Abstract
The lungs of patients with cystic fibrosis (CF) are typically chronically infected with Pseudomonas aeruginosa. We used an immunoproteomics approach to analyze the responses of patients to secreted P. aeruginosa proteins. Extracellular proteins from P. aeruginosa strain PAO1 that had been grown to stationary phase were separated by two-dimensional polyacrylamide gel electrophoresis and analyzed by Western blotting using sera from four chronically infected patients. Sera from all four patients detected multiple extracellular proteins. The identities of selected proteins recognized by antisera were determined. Production of at least four of these proteins (azurin and three proteases: elastase, PrpL, and PasP) is governed by quorum sensing, consistent with active bacterial quorum sensing in the lungs of CF patients. The CF lung is generally thought to be an iron-deficient environment for infecting bacteria, and growing the bacteria in the presence of an iron-chelating agent, ethylene-diamine-di(o-hydroxyphenylacetic acid), enabled detection of additional proteins that were recognized by patient sera. The sera also detected multiple proteins from cells in the logarithmic growth phase, and protein identification suggested that most of these were the result of cell lysis or secretion in membrane vesicles. Comparison with extracellular proteins from a second P. aeruginosa strain, strain Pa4, showed that many proteins recognized by patient sera are common to both strains, although there are also some strain-specific extracellular proteins. Our data show that while there are some differences in the responses of different patients to P. aeruginosa, there are also many similarities, and that an immunoproteomics approach enables the identification of proteins that are made by P. aeruginosa during infection.
Pseudomonas aeruginosa is an opportunistic pathogen that causes a wide range of acute and chronic infections (14, 27). Chronic infections of the lung epithelia in individuals with cystic fibrosis (CF) have been the subject of particular study (28). CF patients have a defect in the ciliated epithelial cells preventing clearance of the lungs that predisposes them to infection by a number of bacteria, in particular, P. aeruginosa (16). However the abnormal composition and mechanical properties of airway secretions do not in themselves explain the propensity for the CF airway to become colonized by P. aeruginosa, and the reasons that the lungs of these patients are prone to infection are not fully understood. It has been known for some time that in the CF lung the bacteria frequently have a mucoid phenotype, associated with the production of copious amounts of extracellular alginate (6, 38), but this is not in itself sufficient to explain the ability of the bacteria to chronically colonize the CF lung. CF patients show normal immune responses to standard immunizations, indicating an absence of an underlying systemic immunodeficiency that would explain the chronic pulmonary infection. However, despite an early and sustained acquired immune response to P. aeruginosa, the host is generally unable to clear P. aeruginosa from the airways (12). There are multiple factors that contribute to the ineffectiveness of the acquired immune response.
In vitro studies have identified a number of cell surface and secreted proteins that are necessary to cause infection (27, 42). These virulence factors interfere with the defense mechanisms of the patient and assist the bacteria in causing disease. Cell surface and secreted proteins are of particular importance, as they interact most directly with host tissues. Proteomic studies suggest that there are similarities, but also significant differences, between the profiles of secreted proteins from different strains grown in vitro (33, 44, 53). These differences are presumably a consequence of genetic differences between strains coupled with adaptation of the bacteria to different environments. A comparison of two genetically related isolates from patients with CF estimated a 5.5% difference in secreted proteins (2). In vitro, production of many virulence factors is regulated by the culture conditions and factors such as the cell population density (quorum sensing) and the concentration of iron (3, 47, 48, 50). Various approaches have been used to attempt to better understand virulence factor production in vivo in the CF lung. They include in vivo expression technology to identify genes that are upregulated in infection (18), microarray approaches to identify genes that are upregulated by using sputa from CF patients (35, 52), and bacteriophage display to identify proteins made during infection (5). The analysis of bacterial signaling molecules (quorum-sensing molecules) in sputum samples from patients indicates that in the CF lung the bacteria grow in biofilms (41), and there is also evidence that in the CF lung the bacteria may exist under conditions of low oxygen (hypoxia) (57).
