Abstract
Ceramide functions as an important second messenger in apoptosis signaling pathways. In this report, we show that treatment of NT-2 neuronal precursor cells with hypoxia/reoxygenation (H/R) resulted in ceramide up-regulation. This elevation in ceramide was primarily due to the actions of acid sphingomyelinase and ceramide synthase LASS 5, demonstrating the action of the salvage pathway. Hypoxia/reoxygenation treatment led to Bax translocation from the cytoplasm to mitochondria and cytochrome c release from mitochondria. Down-regulation of either acid sphingomyelinase or LASS 5-attenuated ceramide accumulation and H/R-induced Bax translocation to mitochondria. Overall, we have demonstrated that ceramide up-regulation following H/R is pertinent to Bax activation to promote cell death.
The pro-apoptotic protein Bax is a member of the Bcl-2 family that plays an important role in apoptosis regulation (1). Physiologically, Bax is involved in neuronal development (2) and spermatogenesis (3, 4). Under pathological conditions such as cerebral and cardiac ischemia/reperfusion (I/R),4 it has been shown that Bax is up-regulated in the afflicted regions of the tissues, presumably to promote neuronal and cardiac cell death (5–7). This protein is primarily a soluble protein in healthy cells (8–11). Upon treatment with a variety of apoptotic stimuli, Bax translocates to mitochondria and is associated with the loss of mitochondrial membrane potential (8–11) and the release of cytochrome c from mitochondrial intermembrane space (12–15). Cytochrome c then initiates the formation of apoptosomes to promote caspase activation and cell death (16).
Currently, two major apoptotic pathways that signal Bax translocation to mitochondria have been identified (17). In the extrinsic pathway, the binding of death ligands such as FAS ligand and tumor necrotic factor α (TNFα) to their respective receptors results in the activation of caspase-8. Caspase-8 then cleaves the BH3-only protein Bid, and the truncated Bid (tBid) activates Bax and causes its translocation to mitochondria (18–20). In the intrinsic pathway, apoptotic stimuli trigger Bax translocation to mitochondria via mechanisms that are independent of caspase-8 and Bid. Although it has been shown that H/R induces Bax translocation to mitochondria and subsequent cytochrome c release into the cytoplasm (21), the molecular trigger for Bax activation is still not known.
Ceramide is a signaling molecule shown to be involved in cellular growth, differentiation, and apoptosis (22). Exposure of rat pheochromocytoma (PC12) cells to oxygen-glucose deprivation (23) and of brain tissues to I/R resulted in ceramide accumulation (24, 25). Ceramide can be generated via the salvage pathway through the action of sphingomyelinases, or the de novo synthetic pathway through the action of ceramide synthases. Currently, five distinct sphingomyelinases have been identified based on their preferred optimal pH for activity, subcellular localization, and dependence on cations (for a review see Ref. 26). Among them, the acid sphingomyelinase (aSMase) and the neutral Mg2+-dependent neutral sphingomyelinase (nSMase) have been shown to be involved in ceramide generation in response to apoptotic stimuli (27–29).
The de novo pathway commences with the action of serine palmitoyl transferase leading to the formation of dihydrosphingosine and then dihydroceramide, which is produced by (dihydro)ceramide synthases (30, 31). Dihydroceramide is then converted into ceramide by dihydroceramide desaturase (32, 33). A homologue of ceramide synthases, also known as longevity assurance factors (LASS/CerS), was first identified in yeast. Its deletion resulted in an increased yeast lifespan (34). Currently, six LASS genes have been identified in mammals, and each of them displays a unique substrate specificity profile for chain length and/or saturation in fatty acid acyl-CoA (35).
Recently, a more complex mechanism of regulation of ceramide levels has become appreciated involving the recycling or salvage pathway. In the salvage pathway, ceramide generated via sphingomyelin hydrolysis is further hydrolyzed by ceramidases to sphingosine, which is then re-acylated via the action of ceramide synthases (LASS/CerS) to regenerate ceramide.
In neuronal cells, H/R induces Bax mitochondrial localization and subsequent cytochrome c release (21). Because ceramide has been suggested to play a role in Bax activation (36, 37), we set out to examine the cross-talk between sphingolipid metabolism and Bax activation following H/R. Using an NT-2 neuronal precursor cell line stably expressing GFP-tagged Bax, we examined the mechanism of ceramide accumulation in these cells and the contribution of the salvage and de novo pathways of ceramide synthesis. In addition, we have determined the roles of these ceramide-producing enzymes in the activation of Bax following H/R.
EXPERIMENTAL PROCEDURES
Materials—NT-2 human neuronal precursor cell line was obtained from ATCC. Fetal bovine serum, Dulbecco's modified Eagle's medium, tissue culture supplements, SuperScript first-strand synthesis system, caspase-3 assay kit, and the Quant-iT RiboGreen RNA assay kit were from Invitrogen. FuGENE 6 transfection reagent was from Roche Applied Science. Maxiprep kits for plasmids extraction, sequence-specific small interfering RNA reagents, minikits for mRNA extraction, and SYBR Green PCR reagents were purchased from Qiagen. Horseradish peroxidase-conjugated sheep anti-mouse immunoglobulins and the ECL Western blot detection kit were from Amersham Biosciences. The anti-cytochrome c antibody was from BD Pharmingen. Pan-caspase inhibitor zVAD-fmk was purchased from Axxora. The reverse transcriptase kit was from Promega. The iQ SYBR Green PCR kit was bought from Bio-Rad. 17C-sphingosine was from Avanti Polar Lipids, Inc. All other chemicals were from either Sigma or Fisher Scientific.
Development of NT-2 Cells Stably Expressing GFP-Bax—NT-2 cells were cultured in DMEM/F12 (1:1) supplemented with 2 mm glutamine and 10% fetal bovine serum and maintained at 37 °C in the presence of 5% CO2. To develop stable clones of NT-2 cells expressing GFP-Bax, NT-2 cells (in a 10-cm plate) were transfected with 12 μg of C3-EGFP-Bax (10) with the FuGENE transfection reagent. A day after transfection, the cells were selected with 0.2 mg/ml G418. After a week of selection, the surviving cells were trypsinized, and GFP-positive cells were sorted by flow cytometry (carried out by the MUSC flow cytometry facility). The cells were subsequently maintained in the culture medium supplemented with 0.2 mg/ml G418. GFP-Bax-stable NT-2 cells were plated onto either 6-well (for Bax localization analysis) or 10-cm (for SMase/LASS activity assays or lipid analysis) plates.
Treatment of NT-2 Cells with H/R and Fluorescence Microscopy—NT-2 cells were grown to 70–80% confluency in 6-well plates. The plates were placed in a hypoxic chamber (Billups-Rothenberg), flushed with 95% N2 and 5% CO2, and incubated for 12 h at 37 °C (38, 39). Plates were then removed from the chamber, and reoxygenation was allowed to proceed for specified time periods. Cell visualization was carried out using an Olympus IX-70 fluorescence microscope using an LCPlanFI ×20 objective lens with a 1.5× intermediate magnification. The images were captured with an Optronics DEI-750D digital imaging camera (39). The percentages of GFP-Bax punctate cells were determined by fluorescence microscopy.
