double mutant DB242 (polαts13 chk1Δ) grown at 25°C was shifted to 36°C for 6 h. Cells were stained with DAPI and calcofluor. (B) Activation of p56chk1:ep in DB232. Thirty micrograms of protein from cell lysates of DB232 carrying polαts13 and epitope-tagged chk1+, DBts13 (polαts13), and NW222 containing epitope-tagged chk1+ were fractionated on 8% SDS-polyacrylamide gels, transferred to polyvinylidene difluoride membranes, and probed with the 12CA5 antibody, and p56chk1:ep was detected using the ECL system (Amersham, Arlington Heights, IL). The unphosphorylated p56chk1:ep is marked by arrow a, and the phosphorylated p56chk1:ep is marked by arrow b. Lanes 1 and 3, Strains NW222 and DB232 (polαts13 p56chk1:ep) treated with MMS; lane 2, DBts13 (polαts13) containing no epitope-tagged chk1+ as a control; lanes 4–7, lysates from DB232 (polαts13 p56chk1:ep) after 0, 2, 4, and 6 h at 36°C; lane 8, phosphatase-treated lysates from DB232 (polαts13 p56chk1:ep) grown at 36°C for 6 h. (C) Activation of Cds1 kinase activity at 36°C. Cells were grown to midlog phase at 25°C and then shifted to 36°C for 3 h. Cds1 kinase activity was measured in wild-type cells, (wt), polαts11, and polαts13 as described in MATERIALS AND METHODS.
Disruption of Replication by polαts Mutants at the Restrictive Temperature Induces Phosphorylation of Chk1 Protein
Previous studies have shown that cells arrested by a cdc mutation in a chk1Δ background enter mitosis inappropriately (Francesconi et al., 1995; Uchiyama et al., 1997). We thus investigated the cell cycle checkpoint responses of polαts mutants at the restrictive temperature. At 36°C, nearly all of the polαts11 chk1Δ and polαts13 chk1Δ double mutants died with a small cell size and classic cut nuclear morphology (Table 4 and Figure 6A). This suggests that at 36°C, severe disruption of replication caused by these two Polαts enzymes requires a functional Chk1 kinase to prevent cells from proceeding to inappropriate mitosis.
We then analyzed the phosphorylation status of Chk1 in the polαts mutant cells at the restrictive temperature. Strains containing either polαts11 or polαts13 and chk1+ tagged with three copies of hemagglutinin epitope (Walworth and Bernards, 1996) were constructed. We tested the phosphorylation status of p56chk1:ep in these polαts strains at the permissive and the restrictive temperatures, using the phosphorylation of p56chk1:ep in MMS-treated cells as a reference for the phosphorylated protein band shift (Figure 6B). As expected, at the permissive temperature, there was no discernible p56chk1:ep phosphorylation in the polαts mutant (Figure 6B, lane 4, and Table 4). In contrast, 2 h after shifting to 36°C, phosphorylation of p56chk1:ep was observed in both polαts11 and polαts13 strains. The levels of p56chk1:ep phosphorylation increased after 4 h and were maintained up to 6 h (Figure 6B, lanes 5–7). It is not yet known whether the phosphorylation of Chk1 protein correlates to an induction of Chk1 kinase activity. Attempts to discern whether the Chk1 kinase activity positively correlated to the phosphorylation of p56chk1:ep by assaying the kinase activity of the anti-hemagglutinin immunoprecipitates were not successful. Immunoprecipitates of DBts13 (polαts13) with no epitope-tagged Chk1 yielded a high background level of nonspecific kinase activity, and this precluded resolution of this question. Efforts to differentiate between the phosphorylation status of p56chk1:ep in the polαts13 strain at the restrictive temperature versus the MMS-treated polαts13 strain using electrofocusing followed by SDS gel electrophoresis also did not yield any informative information.
To further clarify the roles played by Chk1 and Cds1, we also investigated the Cds1 kinase response in polαts mutants at 36°C. The Cds1 kinase activity of DBts11 (polαts11) and DBts13 (polαts13) at the restrictive temperature was induced eightfold higher than in the wild-type integrant DB10 (polα+) cells (Figure 6C). This is not significantly higher than the induction observed at 25°C (Figure 5C). In addition, the double mutant polαts13 cds1Δ at the restrictive temperature has a similar phenotype as the DBts13 (polαts13) single mutant.
