Abstract
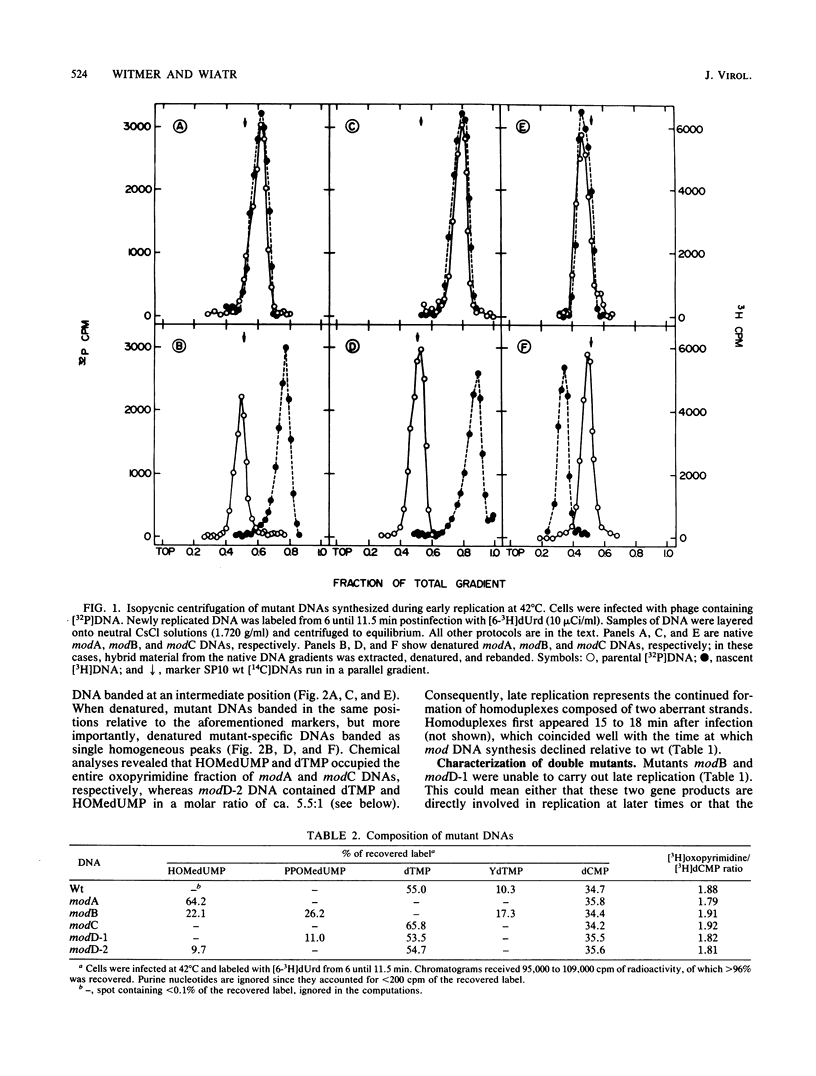
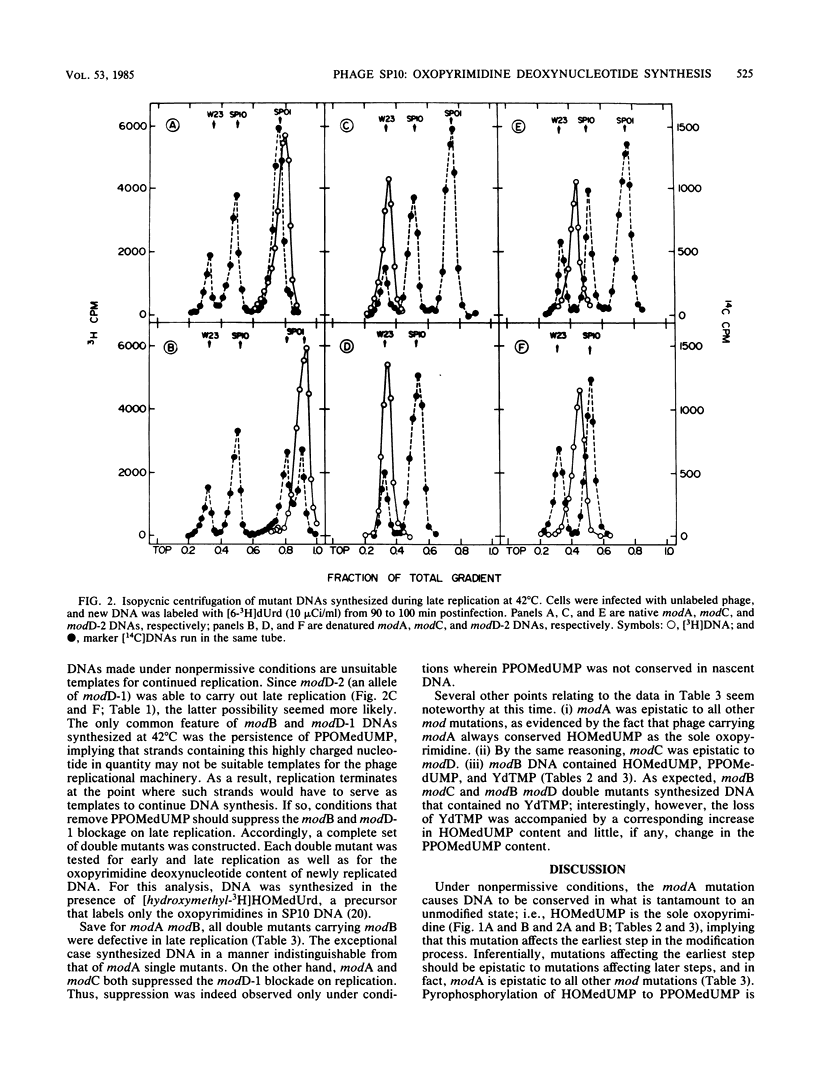
Bacillus subtilis phage SP10 DNA has two oxopyrimidines, thymidine 5'-monophosphate (dTMP) and its hypermodified analog (YdTMP). Published data suggest that both are synthesized by postreplicational modification of 5-hydroxymethyldeoxyuridylate (HOMedUMP) in nascent DNA by the following pathway: HOMedUMP----PPOMedUMP----dTMP (85%) or YdTMP (15%); PPOMedUMP is 5-(hydroxymethyl-O-pyrophosphoryl)deoxyuridylate, the pyrophosphoric acid ester of the C5CH2OH function of HOMedUMP. This paper describes aberrant DNAs synthesized at nonpermissive temperatures by a complementary series of heat-sensitive, modification-defective (mod) mutants. Collectively, these mutants encompass the major steps in the complete modification of nascent SP10 DNA. DNA produced by modA phage retains HOMedUMP as its sole oxopyrimidine, implying that (i) this mutant is defective in the pyrophosphorylation step and (ii) formation of PPOMedUMP is required for any further modification. Furthermore, studies with double mutants indicated that modA is epistatic for all other mod mutants, which supports the hypothesis that modA controls the earliest step in the modification pathway. Since their DNAs contain no YdTMP, modC and modD are defective in hypermodification (i.e., PPOMedUMP----YdTMP). However, dTMP occupies the entire oxopyrimidine fraction of modC DNA, whereas modD DNA has a normal dTMP content, but the now-missing YdTMP is replaced by either PPOMedUMP or a byproduct of abortive hypermodification. It is proposed that the modD mutants are defective in the catalytic aspects of hypermodification and that modC are defective in some regulatory function that promotes hypermodification at the expense of reductive modification (i.e., PPOMedUMP----dTMP). Reductive modification is defective in modB phage, as evidenced by the absence of dTMP. In contrast to the others, modB DNA has a complex oxopyrimidine content: HOMedUMP, ca. 30%; PPOMedUMP, ca. 40%; and YdTMP, ca. 30%. The expanded level of YdTMP suggests that at certain sites, reductive modification and hypermodification are competing reactions. Interestingly, the PPOMedUMP content of modB DNA seemingly reflects the maximum degree to which phage DNA can be pyrophosphorylated, since the loss of YdTMP from modBmodC and modBmodD DNAs results in a unilateral increase in HOMedUMP content.