Immunofluorescence Labeling with an Anti-cytochrome c Antibody—For immunofluorescence labeling, GFP-Bax stable NT-2 cells were subjected to H/R. Control and treated cells were then washed three times with phosphate-buffered saline (PBS), fixed in 6% paraformaldehyde (30 min), and permeabilized with 0.06% saponin in PBS (15 min). The cells were then incubated with the blocking buffer (PBS, 5% fetal bovine serum, and 0.06% saponin) for 30 min and then with 5 μg/ml anti-cytochrome c antibody diluted in the blocking buffer for 2 h. The cells were subsequently washed and incubated with 6 μg/ml Cy3-labeled goat anti-mouse immunoglobulin in the blocking buffer (2 h). After additional washes with PBS and treatment with the antifade reagent, the labeled cells were visualized by fluorescence microscopy.
Down-regulation of Acid Sphingomyelinase and LASS 5 by siRNA Oligonucleotides—The gene silencing of human aSMase and LASS 5 was performed essentially according to a standard protocol (40). The sequences of specific small interfering RNAs are as below: human aSMase: sense (5′-CUC CUU UGG AUG GGC CUG G-3′) and antisense (5′-CCA GGC CCA UCC AAA GGA G-3′) (41); human LASS 5: sense (5′-ACC CUG UGC ACU CUG UAU U-3′) and antisense (5′-AAU ACA GAG UGC ACA GGG U-3′); human LASS 6: sense (5′-CGC UGG UCC UUU GUC UUC A-3′) and antisense (5′-UGA AGA CAA AGG ACC AGC G-3′). The efficiency of gene silencing was assessed by enzymatic activity or by measuring mRNA levels of the target genes using quantitative real-time PCR (qPCR). Briefly, GFP-Bax-stable NT-2 cells were plated onto either 6-well (for Bax localization analysis) or 10-cm (for activity assays or lipid analyses) plates. The cells were then transfected with scrambled control siRNA oligonucleotides or siRNA oligonucleotide duplexes against human aSMase (150 nm), LASS 5 (5 nm), or LASS 6 (5 nm) using the Dharmafect or Hiperfect transfection reagent. At 48-h post-transfection, the cells were replated onto 6-well or 10-cm plates and subjected to H/R for specified times.
Analyses of Endogenous Sphingolipids—Normoxic and H/R-treated cells were collected, fortified with internal standards, extracted with ethyl acetate/isopropyl alcohol/water (60:30:10, v/v/v), evaporated to dryness, and reconstituted in 100 μl of methanol (42, 43). Simultaneous ESI/MS/MS analyses of sphingoid bases, sphingoid base 1-phosphates, ceramides, and sphingomyelins were performed on a Thermo Finnigan TSQ 7000 triple quadrupole mass spectrometer operating in a multiple reaction monitoring positive ionization mode (43). Results were normalized to total phospholipid levels.
Quantitative Real-time PCR Analysis—RNA was extracted from NT-2 cells using the Qiagen RNeasy kit according to the manufacturer's protocol. RNA concentrations were determined by the Quant-iT RiboGreen RNA assay kit. The extracted RNA (1 μg) was used to produce cDNA with SuperScript first-strand synthesis system according to the manufacturer's protocol. The resulting cDNA (500 ng) were analyzed in triplicates by qPCR using the iQ SYBR Green PCR kit and an ABI 7300 Q-PCR system as described by the manufacturer. Primers (final concentration of 400 nm) used for the qPCR analysis are targeted to human ceramide synthase/LASS family members and are as follows: LASS 1 forward: 5′-ACG CTA CGC TAT ACA TGG ACA C-3′ and reverse: 5′-AGG AGG AGA CGA TGA GGA TGA G-3′; LASS 3 forward: 5′-ACA TTC CAC AAG GCA ACC ATT G-3′ and reverse: 5′-CTC TTG ATT CCG CCG ACT CC-3′; LASS 4 forward: 5′-CTT CGT GGC GGT CAT CCT G-3′ and reverse: 5′-TGT AAC AGC AGC ACC AGA GAG-3′; LASS 5 forward: 5′-TGT AAC AGC AGC ACC AGA GAG-3′ and reverse: 5′-GCC AGC ACT GTC GGA TGT C-3′; LASS 6 forward: 5′-GGG ATC TTA GCC TGG TTC TGG-3′ and reverse: 5′-GCC TCC TCC GTG TTC TTC AG-3′. Primers used for aSMase are as follows: forward: 5′-TGG CTC TAT GAA GCG ATG GC-3′ and reverse 5′-TTG AGA GAG ATG AGG CGG AGA-3′. Target mRNA expression was normalized to that of β-actin expression. The β-actin primers used are: forward: 5′-ATT GGC AAT GAG CGG TTC C-3′ and reverse: 5′-GGT AGT TTC GTG GAT GCC ACA-3′. The qPCR results were analyzed using Q-Gene software, which expresses data as mean normalized expression (MNE), which is directly proportional to the amount of the target gene mRNA relative to the reference gene (β-actin) mRNA.
Ceramide Synthase Activity Assay—To evaluate the activity of ceramide synthase in cells, we employed a synthetic 17C-sphingosine as the substrate. NT-2 cells were grown to 70–80% confluency, and subjected to H/R. At the indicated time points, cells were treated with 1 μm 17C-sphingosine for 30 min prior to harvest. Ceramide synthase activity was determined by measuring the accumulation of 17Cn-ceramide using tandem MS on a Thermo Finnigan TSQ 7000 triple quadrupole mass spectrometer (44). Ceramide synthase specific activity was expressed as pmol 17Cn-ceramide/nmol phosphate.
Sphingomyelinase Assays—NT-2 cells were grown to 70–80% confluency, and subjected to H/R for the indicated time periods. The cells were then collected and subjected to in vitro acid and neutral sphingomyelinase enzymatic assays using [14C]sphingomyelin according to the previously described protocol (40, 45).
SDS-Polyacrylamide Gel Electrophoresis and Western Blotting—GFP-Bax-stable NT-2 cells were collected and lysed in the lysis buffer (PBS, 1% Triton X-100, 0.1% SDS, and 25 μg/ml phenymethylsulfonyl fluoride). Cell lysates were normalized to protein concentration using the Bio-Rad protein assay kit. SDS-polyacrylamide gel electrophoresis (12%) and Western blotting analyses were performed as described before (46). For immunoblotting analysis, the blots were probed with hBax IF6 and hBcl-2 8C8 antibodies (8). The ECL detection system was used to visualize the labeled protein bands.
In Vitro Assessment of Cytochrome c Release—Control and siRNA-transfected cells were subjected to H/R. At specified time points, cells were collected and washed in PBS. Cells were then subjected to subcellular fractionation and cytosolic extracts were isolated as described earlier (47). Briefly, cells were lysed on ice for 30 min and then disrupted by repeated passage through a 27-gauge needle. The lysate was then spun at 10,000 × g (10 min, 4 °C) to obtain the cytosolic fraction. Cytosolic fractions were the analyzed by Western blotting with anti-cytochrome c and β-actin antibodies. Equal amounts of proteins were loaded per lane.