CF patients mount a significant humoral response to P. aeruginosa antigens, and there is good evidence that serum antibodies directed against whole-cell P. aeruginosa lysates or specific P. aeruginosa antigens are the first markers of P. aeruginosa infection in young children with CF (7, 10, 54). Patients with chronic P. aeruginosa infection have high concentrations of antibodies directed against multiple P. aeruginosa protein antigens, including cell surface and secreted exoproducts, indicating that these are made during infection (1, 21). Immunoglobulin G (IgG) and its subclasses, which comprise 80% of the antibodies in serum and are important in the secondary response in CF patients, such as opsonization of bacterial antigens, are of particular significance. IgGs are produced in very high titer against specific P. aeruginosa antigens during a chronic infection and can be used as diagnostic indicators of the progression of the infection (7, 8, 36, 37).
Sera from patients infected with other bacterial species have been used to identify proteins that are produced during infection, using two-dimensional gel electrophoresis (2-DE) and Western blotting from cultures grown in the laboratory, and this has been termed “immunoproteomics” (17, 51). The simultaneous resolution of multiple immunogenic antigens within a sample is a significant advantage of Western blotting compared to the majority of other immunochemical methods (25). Secreted proteins are of particular interest because of their key roles in host-pathogen interactions and as immunogens (51). A comprehensive proteomic analysis of proteins that are secreted by P. aeruginosa in laboratory growth media has been carried out (32).
The aim of this research was to draw correlations between extracellular proteins made by P. aeruginosa in laboratory culture and those that are synthesized during infection in the CF lung, as shown through recognition by antibodies in the sera of CF patients. The growth phase and the effect of iron were examined as potential regulators of the extracellular proteome in laboratory culture.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains that were used were P. aeruginosa PAO1 (45), the isogenic mutant strain PAO1pvdS (34), and P. aeruginosa Pa4 (30). Bacteria were grown in LB broth (39) supplemented with the iron chelator ethylene-diamine-di(o-hydroxyphenylacetic-acid) (EDDHA) (Sigma; 200 μg/ml) as required. Cultures were grown at 37°C with orbital shaking (200 rpm).
Preparation of extracellular-protein samples.
Extracellular proteins for 2-DE were prepared from P. aeruginosa cultures grown in 250 ml LB broth to an optical density at 600 nm of 0.8 (logarithmic phase) or 3.5 (early stationary phase). Following centrifugation, the supernatants were collected, and centrifugation was repeated twice more. The supernatants were filtered through a 0.2-μm Millex filter (Millipore) at 4°C. Proteins were concentrated by trichloroacetic acid as described previously (20). The protein pellets were air dried and resuspended in rehydration buffer (see below) for 2-DE analysis or 40 mM Tris for Bradford protein assay (Bio-Rad).
2-DE and Western blotting.
Proteins were solubilized in rehydration buffer, which was comprised of 5 M urea, 2 M thiourea, 2% (wt/vol) 3-[(3-choloamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2% (wt/vol) [N-decyl-N,N-dimethyl-3-ammonio-1-propane sulfonate] sulfobetaine 3-10, 40 mM Tris-HCl, 2 mM tributyl phosphine, 1% (vol/vol) immobilized pH gradient (IPG) buffer, and 0.005% bromophenol blue (32). The protein sample was briefly mixed and centrifuged (60,000 × g; 10 min; 18°C) to remove any insoluble material prior to isoelectric focusing. Isoelectric focusing (first dimension) was performed with the Ettan IPGphor system and Immobiline DryStrip gels (GE Healthcare). Proteins (350 to 400 μg) were loaded by in-gel rehydration on 7-cm pH 3 to 10 or 4 to 7 linear IPG strips with a final volume of 125 μl. Following in-gel rehydration for 10 to 16 h at 20°C under mineral oil, the proteins were focused for 40 kV h at 20°C. Following isoelectric focusing, the separated proteins were reduced, alkylated, and detergent exchanged using a two-step protocol as described previously (3). The strips were blotted dry and stored at −80°C until the second-dimension electrophoresis (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) was carried out. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis, the first-dimension strips were embedded on top of 12.5% acrylamide gels, and electrophoresis was carried out at 170 V using the Mini-Protean II apparatus (Bio-Rad). Protein molecular weight markers (Bio-Rad) with or without dye labels were utilized as required. Proteins were visualized by staining them with Coomassie brilliant blue R-250 as described elsewhere (31).