Thus, at the restrictive temperature, Chk1 and not Cds1 plays a major role in preventing the cells from entering inappropriate mitosis. Interestingly, despite the phosphorylation of Chk1 protein in these mutant cells at the restrictive temperature, a population of the cells still enter inappropriate mitosis after 4 h (Figure 3, C and H). The possible reasons for these phenotypes are discussed below.
DISCUSSION
In this study we investigated cell cycle responses induced by mutations in Polα. We report 1) the initiation DNA structure synthesized by Polα is required to bring about the S-M phase checkpoint; 2) polαts mutants in cdc20 (polε) background arrest the cell cycle with a cdc phenotype, not a polαts-like phenotype; and 3) during S phase progression, different degrees of replication defects caused by Polα mutations induce different downstream cell cycle surveillance kinases.
The Catalytic Function of Polα Is Required to Generate the Signal to Bring about the Replication Checkpoint
Genetic evidence has indicated that initiation of S phase generates a signal activating the S phase to mitosis checkpoint (Kelly et al., 1993b; Li and Deshaies, 1993). In this study, we generated a catalytically dead but structurally intact Polα mutant, Polα(D984N), to dissect the nature of the signal that is generated at the initiation of S phase. Previous mutational studies indicate that mutation of Asp984 to Asn completely abolishes the catalytic function of Polα without affecting the mutant protein’s structure and stability or its ability to assemble into the Polα–primase complex (Copeland and Wang, 1993a,b). Our results showed that the catalytically dead Polα mutant when overexpressed had a dominant negative effect on vegetative cell growth (Figure 2C). This further indicates that the mutant Polα(D984N) protein is physically stable and can be assembled into the replication complex. Thus, germinating spores with an endogenous polαΔ containing the mutant polα(D984N) on a plasmid have a stable and structurally intact but catalytically inactive Polα that is unable to synthesize DNA. Results of germinating spores indicate that cells harboring the polα(D984N) mutant enter mitosis with aberrant nuclear morphology in the absence of DNA synthesis (Figure 2B). The phenotype of the germinating spores is identical to that of the cells with polαΔ (Figure 1B) or cdc18Δ (Kelly et al., 1993a). This strongly suggests that it is the initiation DNA structure synthesized by a functional Polα, and not the physical presence of Polα in the replication complex, that is required for the S phase to mitosis checkpoint. However, it is not yet known whether the signal is the initiation DNA structure itself or the subsequent events that are dependent on the formation of the initiation DNA structure.
polα Mutant in cdc20 Background Arrests the Cell Cycle with a cdc Phenotype
We found that polαts mutant in cdc20 background arrests the cell cycle with a cdc20-like phenotype instead of a polαts-like phenotype (Figure 4). Although Polε (Cdc20 or POL2) is essential for chromosomal replication (Morrison et al., 1990; Campbell, 1993; D’Urso and Nurse, 1997), the precise role of Polε in DNA replication has not yet been resolved. POL2 of budding yeast (Polε of budding yeast) has been shown to function as an S phase sensor (Navas et al., 1995, 1996). In contrast, fission yeast Polε does not have a role in the replication checkpoint, although it is required early in S phase (D’Urso and Nurse, 1997). Because the two yeasts have different cell cycle setups, fission yeast Polε is different from budding yeast POL2 in cell cycle checkpoint signaling function. Finding that Polε does not have a role in coordinating S phase to mitosis (D’Urso and Nurse, 1997) and double mutant polαts13 cdc20 arrests with the cdc20-like phenotype, not the polαts phenotype, may be explained as follows.