Caspase-3 Activity Assay—At various time points post hypoxia, cells were harvested, and caspase-3 activity was determined using the EnzChek Caspase-3 assay kit 2 (Molecular Probes) according to the manufacturer's instructions.
Statistical Analysis—Data expressed as the means ± S.E. on a minimum of three independent experiments were analyzed by Student's t test. A p value of 0.05 or less is considered as statistically significant and marked with an asterisk.
RESULTS
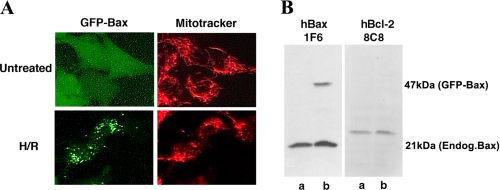
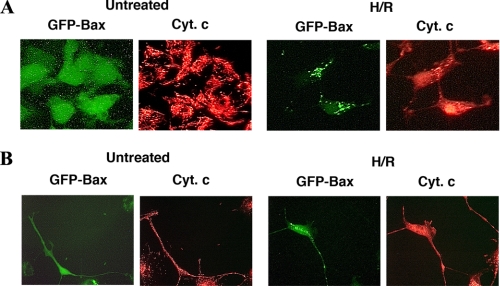
Hypoxia/Reoxygenation Induced Bax Translocation from the Cytoplasm to Mitochondria—To examine Bax activation in H/R-treated NT-2 neuronal precursor cells, we generated a cell line stably expressing GFP-Bax for rapid assessment of Bax subcellular localization. These cells were developed by transfecting NT-2 cells with an EGFP-Bax construct, and the cells were selected with G418. The expression of GFP-Bax protein in these cells was confirmed by fluorescence microscopy (Fig. 1A) and by Western blotting analysis with an anti-Bax antibody (Fig. 1B). Western blotting analysis indicated that GFP-Bax-stable cells express equivalent levels of endogenous Bax and Bcl-2 as their parental cells (Fig. 1B). Fluorescence microscopy analysis indicated that under the normoxic condition, almost all the cells had Bax localized to the cytoplasm (Fig. 1A). When these cells were subjected to H/R treatment, there was a shift in the fluorescence pattern of GFP-Bax from a diffuse cytoplasmic state to a punctate membrane-bound state. The punctate GFP-Bax was localized to mitochondria, as determined by staining with Mitotracker, a dye that specifically labeled mitochondria (Fig. 1A). After a 12-h hypoxia treatment, ∼10% of the cells displayed a punctate GFP-Bax localization pattern (data not shown). The percentage of GFP-Bax punctate cells increased following reoxygenation (up to 50% at 24 h post reoxygenation) (Figs. 6D and 8C). Interestingly, this translocation induced by H/R was not blocked by the addition of the pan-caspase inhibitor zVAD-fmk (data not shown), indicating that this redistribution process is caspase-independent. As reported previously, Bax translocation to mitochondria leads to the release of cytochrome c (14, 48, 49) and cell death. By immunofluorescence labeling with an anti-cytochrome c antibody, we found that cytochrome c was retained in the mitochondria of undifferentiated (Fig. 2A) and retinoic acid-differentiated GFP-Bax-stable NT-2 cells (Fig. 2B). Following H/R treatment, cytoplasmic cytochrome c became visible in cells that had GFP-Bax localized to the mitochondria (Fig. 2).
FIGURE 1.
Hypoxia/reoxygenation induces Bax translocation from the cytoplasm to mitochondria in GFP-Bax-stable NT-2 cells. A, untreated and hypoxia (12 h)/reoxygenation (24 h)-treated GFP-Bax-stable NT-2 cells were stained with Mitotracker and visualized by fluorescence microscopy. B, cell lysates prepared from untransfected NT-2(lanes a) or GFP-Bax-stable NT-2 cells (lanes b) were subjected to Western blotting analyses with anti-Bax 1F6 and anti-Bcl-2 8C8 monoclonal antibodies.
FIGURE 6.
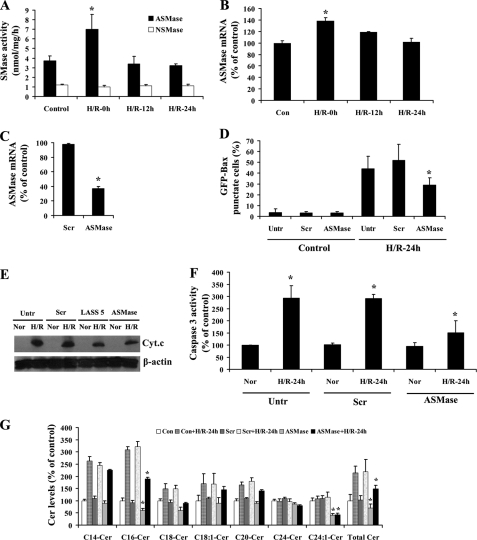
Acid sphingomyelinase is involved in H/R-induced ceramide accumulation and Bax translocation to mitochondria. A, GFP-Bax-stable NT-2 cells were subjected to hypoxia (12 h) and varying lengths of reoxygenation (0, 12, and 24 h). Acid and neutral sphingomyelinase activity assays were carried out on control untreated and H/R-treated cells. B, acid SMase mRNA levels were determined in GFP-Bax-stable NT-2 cells following H/R treatment by qPCR. C, GFP-Bax-stable NT-2 cells were transfected with either control scrambled or aSMase siRNA oligonucleotides. The decrease in aSMase mRNA level was determined by qPCR. D, GFP-Bax-stable NT-2 cells transfected with either control scrambled or aSMase siRNA oligonucleotides were subjected to H/R. The percentages of GFP-Bax punctate cells were visualized by fluorescence microscopy and quantitated from four separate visual fields. The results were averaged from three independent studies. E, untransfected and GFP-Bax-stable NT-2 cells transfected with control scrambled, LASS 5, or aSMase siRNA oligonucleotides were subjected to normoxia or H/R (24 h). The cells were then subjected to subcellular fractionation, and the isolated cytosolic extracts were analyzed by Western blotting with anti-cytochrome c antibody. The blot was probed with anti-β actin antibody to show equal loading. F, GFP-Bax-stable NT-2 cells were transfected with control scrambled or aSMase siRNA oligonucleotides. Untransfected and transfected cells were subjected to either normoxia or H/R (24 h). Caspase-3 activities of the normoxia and H/R-treated cells were then determined. The results were averaged from three independent studies and expressed as a percentage of the untreated cells. G, GFP-Bax-stable NT-2 cells were transfected with control scrambled or aSMase siRNA oligonucleotides. Untransfected (Con) and transfected cells were subjected to normoxia or hypoxia (12 h)/reoxygenation (24 h) (H/R-24h). The levels of various ceramide species were determined by tandem MS and expressed as a percentage of the control untreated cells. The results were averaged from three independent studies.
FIGURE 8.