For Western analysis, proteins were transferred to nitrocellulose (0.45 μm; Bio-Rad) by electroblotting at 100 V for 1 h with transfer buffer (25 mM Tris-HCl [pH 8.3], 192 mM glycine, 20% [wt/vol] methanol) using a Mini-Trans-Blot electrophoretic transfer cell (Bio-Rad). Sera were prepared from four patients attending the CF clinic at Dunedin Hospital who were infected with P. aeruginosa and from two control (non-CF) individuals with no history of Pseudomonas infection. Collection and use of sera for this research was approved by the Ethics Committee of the Otago Health Board. Immunodetection was carried out by an adaptation of an existing method (19). All incubation steps were carried out at ambient temperature on a rocking platform. Membranes were incubated in blocking buffer for at least 1 hour. The membranes were then incubated overnight with serum (1/1,000 dilution in blocking solution). The sera were decanted, and the membranes were rinsed for 20 min in Tris-buffered saline containing Tween 20 (0.05%) with three changes of buffer. Goat anti-human polyvalent IgA, IgG, and IgM conjugated to alkaline phosphatase (Sigma) were prepared in the same buffer (1/40,000 dilution) and incubated with the membranes for 2 h. The solution was decanted, and the membranes were rinsed three times in Tris-buffered saline for 20 min each time. The membranes were incubated with p-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indoyl-phosphate p-toluidine salt (BCIP) until color development had reached the desired level (2 to 3 min unless otherwise stated) and then rinsed in H2O to halt further color development.
Data analysis.
To align Coomassie-stained gels with Western blots, gel images were acquired with a GS-800 scanner (Bio-Rad) using an appropriate filter. Exported TIFF (tagged-image file format) files were printed onto transparency film. Estimates of the pI (isoelectric point) were made by measuring the length of the gel, and the protein spot position was expressed as a percentage of the gel length. The percentage was plotted using a graph of pH as a function of distance (GE Healthcare), and molecular weight was estimated from a standard curve generated from the protein markers on the 2-D gel. Printed transparencies of the Coomassie-stained proteins were overlaid onto the Western (nitrocellulose) membranes to allow direct comparison of proteins detected by the different methods. Proteins that were visible on all the Western blots were used as references for alignment. Protein detection and Western analysis were carried out at least twice for each sample, and the results, i.e., which proteins were detected, were reproducible.
Protein identification using peptide mass mapping.
Selected protein spots were excised from the 2-DE gels using a sterile scalpel and placed into microcentrifuge tubes. The protein spots were processed to yield tryptic peptides of a sufficient concentration for peptide mass mapping, as described previously (32). Peptide mass maps of tryptic peptides were generated by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry using an Applied Biosystems Voyager DE-STR. Peptides in 0.8 μl of matrix solution (α-cyano-4-hydroxycinnamic acid [Sigma, St. Louis, MO], 8 mg/ml in 70% [vol/vol] acetonitrile/1% [vol/vol] formic acid) were spotted directly onto a MALDI-TOF target plate. Spectra were obtained in reflectron mode using an accelerating voltage of 20 kV. Mass calibration was performed using trypsin autolysis peaks, 2,211.11 Da and 842.51 Da, as internal standards. The resulting data were used to perform searches of the SWISS-PROT and TrEMBL databases using the program MASCOT. The identification parameters included peptide mass accuracy within 0.08 Da and one possible missed tryptic cleavage per peptide, with the methionine sulfoxide and cysteine-acrylamide modifications checked. Identifications were based on the MASCOT score, observed pI and mass (kDa), number of matching peptide masses, and total percentage of the amino acid sequence that those peptides covered. Generally, a peptide match with at least 25% total sequence coverage was required for confidence in the identification, but very low- and high-mass proteins, and those resulting from protein fragmentation, might not always meet this criterion.
RESULTS
Detection of proteins from logarithmic-phase cultures by CF patient sera.