Recent studies in S. cerevisiae have shown that entry into S phase requires establishment of the prereplication complex that contains Orc, Cdc6p (homologue of S. pombe Cdc18p), and MCM proteins (Diffley et al., 1994; Santocanale and Diffley, 1996; Newlon, 1997). Recognition of an ORC–origin complex by Cdc6p (Cdc18p) results in the recruitment of MCMs and the formation of a prereplication complex (Aparicio et al., 1997; Donovan et al., 1997; Tanaka et al., 1997). Subsequent activation of the prereplication complex leads to an unwound DNA structure at the origin and the recruitment of replication proteins such as Polε (Aparicio et al., 1997; Newlon, 1997). It is possible that the prereplication complex induces the formation of an active replication complex by first recruiting Polε followed by Polα–primase. However, the presence of Polε in the replication complex is not a prerequisite for recruiting Polα into the replication complex. Because Polε does not have the ability to synthesize an initiation DNA structure, which is required to generate the signal to coordinate replication with mitosis, Polε, despite being recruited into the replication complex before Polα, does not play a role in coordinating S phase to mitosis. In contrast, proteins Cdc30 (Orp1) and Cdc18, which are responsible for the assembly of the Polα-containing replication complex, and Polα, which directly participates in initiation DNA structure synthesis, play essential roles to bring about the S phase to mitosis checkpoint.
There is an alternate explanation for the observation that double mutant polαts13 cdc20 at 36°C exhibit the cdc20 phenotype rather than the polαts13-like phenotype. It is possible that a functional Polε(cdc20+) is a prerequisite for cells carrying the polαts allele to exhibit the polαts-like phenotype at the restrictive temperature. A recent study has suggested that POL2 (Polε) may be involved in the formation of elevated Holiday junction (xDNA) levels (Zou and Rothstein, 1997). The elevated levels of recombinogenic xDNA may somehow cause cells carrying the polαts13 allele to have the observed aberrant nuclear morphology at 36°C. In a cdc20 background with a defective Polε, cells may have reduced levels of recombinogenic xDNA structure, as suggested in the budding yeast pol2 mutant, thus double mutant polαts13 cdc20 at 36°C exhibit the cdc20 phenotype rather than the polαts13-like phenotype.
Replication Perturbation at 25°C and Replication Disruption at 36°C Caused by Polαts Mutations Induce Different Downstream Cell Cycle Surveillance Kinases
At the permissive temperature, DBts11 (polαts11) and DBts13 (polαts13) both have a slightly elongated cell size. This indicates that the mutant cells at the permissive temperature have a replication perturbation but not enough to compromise the growth rate of the cells. We found that the mutant cells require the function of checkpoint Rads, Cds1, and Rqh1 to maintain viability and growth even at the permissive temperature (Table 4). Furthermore, Cds1 kinase is activated in both polαts mutant strains at 25°C (Figure 5C). We reason that the replication perturbation is sensed by the checkpoint Rads, which then activates the downstream effector Cds1 kinase but not Chk1. The activated Cds1 helps maintain a functionally productive replication status in the mutant cells, resulting in a normal growth rate.
Both polαts11 or polαts13 are synthetic lethal with rqh1Δ (Table 4). The replication perturbation caused by either Polαts11 or Polαts13 enzyme at 25°C may result in elevated levels of recombinogenic lesions that require checkpoint Rads to activate the Rqh1-dependent recovery process to prevent inappropriate recombination to maintain viability of the cells.
At 36°C, Cds1 kinase activity is not significantly enhanced over that at 25°C. Also, the phenotype of the double mutant polαts13 cds1Δ is identical to that of the polαts13 single mutant. This suggests that Cds1 does not play a significant role at 36°C. Because replication perturbation by polαts13 is further exacerbated at 36°C, Cds1 may no longer be able to maintain a productive replication status of the mutant cells.
In contrast to Cds1, Chk1, which is not phosphorylated at 25°C in polαts mutants, is phosphorylated in these mutants at 36°C (Figure 6B). In addition, at 25°C, the double mutant polαts chk1Δ grows at the same rate as the polαts mutant cells, whereas at 36°C nearly all of the double mutants of polαts chk1Δ die with small cell size and cut nuclear morphology (Figure 6A). These results indicate that at 36°C, Chk1 but not Cds1 plays a major role in preventing the polαts mutant cells from proceeding into mitotic catastrophe.