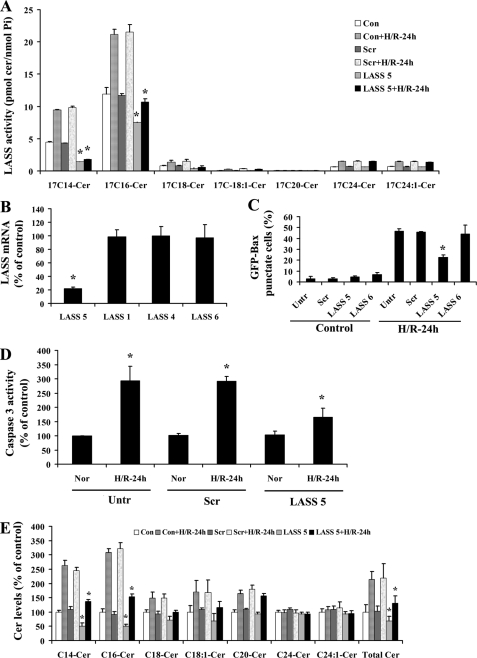
Down-regulation of LASS 5 attenuated H/R-induced Bax translocation to mitochondria. A, GFP-Bax-stable NT-2 cells were transfected with control scrambled or LASS 5 siRNA oligonucleotides. Untransfected and transfected cells were subjected to normoxia or hypoxia (12 h)/reoxygenation (24 h). Ceramide synthase activities of the control and H/R-treated cells were then measured. B, GFP-Bax-stable NT-2 cells were transfected with LASS 5 siRNA oligonucleotides. A qPCR analysis of various LASS isoforms was carried out. C, GFP-Bax-stable NT-2 cells transfected with control scrambled, LASS 5, or LASS 6 siRNA oligonucleotides were subjected to H/R and then visualized by fluorescence microscopy. GFP-Bax punctate cells were quantitated from four separate visual fields. The results were averaged from three independent studies. D, GFP-Bax-stable NT-2 cells were transfected with control scrambled or LASS 5 siRNA oligonucleotides. Untransfected and transfected cells were subjected to either normoxia or H/R (24 h). Caspase-3 activities of the normoxia and H/R-treated cells were then determined. The results were averaged from 3 independent studies and expressed as a percentage of the untreated cells. E, GFP-Bax-stable NT-2 cells were transfected with control scrambled (Scr) or LASS 5 siRNA oligonucleotides. Untransfected (Con) and transfected cells were subjected to normoxia or hypoxia (12 h)/reoxygenation (24 h). The levels of various ceramide species were determined by tandem MS and expressed as a percentage of the control untransfected cells. The results were averaged from three independent studies.
FIGURE 2.
Hypoxia/reoxgenation-induced Bax translocation to mitochondria in NT-2 cells was associated with the release of cytochrome c. Undifferentiated (A) and retinoic acid-differentiated (B) GFP-Bax-stable NT-2 cells were subjected to hypoxia (12 h)/reoxygenation (24 h). The treated cells were then stained with an anti-cytochrome c antibody and visualized by fluorescence microscopy.
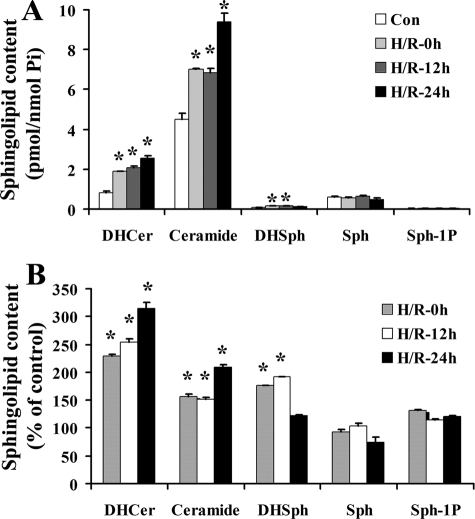
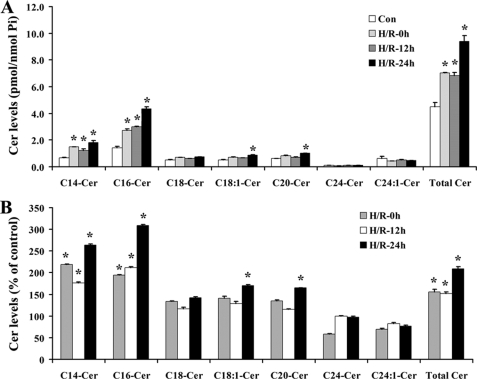
H/R Triggered Accumulation of Ceramide in NT-2 Cells—Because ceramide is implicated in apoptosis signaling, we investigated whether H/R treatment affected ceramide content in NT-2 cells. NT-2 cells treated with H/R were collected at 0, 12, and 24 h after reoxygenation. Lipid extraction and mass spectrometry analyses were then carried out on the collected cells. As shown in Fig. 3, A and B, dihydroceramide, ceramide, and dihydrosphingosine levels were elevated in NT-2 cells after H/R. On the other hand, sphingosine and sphingosine-1-phosphate levels in these cells were unchanged. Following hypoxia, ceramide levels were found to increase, and they remained elevated upon extended reoxygenation. Analyses of the various ceramide species revealed that H/R induced a robust elevation in C14:0- and C16:0-ceramides, a small increase in C18:0-, C18:1-, and C20:0-ceramides, and no increase in C24:0- and C24:1-ceramides (Fig. 4, A and B). These results demonstrate the significant effects of H/R on ceramide metabolism. They also suggest that H/R treatment may induce ceramide accumulation in NT-2 cells through up-regulation of ceramide synthesis rather than activation of neutral sphingomyelinase because the former has been associated with preferential accumulation of C16-ceramide whereas the latter is associated with C24-ceramide.
FIGURE 3.
Hypoxia/reoxygenation triggers accumulation of ceramide metabolites in NT-2 cells. GFP-Bax-stable NT-2 cells were subjected to hypoxia (12 h) and varying lengths of reoxygenation (0, 12, and 24 h). Lipids were extracted from the collected cells and dihydroceramide (DHCer), ceramide (Cer), dihydrosphingosine (DHSph), sphingosine (Sph), and sphingosine-1-phosphate (Sph-1P) contents were analyzed by HPLC and tandem MS and expressed either as pmol/nmol phosphate (A) or as a percentage of the control untreated cells (B;*, p ≤ 0.05, compared with corresponding control).
FIGURE 4.
Hypoxia/reoxygenation triggers accumulation of various ceramide species in NT-2 cells. GFP-Bax-stable NT-2 cells were subjected to 12 h hypoxia and varying lengths of reoxygenation (0, 12, and 24 h). The levels of various ceramide species were determined by tandem MS and expressed either as pmol/nmol phosphate (A) or as a percentage of the control untreated cells (B).
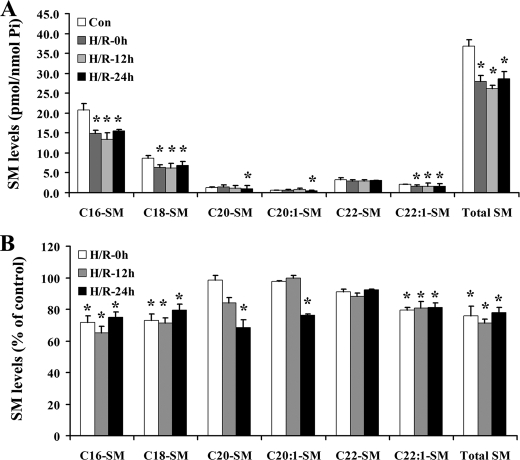
Acid Sphingomyelinase and the Ceramide Salvage Pathway Are Involved in Hypoxia/Reoxygenation-induced Ceramide Accumulation and Bax Translocation—Because ceramide can be generated from the hydrolysis of sphingomyelin (SM), SM levels were examined following H/R (Fig. 5). GFP-Bax-stable NT-2 cells were subjected to H/R, and SM species from the control and H/R-treated cells were determined by MS analyses. The results indicated that almost all following hypoxia and species of SM were decreased in NT-2 cells following hypoxia and remained mostly depressed upon extended reoxygenation.