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown N. C. 6-(p-hydroxyphenylazo)-uracil: a selective inhibitor of host DNA replication in phage-infected Bacillus subtilis. Proc Natl Acad Sci U S A. 1970 Nov;67(3):1454–1461. doi: 10.1073/pnas.67.3.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella E., Markewych O., Dosmar M., Witmer H. Production and expression of dTMP-enriched DNA of bacteriophage SP15. J Virol. 1978 Dec;28(3):753–766. doi: 10.1128/jvi.28.3.753-766.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Specificity of the bacteriophage Mu mom+ -controlled DNA modification. J Virol. 1980 Apr;34(1):277–279. doi: 10.1128/jvi.34.1.277-279.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski A. M., Bose R. J., Warren R. A. 5-(4-Aminobutylaminomethyl)uracil, an unusual pyrimidine from the deoxyribonucleic acid of bacteriophage phiW-14. Biochemistry. 1973 Jan 2;12(1):151–157. doi: 10.1021/bi00725a025. [DOI] [PubMed] [Google Scholar]
- Lembach K. J., Buchanan J. M. The relationship of protein synthesis to early transcriptive events in bacteriophage T4-infected Escherichia coli B. J Biol Chem. 1970 Apr 10;245(7):1575–1587. [PubMed] [Google Scholar]
- Maltman K. L., Neuhard J., Lewis H. A., Warren R. A. Synthesis of thymine and alpha-putrescinylthymine in bacteriophage phi W-14-infected Pseudomonas acidovorans. J Virol. 1980 May;34(2):354–359. doi: 10.1128/jvi.34.2.354-359.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltman K. L., Neuhard J., Warren R. A. 5-[(Hydroxymethyl)-O-pyrophosphoryl]uracil, an intermediate in the biosynthesis of alpha-putrescinylthymine in deoxyribonucleic acid of bacteriophage phi W-14. Biochemistry. 1981 Jun 9;20(12):3586–3591. doi: 10.1021/bi00515a043. [DOI] [PubMed] [Google Scholar]
- Markewych O., Boghosian A., Dosmar M., Ende D., Witmer H. SP-10 bacteriophage-specific nucleic acid and enzyme synthesis in Bacillus subtilis W23. J Virol. 1977 Jan;21(1):84–95. doi: 10.1128/jvi.21.1.84-95.