Chk1 phosphorylation has been shown to be induced by DNA damage but not by hydroxyurea (Walworth and Bernards, 1996). Using a polδts mutant to arrest S phase at 37°C, Francesconi et al. (1997) have isolated two chk1 alleles, chk1-1 and chk1-2, that maintain the DNA damage checkpoint but fail to prevent mitotic catastrophe. Our finding that Chk1 is phosphorylated only at 36°C and not at 25°C in the polαts mutants cells (Figure 6B) together with the finding by Francesconi et al. (1997) indicate that a disrupted replication structure may be the signal that activates Chk1. Disrupted DNA replication caused by either polαts or polδts mutants at 36°C could yield a DNA structure similar to a damaged DNA structure. This signal is then detected by the checkpoint Rads, which phosphorylate Chk1 to delay mitotic entry.
Why Does a Population of polαts Cells Enter Aberrant Mitosis Despite Activation of Chk1?
Despite phosphorylation of Chk1 at 36°C, polαts mutants arrest with heterogeneous cell size, and a population of cells exhibit mixed aberrant nuclear morphology. There are several possible reasons for these phenotypes. Mutant cells that arrest during DNA synthesis could yield a DNA structure that activates Chk1, resulting in cells arresting with cdc phenotype. Meanwhile, mutant cells that have lost the capacity to synthesize the initiation DNA structure enter inappropriate mitosis. Alternately, Chk1 may delay mitosis in polαts cells until the DNA ends are repaired to a state that no longer can generate a checkpoint signal. However, the repaired DNA ends are in a state that is no longer compatible for mitosis, and, hence, improper nuclear segregation takes place.
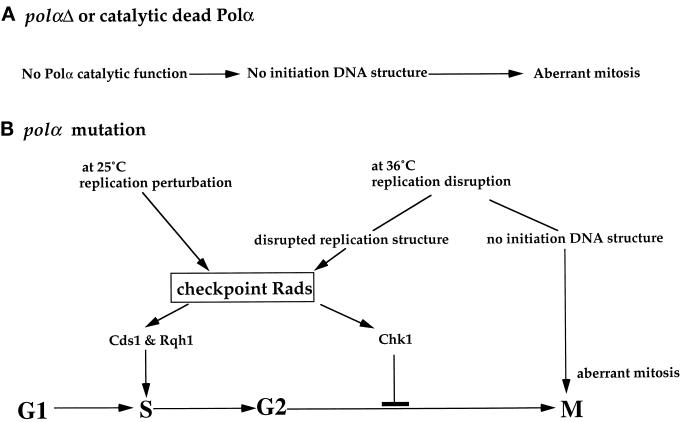
In conclusion, our studies show that mutations in Polα can induce different cell cycle surveillance responses. A summary is presented in Figure 7.
Figure 7.
Mutational effects of polα+ on cell cycle events. A summary of the mutational effects of polα+ on cell cycle surveillance responses is depicted, and details are described in the text (see DISCUSSION).
ACKNOWLEDGMENTS
We thank A.M. Carr for communication of results before publication and providing us the rad/hus strains and cds1Δ strain, S. Forsburg for most of the parental cdc strains, T. Enoch for rqh1Δ strain, and members of our lab for helpful discussions. This work was supported by a grant from National Institutes of Health (CA54415).