FIGURE 5.
Sphingomyelin levels are decreased in NT-2 cells following H/R treatment. GFP-Bax-stable NT-2 cells treated with hypoxia (12 h)/reoxygenation (24 h) were collected and sphingomyelin species were analyzed by lipid extraction and tandem MS. Sphingomyelin levels were expressed either as pmol/nmol phosphate (A) or as a percentage of the control untreated cells (B).
The finding that sphingomyelin level was decreased following H/R prompted us to investigate the action of sphingomyelinases, namely acid sphingomyelinase (aSMase) and neutral sphingomyelinase (nSMase). GFP-Bax-stable NT-2 cells were subjected to hypoxia (12 h) and varying lengths of reoxygenation (0, 12, and 24 h). The cells were then collected and aSMase and nSMase activities were measured. As shown in Fig. 6A, aSMase activity was elevated following hypoxia, and it decreased back to the basal level following reoxygenation. In contrast, the activity of nSMase was not affected by the H/R treatment. Analyses by qPCR indicated that aSMase mRNA level was elevated following hypoxia and dropped upon extended reoxygenation (Fig. 6B).
To examine the role of aSMase in H/R-induced Bax translocation, GFP-Bax-stable NT-2 cells were transfected with control scrambled or aSMase siRNA oligonucleotides. A knockdown in aSMase mRNA expression was confirmed by qPCR (Fig. 6C). As shown in Fig. 6D, knockdown of aSMase led to a significant decrease in Bax translocation to mitochondria following H/R. In addition, we found that aSMase knockdown attenuated H/R-induced cytochrome c release and caspase-3 activation (Fig. 6, E and F). Moreover, down-regulation of aSMase decreased H/R-induced ceramide elevation, particularly C16- and C18-ceramides (Fig. 6G). These results suggest that elevated aSMase activity during hypoxia is important for ceramide up-regulation and for subsequent Bax translocation to mitochondria.
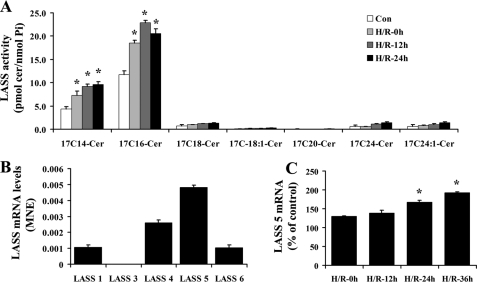
LASS 5 Is Involved in H/R-induced Ceramide Accumulation and Bax Translocation—The salvage pathway also involves the action of ceramide synthase (50, 51). To determine the contributory role of the salvage pathway to H/R-induced ceramide up-regulation, we examined the activity of ceramide synthase (LASS). GFP-Bax-stable NT-2 cells were subjected to hypoxia and varying lengths of reoxygenation. The cells were then labeled with synthetic 17C-sphingosine for 30 min. The incorporation of 17C-sphingosine into ceramide was determined by lipid extraction and tandem MS analyses. As shown in Fig. 7A, 17Cn-ceramide synthesis was elevated by hypoxia, and it continued to be elevated post-reoxygenation. Specifically, 17C14- and 17C16-ceramides were found to be the predominant species synthesized.
FIGURE 7.
Ceramide synthase LASS 5 was up-regulated following H/R in NT-2 cells. A, GFP-Bax-stable NT-2 cells were subjected to 12 h hypoxia followed by varying lengths of reoxygenation (0, 12, and 24 h). Ceramide synthase activities (pmol 17C-ceramide/nmol phosphate) of the collected cells were determined by tandem MS. B, mRNA expression levels of various ceramide synthases (LASS) in NT-2 cells were determined by qPCR. C, mRNA expression level of LASS 5 in NT-2 subjected to hypoxia and varying lengths of reoxygenation were determined by qPCR and expressed as a percentage of the control untreated cells.
Each of the six known LASSes displays unique substrate specificity for fatty acyl chain length and/or saturation (35). Because C14:0- and C16:0-ceramide species were increased the most following H/R, we hypothesized that LASS 5 and/or LASS 6 may be the source of the ceramide up-regulation since they share substrate preference for C16-ceramide. To identify the LASS isoform that is responsible for the up-regulated ceramide synthesis, LASS gene expression in NT-2 cells was first examined. Analyses by qPCR revealed that LASS 1, LASS 4, LASS 5 and LASS 6 are expressed in NT-2 cells, with LASS 5 being the most abundant, followed by LASS 4, LASS 1, and LASS 6 (Fig. 7B). Following hypoxia, we found that LASS 5 mRNA level was elevated, and it continued to rise upon extended reoxygenation (Fig. 7C). On the other hand, there was little change in expression of LASS 6 or other LASS genes (data not shown). To test whether LASS 5 is responsible for H/R-induced ceramide up-regulation, GFP-Bax-stable NT-2 cells were transfected with sequence-specific siRNA oligonucleotides to down-regulate LASS 5 gene expression. The down-regulation of LASS 5 expression was first confirmed by ceramide synthase activity assay. In the study, it was found that the H/R-induced increase in ceramide synthase activity was significantly inhibited in LASS 5 siRNA oligonucleotide-transfected cells (Fig. 8A). On the other hand, transfection with the LASS 6 siRNA oligonucleotides had no effect on ceramide synthase activity (data not shown). Transfection with LASS 5 siRNA oligonucleotides only decreased the LASS 5 mRNA level without affecting the mRNA levels of other LASSes including 1, 4, and 6, as determined by qPCR (Fig. 8B).
Because physiological up-regulation of ceramide has been implicated in Bax activation (52–54), we set out to determine whether a knockdown in LASS 5 could attenuate H/R-induced Bax translocation to mitochondria. GFP-Bax-stable NT-2 cells were transfected with either control scrambled or LASS 5-specific siRNA oligonucleotides. The transfected cells were then subjected to hypoxia (12 h)/reoxygenation (24 h) and visualized by fluorescence microscopy. As shown in Fig. 8C, LASS 5 knockdown partially inhibited H/R-induced Bax localization to mitochondria. In contrast, down-regulation of LASS 6 had no effect on the Bax subcellular distribution. In addition, it was found that knockdown of LASS 5 could attenuate H/R-induced cytochrome c release and caspase-3 activation (Figs. 6E and 8D). Moreover, LASS 5 knockdown decreased primarily the H/R-induced up-regulation of C14- and C16-ceramides (Fig. 8E). These results suggest that LASS 5 is important for ceramide up-regulation following H/R and that its down-regulation would attenuate Bax activation.