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markewych O., Casella E., Dosmar M., Witmer H. Deoxythymidine nucleotide metabolism in Bacillus subtilis W23 infected with bacteriophage SP1Oc: preliminary evidence that dTMP in SP10c DNA is synthesized by a novel, bacteriophage-specific mechanism. J Virol. 1979 Jan;29(1):61–68. doi: 10.1128/jvi.29.1.61-68.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhard J., Maltman K. L., Warren R. A. Bacteriophage phi W-14-infected Pseudomonas acidovorans synthesizes hydroxymethyldeoxyuridine triphosphate. J Virol. 1980 May;34(2):347–353. doi: 10.1128/jvi.34.2.347-353.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M. M., Brown N. C. Inhibition of a discrete bacterial DNA polymerase by 6-(p-hydroxyphenylazo)-uracil and 6-(p-hydroxyphenylazo-)-isocytosine. Nat New Biol. 1972 Nov 15;240(98):80–82. doi: 10.1038/newbio240080a0. [DOI] [PubMed] [Google Scholar]
- Pogolotti A. L., Jr, Santi D. V. Model studies of the thymidylate synthetase reaction. Nucleophilic displacement of 5-p-nitrophenoxymethyluracils. Biochemistry. 1974 Jan 29;13(3):456–466. doi: 10.1021/bi00700a010. [DOI] [PubMed] [Google Scholar]
- Walker M. S., Mandel M. Biosynthesis of 5-(4'5'-dihydroxypentyl) uracil as a nucleoside triphosphate in bacteriophage SP15-infected Bacillus subtilis. J Virol. 1978 Feb;25(2):500–509. doi: 10.1128/jvi.25.2.500-509.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. A. Modified bases in bacteriophage DNAs. Annu Rev Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- Wiatr C. L., Witmer H. J. Selective protection of 5' ... GGCC ... 3' and 5' ... GCNGC ... 3' sequences by the hypermodified oxopyrimidine in Bacillus subtilis bacteriophage SP10 DNA. J Virol. 1984 Oct;52(1):47–54. doi: 10.1128/jvi.52.1.47-54.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witmer H., Franks M. DNA Synthesis and Gene Expression in Bacillus subtilis Infected with Wild-Type and Hypermodification-Defective Bacteriophage SP10. J Virol. 1982 May;42(2):636–648. doi: 10.1128/jvi.42.2.636-648.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witmer H. Synthesis of deoxythymidylate and the unusual deoxynucleotide in mature DNA of Bacillus subtilis bacteriophage SP10 occurs by postreplicational modification of 5-hydroxymethyldeoxyuridylate. J Virol. 1981 Aug;39(2):536–547. doi: 10.1128/jvi.39.2.536-547.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]