REFERENCES
- Al-Khodairy F, Carr AM. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJ, Lehmann AR, Carr AM. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Brun C, Dubey DD, Huberman JA. pDblet, a stable autonomously replicating shuttle vector for Schizosaccharomyces pombe. Gene. 1995;164:173–177. doi: 10.1016/0378-1119(95)00497-t. [DOI] [PubMed] [Google Scholar]
- Burke JD, Gould KL. Molecular cloning and characterization of the Schizosaccharomyces pombe his3 gene for use as a selectable marker. Mol Gen Genet. 1994;242:169–176. doi: 10.1007/BF00391010. [DOI] [PubMed] [Google Scholar]
- Campbell JL. Yeast DNA replication. J Biol Chem. 1993;268:25261–25264. [PubMed] [Google Scholar]
- Carr AM. Checkpoints take the next step. Science. 1996;271:314–315. doi: 10.1126/science.271.5247.314. [DOI] [PubMed] [Google Scholar]
- Carr AM, Hoekstra MF. The cellular responses to DNA damage. Trends Cell Biol. 1995;5:32–40. doi: 10.1016/s0962-8924(00)88934-5. [DOI] [PubMed] [Google Scholar]
- Copeland WC, Wang TS-F. Mutational analysis of the human DNA polymerase α. The most conserved region in α-like DNA polymerases is involved in metal-specific catalysis. J Biol Chem. 1993a;268:11028–11040. [PubMed] [Google Scholar]
- Copeland WC, Wang TS-F. Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J Biol Chem. 1993b;268:26179–26189. [PubMed] [Google Scholar]
- Correa-Bordes J, Nurse P. p25rum1 orders S phase and mitosis by acting as an inhibitor of the p34cdc2 mitotic kinase. Cell. 1995;83:1001–1009. doi: 10.1016/0092-8674(95)90215-5. [DOI] [PubMed] [Google Scholar]
- D’Urso G, Grallert B, Nurse P. DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J Cell Sci. 1995;108:3109–3118. doi: 10.1242/jcs.108.9.3109. [DOI] [PubMed] [Google Scholar]
- D’Urso G, Nurse P. Schizosaccharomyces pombe cdc20+ encodes DNA polymerase ε and is required for chromosomal replication but not for the S phase checkpoint. Proc Natl Acad Sci USA. 1997;94:12491–12496. doi: 10.1073/pnas.94.23.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Poch O, Tordo N, Morase D, Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two step in assembly of complexes at yeast replication origin in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Diffley JFX. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto prereplicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Enoch T, Carr AM, Nurse P. Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 1992;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- Enoch T, Carr A, Nurse P. Checkpoint check. Nature. 1993;361:26. doi: 10.1038/361026b0. [DOI] [PubMed] [Google Scholar]
- Enoch T, Nurse P. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell. 1990;60:665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- Forsburg SL. Regulation of S phase in the fission yeast Schizosaccharomyces pombe. In: Blow JJ, editor. Eukaryotic DNA Replication. Oxford: Oxford University Press; 1996. pp. 197–228. [Google Scholar]
- Forsburg SL, Nurse P. The fission yeast cdc19+ gene encodes a member of the MCM family of replication proteins. J Cell Sci. 1994;107:2779–2788. doi: 10.1242/jcs.107.10.2779. [DOI] [PubMed] [Google Scholar]
- Francesconi S, Grenon M, Bouvier D, Baldacci G. p56chk1 protein kinase is required for the DNA replication checkpoint at 37°C in fission yeast. EMBO J. 1997;16:1332–1341. doi: 10.1093/emboj/16.6.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi S, Park H, Wang TS. Fission yeast with DNA polymerase δ temperature-sensitive alleles exhibits cell division cycle phenotype. Nucleic Acids Res. 1993;21:3821–3828. doi: 10.1093/nar/21.16.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi S, Recondo A-MD, Baldacci G. DNA polymerase δ is required for the replication feedback control of cell cycle progression in Schizosaccharomyces pombe. Mol Gen Genet. 1995;246:561–569. doi: 10.1007/BF00298962. [DOI] [PubMed] [Google Scholar]
- Grallert B, Nurse P. The ORC homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- Griffiths DJF, Barbet NC, McCready S, Lehmann AR, Carr AM. Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Loprieno N. Handbook of Genetics. I. R.C. King, New York: Plenum Press; 1974. Schizosaccharomyces pombe; pp. 395–446. [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–960. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Hofmann JX, Beach D. cdt1 is an essential target of the cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J. 1994;13:425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Braithwaite DK. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991;19:4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993a;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Nurse P, Forsburg SL. Coupling DNA replication to the cell cycle. Cold Spring Harb Symp Quant Biol. 1993b;58:637–644. doi: 10.1101/sqb.1993.058.01.071. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotype selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Li JJ, Deshaies RJ. Exercising self-restraint: discouraging illicit acts of S and M in eukaryotes. Cell. 1993;74:223–226. doi: 10.1016/0092-8674(93)90413-k. [DOI] [PubMed] [Google Scholar]
- Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Lindsay HD, Griffiths DJF, Edwards R, Murray JM, Christensen PU, Walworth N, Carr AM. S-phase specific activation of Cds1 kinase defines a subpathway of the checkpoint response in S. pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Weinert T. From DNA damage to cell cycle arrest and suicide: a budding yeast perspective. Curr Opin Genet Dev. 1996;6:4–11. doi: 10.1016/s0959-437x(96)90003-9. [DOI] [PubMed] [Google Scholar]
- Maiorano D, van Assendelft GB, Kearsey SE. Fission yeast cdc21, a member of the MCM protein family, is required for onset of S phase and is located in the nucleus throughout the cell cycle. EMBO J. 1996;15:861–872. [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrison A, Araki H, Clark AB, Hamatake RK, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- Murray JM, Lindsay HD, Munday CA, Carr AM. Role of Schizosaccharomyces pombe RecQ homolog, recombination, and checkpoint genes in UV damage tolerance. Mol Cell Biol. 1997;17:6868–6875. doi: 10.1128/mcb.17.12.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzi FM, Brown GW, Kelly TJ. cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzi FM, Kelly TJ. Orp1, a member of the Cdc18/Cdc6 family of S-phase regulators, is homologous to a component of the origin recognition complex. Proc Natl Acad Sci USA. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Nurse P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1981;182:119–124. doi: 10.1007/BF00422777. [DOI] [PubMed] [Google Scholar]
- Navas TA, Sanchez Y, Elledge SJ. RAD9 and DNA polymerase e form parallel sensory branches for transducing the DNA damage checkpoint signals in Saccharomyces cerevisiae. Genes Dev. 1996;10:2632–2643. doi: 10.1101/gad.10.20.2632. [DOI] [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ. DNA polymerase e links DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- Newlon CS. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- Nurse P. Ordering S phase and M phase in the cell cycle. Cell. 1994;79:547–550. doi: 10.1016/0092-8674(94)90539-8. [DOI] [PubMed] [Google Scholar]
- O’Connell MJ, Raleigh JM, Verkade HM, Nurse P. Chk1 is a wee1 kinase in the G2 DNA damage checkpoint inhibiting cdc2 by Y15 phosphorylation. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- Paulovich AG, Toczyski DP, Hartwell LH. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- Rao PN, Johnson RT. Mammalian cell fusion studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rowley R, Subramani S, Young PG. Checkpoint controls in Schizosaccharomyces pombe: rad1. EMBO J. 1992;11:1335–1342. doi: 10.1002/j.1460-2075.1992.tb05178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Fantes P, Sutani T, McInerny C, Creanor J, Yanagida M. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 1994;13:5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- Santocanale C, Diffley JFX. ORC- and CDC6-dependent complex at active and inactive chromosomal replication origin in Saccharomyces cerevisiae. EMBO J, 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Sheldrick KS, Carr AM. Feedback controls and G2 checkpoints: fission yeast as a model system. Bioessays. 1993;15:775–782. doi: 10.1002/bies.950151202. [DOI] [PubMed] [Google Scholar]
- Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an MCM protein onto DNA replication origin is regulated by cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Uchiyama M, Galli I, Griffiths DJF, Wang TS-F. A novel mutant allele of Schizosaccharomyces pombe rad26 defective in monitoring S phase progression to prevent premature mitosis. Mol Cell Biol. 1997;17:3103–3115. doi: 10.1128/mcb.17.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- Wang TS-F. Eukaryotic DNA polymerases. Annu Rev Biochem. 1991;60:513–552. doi: 10.1146/annurev.bi.60.070191.002501. [DOI] [PubMed] [Google Scholar]
- Wang TS-F. Cellular DNA polymerases. In: DePamphilis ML, editor. DNA Replication in Eukaryotic Cells. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 461–493. [Google Scholar]
- Waseem NH, Labib K, Nurse P, Lane DP. Isolation and analysis of the fission yeast gene encoding polymerase delta accessory protein PCNA. EMBO J. 1992;11:5111–5120. doi: 10.1002/j.1460-2075.1992.tb05618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Gibson S, Tye BK. MCM2 and MCM3, two proteins important for ARS activity, are related in structure and function. Genes Dev. 1991a;4:968–977. doi: 10.1101/gad.5.6.944. [DOI] [PubMed] [Google Scholar]
- Yan H, Nerchant AM, Tye BK. Cell-cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev. 1991b;7:2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]
- Zou H, Rothstein R. Holiday junctions accumulate in replication mutants via a RecA homolog-independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]