DISCUSSION
Apoptosis plays an important role in neuronal cell death following cerebral ischemia. Bax is a key regulator of apoptosis and its activation and localization to mitochondria triggers cytochrome c release and the initiation of the caspase proteolytic pathway. The molecular signaling pathway that regulates Bax activation following H/R is not known. In this report, we show that in NT-2 cells, ceramide up-regulation following H/R is an important trigger for Bax activation. We found that the salvage pathway of ceramide synthesis is involved in ceramide up-regulation following H/R. In particular, we have shown that acid sphingomyelinase and ceramide synthase LASS 5 are the key enzymes responsible for this up-regulation. In addition, we have demonstrated that these ceramide-producing enzymes are important for Bax activation. A knockdown of these enzymes significantly attenuated the extent of Bax localization to mitochondria.
Ceramide could be produced from the hydrolysis of sphingomyelin by sphingomyelinases. Mammalian cells utilize two separate forms of sphingomyelinase, an acidic and a neutral isoform, for ceramide generation (55). It was reported that sphingomyelinase activation and sphingomyelin hydrolysis led to endogenous ceramide accumulation following severe and lethal cerebral ischemia reperfusion (56, 57). Sphingomyelinases appear to play a role in ischemic neuronal cell death as a knock out in acid sphingomyelinase decreased I/R-induced brain tissue damage (24). In cardiomyocytes, neutral sphingomyelinase activation is an early response to H/R (58). However, in another study, it was reported that sphingomyelin hydrolysis was not detected in I/R-treated brain tissues (42). In our system with NT-2 neuronal precursor cells, we found that treatment with H/R resulted in a decrease in overall sphingomyelin content of the cells. Analyses by qPCR indicated that aSMase mRNA level was increased with hypoxia treatment. Consistently, enzyme activity analyses indicated that acid but not neutral sphingomyelinase activity was up-regulated. Interestingly, aSMase activity returned to the basal level following prolonged reoxygenation. Knockdown of aSMase resulted in significant attenuation of the induced levels of ceramide. Taken together, these results suggest that in NT-2 cells, hypoxic condition triggers the activation of aSMase.
Ceramide produced by de novo synthesis has been reported to serve as an apoptotic signaling molecule (50, 59). De novo synthesized ceramide was shown to increase after a brief exposure of cultured neuronal cells to hypoxia, oxygen/glucose deprivation, or tumor necrosis factor α (60). Unfortunately, many studies have relied solely on the use of fumonisin B1, an inhibitor of (dihydro)ceramide synthases to implicate the involvement of the de novo pathway. However, this inhibitor also inhibits ceramide generated from the salvage pathway. In the current study, we found that ceramide synthesis was up-regulated following H/R in NT-2 cells. In addition to ceramide, we found that dihydroceramide level was elevated following H/R. However, this elevated level of dihydroceramide did not seem to significantly contribute to ceramide production, as a knockdown of dihydroceramide desaturase did not affect ceramide production following H/R.5 Thus, it would appear that the synthesis of ceramide from acylation of sphingosine is the primary route for the salvaged synthesis of ceramide.
In a previous study, it was shown that LASS 6, which preferentially generates C16:0- and C18:0-ceramide, plays an important role in I/R-induced ceramide accumulation (42). However, it was proposed that this elevated ceramide synthase activity is mediated through a post-translational modification mechanism, as the protein level of LASS 6 did not increase following I/R. In our study with NT-2 cells, C14:0-, C16:0-ceramides were the major ceramide species found to be elevated following H/R. In addition, we found that for LASS 5, which preferentially generates C14:0-, C16:0-, C18:0-, and C18:1-ceramide (35, 61), its mRNA expression level was elevated following H/R. Similarly, analysis of ceramide synthase activity indicated that C14:0-, C16:0-ceramide increased significantly and preferentially following H/R. On the other hand, we did not detect a comparable increase in the mRNA expression level of LASS 6 (data not shown). Moreover, a knockdown in LASS 5 significantly reduced H/R-induced increase in ceramide synthase activity. Based on these results, it appears that LASS 5 is also important at promoting ceramide up-regulation following H/R.
Bax translocation to mitochondria constitutes a key step in apoptosis signaling pathways. It has been reported that addition of exogenous C2- and C6-ceramide enhanced Bax localization to mitochondria in HL-60 cells (62). In addition, it was shown that UV irradiation could activate sphingomyelinases resulting in ceramide up-regulation and Bax conformational change (63). This elevated ceramide level was attributed to the activation of aSMase (41). Moreover, it was shown that expression of mitochondrial-targeting bacterial sphingomyelinase could trigger Bax translocation to mitochondria (36). In NT-2 cells subjected to H/R, we found that ceramide up-regulation is linked to Bax translocation to mitochondria. Down-regulation of either aSMase or LASS 5 could attenuate H/R-induced Bax translocation to mitochondria. On the other hand, down-regulation of nSMase5 and LASS 6 had no effect on Bax localization.
Mitochondria have been implicated in ceramide-induced apoptosis. They are closely associated with smooth membranous sacs that resemble the ER membranes (64), and it has been suggested that membrane contact between the ER and mitochondria may enable rapid lipid exchange between these two membranous compartments (65, 66). Thus, H/R-induced up-regulation of aSMase and LASS 5 may lead to ceramide accumulation in mitochondria. The mitochondrial pool of ceramide is believed to play a role in TNFα-induced Bax translocation, cytochrome c release and cell death (36). Akt might be a target of ceramide, as inhibition of Akt by ceramide could lead to inhibition of the anti-apoptotic protein Bcl-XL by Bad (67, 68). Increasing the Bax/Bcl-XL or the Bax/Bcl-2 ratio might be another mechanism of ceramide-induced apoptosis (54, 69). However, in our study, we observed no change in the levels of Akt, Bcl-2 and Bcl-XL (data not shown).
Currently, the precise mechanism by which ceramide activates Bax is unclear. It has been shown that nonionic detergents could lead to a change in Bax conformation and oligomeric state (9, 46). It has been proposed that ceramide could potentially mimic the property of nonionic detergents in altering Bax conformation and lead to its activation (63). Alternatively, it has been reported that ceramide derived from aSMase in the lysosome could activate cathepsin D (70), which in turn could lead to Bax activation by a yet unknown mechanism (71). Taken together, it appears that ceramide up-regulation induced by H/R serves as a trigger for Bax activation. In the future, it would be important to decipher the molecular mechanisms of ceramide signaling in cell death regulation.
This work was supported, in whole or in part, by National Institutes of Health Grants NS40932 (to Y.-T. H.) and CA97132 (to Y. A. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: I/R, ischemia/reperfusion; H/R, hypoxia/reoxygenation; GFP, green fluorescent protein; PBS, phosphate-buffered saline; qPCR, quantitative PCR; siRNA, short interfering RNA.
J. Jin, J. M. Kraveka, and Y. T. Hsu, unpublished data.
References
- 1.Oltvai, Z. N., Milliman, C. L., and Korsmeyer, S. J. (1993) Cell 74 609-619 [DOI] [PubMed] [Google Scholar]
- 2.Roth, K. A., and D'Sa, C. (2001) Ment. Retard. Dev. Disabil. Res. Rev. 7261 -266 [DOI] [PubMed] [Google Scholar]
- 3.Print, C. G., and Loveland, K. L. (2000) Bioessays 22423 -430 [DOI] [PubMed] [Google Scholar]
- 4.Knudson, C. M., Tung, K. S., Tourtellotte, W. G., Brown, G. A., and Korsmeyer, S. J. (1995) Science 270 96-99 [DOI] [PubMed] [Google Scholar]
- 5.Cheng, W., Kajstura, J., Nitahara, J. A., Li, B., Reiss, K., Liu, Y., Clark, W. A., Krajewski, S., Reed, J. C., Olivetti, G., and Anversa, P. (1996) Exp. Cell Res. 226316 -327 [DOI] [PubMed] [Google Scholar]
- 6.Krajewski, S., Mai, J. K., Krajewska, M., Sikorska, M., Mossakowski, M. J., and Reed, J. C. (1995) J. Neurosci. 156364 -6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misao, J., Hayakawa, Y., Ohno, M., Kato, S., Fujiwara, T., and Fujiwara, H. (1996) Circulation 941506 -1512 [DOI] [PubMed] [Google Scholar]
- 8.Hsu, Y. T., Wolter, K. G., and Youle, R. J. (1997) Proc. Natl. Acad. Sci. U. S. A. 943668 -3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu, Y. T., and Youle, R. J. (1998) J. Biol. Chem. 27310777 -10783 [DOI] [PubMed] [Google Scholar]
- 10.Wolter, K. G., Hsu, Y. T., Smith, C. L., Nechushtan, A., Xi, X. G., and Youle, R. J. (1997) J. Cell Biol. 1391281 -1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross, A., Jockel, J., Wei, M. C., and Korsmeyer, S. J. (1998) EMBO J. 173878 -3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smaili, S. S., Hsu, Y. T., Sanders, K. M., Russell, J. T., and Youle, R. J. (2001) Cell Death Differ. 8 909-920 [DOI] [PubMed] [Google Scholar]
- 13.De Giorgi, F., Lartigue, L., Bauer, M. K., Schubert, A., Grimm, S., Hanson, G. T., Remington, S. J., Youle, R. J., and Ichas, F. (2002) Faseb. J. 16 607-609 [DOI] [PubMed] [Google Scholar]
- 14.Manon, S., Chaudhuri, B., and Guerin, M. (1997) FEBS Lett. 41529 -32 [DOI] [PubMed] [Google Scholar]
- 15.Eskes, R., Antonsson, B., Osen-Sand, A., Montessuit, S., Richter, C., Sadoul, R., Mazzei, G., Nichols, A., and Martinou, J. C. (1998) J. Cell Biol. 143217 -224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, P., Nijhawan, D., Budihardjo, I., Srinivasula, S. M., Ahmad, M., Alnemri, E. S., and Wang, X. (1997) Cell 91479 -489 [DOI] [PubMed] [Google Scholar]
- 17.Putcha, G. V., Harris, C. A., Moulder, K. L., Easton, R. M., Thompson, C. B., and Johnson, E. M., Jr. (2002) J. Cell Biol. 157441 -453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desagher, S., Osen-Sand, A., Nichols, A., Eskes, R., Montessuit, S., Lauper, S., Maundrell, K., Antonsson, B., and Martinou, J. C. (1999) J. Cell Biol. 144891 -901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eskes, R., Desagher, S., Antonsson, B., and Martinou, J. C. (2000) Mol. Cell. Biol. 20 929-935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez, D., and White, E. (2000) Mol. Cell 653 -63 [PubMed] [Google Scholar]
- 21.Northington, F. J., Ferriero, D. M., Flock, D. L., and Martin, L. J. (2001) J. Neurosci. 211931 -1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruvolo, P. P. (2003) Pharmacol. Res. 47383 -392 [DOI] [PubMed] [Google Scholar]
- 23.Larsen, E. C., Hatcher, J. F., and Adibhatla, R. M. (2007) Neuroscience 146946 -961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu, Z. F., Nikolova-Karakashian, M., Zhou, D., Cheng, G., Schuchman, E. H., and Mattson, M. P. (2000) J. Mol. Neurosci. 1585 -97 [DOI] [PubMed] [Google Scholar]
- 25.Hannun, Y. A., and Obeid, L. M. (2002) J. Biol. Chem. 27725847 -25850 [DOI] [PubMed] [Google Scholar]
- 26.Marchesini, N., and Hannun, Y. A. (2004) Biochem. Cell Biol. 8227 -44 [DOI] [PubMed] [Google Scholar]
- 27.Goni, F. M., and Alonso, A. (2002) FEBS Lett. 53138 -46 [DOI] [PubMed] [Google Scholar]
- 28.Gulbins, E., and Kolesnick, R. (2002) Subcell. Biochem. 36229 -244 [DOI] [PubMed] [Google Scholar]
- 29.Levade, T., and Jaffrezou, J. P. (1999) Biochim. Biophys. Acta 1438 1-17 [DOI] [PubMed] [Google Scholar]
- 30.Merrill, A. H., Jr., and Jones, D. D. (1990) Biochim. Biophys. Acta 1044 1-12 [DOI] [PubMed] [Google Scholar]
- 31.Wang, E., and Merrill, A. H., Jr. (2000) Methods Enzymol. 31115 -21 [DOI] [PubMed] [Google Scholar]
- 32.Michel, C., van Echten-Deckert, G., Rother, J., Sandhoff, K., Wang, E., and Merrill, A. H., Jr. (1997) J. Biol. Chem. 27222432 -22437 [DOI] [PubMed] [Google Scholar]
- 33.Ternes, P., Franke, S., Zahringer, U., Sperling, P., and Heinz, E. (2002) J. Biol. Chem. 27725512 -25518 [DOI] [PubMed] [Google Scholar]
- 34.D'Mello, N., P., Childress, A. M., Franklin, D. S., Kale, S. P., Pinswasdi, C., and Jazwinski, S. M. (1994) J. Biol. Chem. 26915451 -15459 [PubMed] [Google Scholar]
- 35.Mizutani, Y., Kihara, A., and Igarashi, Y. (2005) Biochem. J. 390263 -271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birbes, H., Luberto, C., Hsu, Y. T., El Bawab, S., Hannun, Y. A., and Obeid, L. M. (2005) Biochem. J. 386445 -451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh, H. L., Seok, J. Y., Kwon, C. H., Kang, S. K., and Kim, Y. K. (2006) Neurotoxicology 27 31-38 [DOI] [PubMed] [Google Scholar]
- 38.Lee, M., Hwang, J. T., Lee, H. J., Jung, S. N., Kang, I., Chi, S. G., Kim, S. S., and Ha, J. (2003) J. Biol. Chem. 27839653 -39661 [DOI] [PubMed] [Google Scholar]
- 39.Hou, Q., and Hsu, Y. T. (2005) Am. J. Physiol. Heart Circ. Physiol. 289H477 -H487 [DOI] [PubMed] [Google Scholar]
- 40.Zeidan, Y. H., Pettus, B. J., Elojeimy, S., Taha, T., Obeid, L. M., Kawamori, T., Norris, J. S., and Hannun, Y. A. (2006) J. Biol. Chem. 28124695 -24703 [DOI] [PubMed] [Google Scholar]
- 41.Zeidan, Y. H., and Hannun, Y. A. (2007) J. Biol. Chem. 28211549 -11561 [DOI] [PubMed] [Google Scholar]
- 42.Yu, J., Novgorodov, S. A., Chudakova, D., Zhu, H., Bielawska, A., Bielawski, J., Obeid, L. M., Kindy, M. S., and Gudz, T. I. (2007) J. Biol. Chem. 28225940 -25949 [DOI] [PubMed] [Google Scholar]
- 43.Bielawski, J., Szulc, Z. M., Hannun, Y. A., and Bielawska, A. (2006) Methods 39 82-91 [DOI] [PubMed] [Google Scholar]
- 44.Schulz, A., Mousallem, T., Venkataramani, M., Persaud-Sawin, D. A., Zucker, A., Luberto, C., Bielawska, A., Bielawski, J., Holthuis, J. C., Jazwinski, S. M., Kozhaya, L., Dbaibo, G. S., and Boustany, R. M. (2006) J. Biol. Chem. 2812784 -2794 [DOI] [PubMed] [Google Scholar]
- 45.Marchesini, N., Luberto, C., and Hannun, Y. A. (2003) J. Biol. Chem. 27813775 -13783 [DOI] [PubMed] [Google Scholar]
- 46.Hsu, Y. T., and Youle, R. J. (1997) J. Biol. Chem. 27213829 -13834 [DOI] [PubMed] [Google Scholar]
- 47.Zeidan, Y. H., Wu, B. X., Jenkins, R. W., Obeid, L. M., and Hannun, Y. A. (2008) Faseb J. 22 183-193 [DOI] [PubMed] [Google Scholar]
- 48.Antonsson, B., Montessuit, S., Sanchez, B., and Martinou, J. C. (2001) J. Biol. Chem. 27611615 -11623 [DOI] [PubMed] [Google Scholar]
- 49.Mikhailov, V., Mikhailova, M., Pulkrabek, D. J., Dong, Z., Venkatachalam, M. A., and Saikumar, P. (2001) J. Biol. Chem. 27618361 -18374 [DOI] [PubMed] [Google Scholar]
- 50.Bose, R., Verheij, M., Haimovitz-Friedman, A., Scotto, K., Fuks, Z., and Kolesnick, R. (1995) Cell 82 405-414 [DOI] [PubMed] [Google Scholar]
- 51.Garzotto, M., White-Jones, M., Jiang, Y., Ehleiter, D., Liao, W. C., Haimovitz-Friedman, A., Fuks, Z., and Kolesnick, R. (1998) Cancer Res. 582260 -2264 [PubMed] [Google Scholar]
- 52.Pastorino, J. G., Tafani, M., Rothman, R. J., Marcinkeviciute, A., Hoek, J. B., and Farber, J. L. (1999) J. Biol. Chem. 27431734 -31739 [DOI] [PubMed] [Google Scholar]
- 53.von Haefen, C., Wieder, T., Gillissen, B., Starck, L., Graupner, V., Dorken, B., and Daniel, P. T. (2002) Oncogene 214009 -4019 [DOI] [PubMed] [Google Scholar]
- 54.Kim, W. H., Ghil, K. C., Lee, J. H., Yeo, S. H., Chun, Y. J., Choi, K. H., Kim, D. K., and Kim, M. Y. (2000) Cancer Lett. 15139 -48 [DOI] [PubMed] [Google Scholar]
- 55.Wiegmann, K., Schutze, S., Machleidt, T., Witte, D., and Kronke, M. (1994) Cell 781005 -1015 [DOI] [PubMed] [Google Scholar]
- 56.Nakane, M., Kubota, M., Nakagomi, T., Tamura, A., Hisaki, H., Shimasaki, H., and Ueta, N. (2000) Neurosci. Lett. 29689 -92 [DOI] [PubMed] [Google Scholar]
- 57.Kubota, M., Narita, K., Nakagomi, T., Tamura, A., Shimasaki, H., Ueta, N., and Yoshida, S. (1996) Neurol. Res. 18337 -341 [DOI] [PubMed] [Google Scholar]
- 58.O'Brien, N. W., Gellings, N. M., Guo, M., Barlow, S. B., Glembotski, C. C., and Sabbadini, R. A. (2003) Circ. Res. 92589 -591 [DOI] [PubMed] [Google Scholar]
- 59.Perry, D. K. (2002) Biochim. Biophys. Acta 1585146 -152 [DOI] [PubMed] [Google Scholar]
- 60.Ginis, I., Schweizer, U., Brenner, M., Liu, J., Azzam, N., Spatz, M., and Hallenbeck, J. M. (1999) Am. J. Physiol. 276C1171 -C1183 [DOI] [PubMed] [Google Scholar]
- 61.Spassieva, S., Seo, J. G., Jiang, J. C., Bielawski, J., Alvarez-Vasquez, F., Jazwinski, S. M., Hannun, Y. A., and Obeid, L. M. (2006) J. Biol. Chem. 28133931 -33938 [DOI] [PubMed] [Google Scholar]
- 62.Kim, H. J., Mun, J. Y., Chun, Y. J., Choi, K. H., and Kim, M. Y. (2001) FEBS Lett. 505264 -268 [DOI] [PubMed] [Google Scholar]
- 63.Kashkar, H., Wiegmann, K., Yazdanpanah, B., Haubert, D., and Kronke, M. (2005) J. Biol. Chem. 28020804 -20813 [DOI] [PubMed] [Google Scholar]
- 64.Pickett, C. B., Montisano, D., Eisner, D., and Cascarano, J. (1980) Exp. Cell Res. 128343 -352 [DOI] [PubMed] [Google Scholar]
- 65.Daum, G., and Vance, J. E. (1997) Prog. Lipid Res. 36103 -130 [DOI] [PubMed] [Google Scholar]
- 66.Voelker, D. R. (2005) Trends Biochem. Sci. 30396 -404 [DOI] [PubMed] [Google Scholar]
- 67.Zhou, H., Summers, S. A., Birnbaum, M. J., and Pittman, R. N. (1998) J. Biol. Chem. 27316568 -16575 [DOI] [PubMed] [Google Scholar]
- 68.Zhou, H., Li, X. M., Meinkoth, J., and Pittman, R. N. (2000) J. Cell Biol. 151483 -494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sawada, M., Nakashima, S., Banno, Y., Yamakawa, H., Hayashi, K., Takenaka, K., Nishimura, Y., Sakai, N., and Nozawa, Y. (2000) Cell Death Differ. 7761 -772 [DOI] [PubMed] [Google Scholar]
- 70.Heinrich, M., Wickel, M., Schneider-Brachert, W., Sandberg, C., Gahr, J., Schwandner, R., Weber, T., Saftig, P., Peters, C., Brunner, J., Kronke, M., and Schutze, S. (1999) EMBO J. 185252 -5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bidere, N., Lorenzo, H. K., Carmona, S., Laforge, M., Harper, F., Dumont, C., and Senik, A. (2003) J. Biol. Chem. 27831401 -31411 [DOI] [PubMed] [Google Scholar]