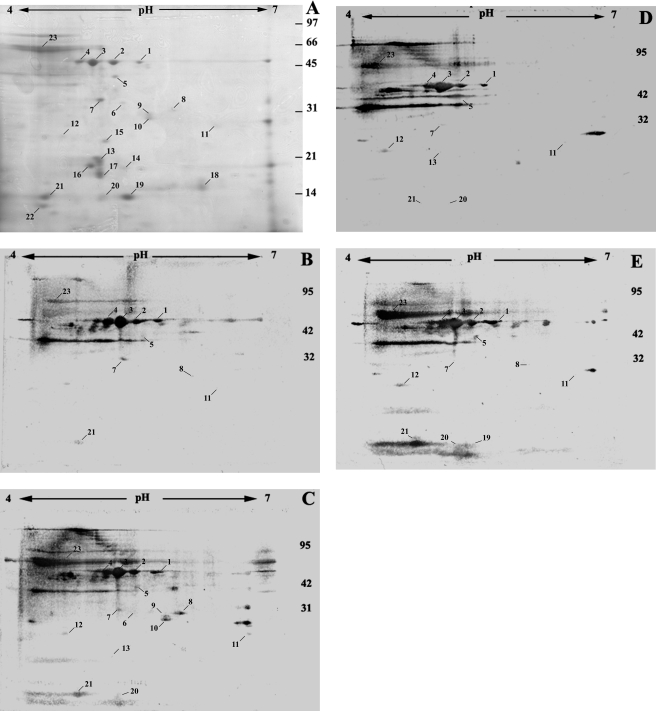
Extracellular proteins from P. aeruginosa strain PAO1 cells isolated during logarithmic-phase growth were separated by 2-DE across pH 3 to 10 to provide an overview of the extracellular proteome. At least 55 proteins were visualized using Coomassie R250 stain (data not shown). The majority (31) of those proteins had pIs between pH 4 and 7. To identify proteins that were also expressed in vivo, proteins from a replicate gel were probed for antibody interactions using serum from a CF patient (number 2) infected with P. aeruginosa. Using this method, the majority of proteins detected by the serum also had pIs between pH 4 and 7 (data not shown). Proteins were therefore separated across pH 4 to 7 in subsequent 2-DE analyses to improve the resolution of the subproteome through increased separation of proteins with similar pIs.
Secreted proteins were tested for antibody interactions using sera from four CF patients (Fig. 1) and two uninfected controls. CF patient sera detected different (between 10 and 16) proteins that could also be visualized with Coomassie stain (data not shown). Different titers of specific antibodies existed in each patient, as indicated by different signal intensities on the blots. Control sera weakly detected 6 of the 23 proteins, but only after prolonged development (data not shown), indicating that the proteins detected by patient sera were a consequence of infection by P. aeruginosa. Control incubations with the secondary antibody alone failed to yield any detected signal (data not shown).
FIG. 1.
Extracellular proteins from exponentially growing P. aeruginosa PAO1. Extracellular proteins from exponential-phase cells were separated on 2-DE gels (pH gradient for isoelectric focusing, pH 4 to 7). (A) Proteins detected by Coomassie staining. (B to E) Proteins detected by sera from CF patients (1 to 4) following Western blotting. The numbers represent the protein spots listed in Table 1 and the text. The approximate positions of molecular mass markers (kDa) are indicated.
Protein identities for nine spots that were detected strongly by at least one of the four CF patient sera were determined via MALDI-TOF mass spectrometry following in-gel tryptic digestion (Table 1). GroEL is a cytoplasmic protein, and antibodies to this protein in CF sera have been reported previously (46). PA0624 and PA0634 are also predicted to be cytoplasmic (26). BraC and SpeB are thought to be located in the periplasm (22, 26), and the FliC protein is part of the bacterial flagellum. The subcellular location of EftA is not certain, nor is that of PA0974, although it is predicted to have a type I signal sequence.
TABLE 1.
Protein identities determined for PAO1 proteinsa
| Spot no.b | PA no.c | Protein | Functiond | No. of matching peptides | Sequence coverage (%) | pIe | Mass (kDa)e | Immunoblotf
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||||||||
| Logarithmic-phase proteins | |||||||||||
| 1 | PA1092 | FliC | Flagellin type B | 19 | 43.6 | 5.4 | 49.2 | + | + | + | + |
| 2 | PA1092 | FliC | Flagellin type B | 18 | 43.6 | 5.4 | 49.2 | + | + | + | + |
| 5 | PA1074 | BraC | Amino acid binding protein BraC | 17 | 51.6 | 5.16 | 36.9 | + | + | + | + |
| 7 | PA2951 | EtfA | Electron transfer flavoprotein, α subunit | 9 | 45.3 | 4.98 | 31 | + | + | + | + |
| 8 | PA0974 | PA0974 | Unknown | 12 | 40.9 | 7.80 | 29 | + | + | − | + |
| 11 | PA0277 | SpeB | Agmatine ureohydrolase | 9 | 30.2 | 6.08 | 26.6 | + | + | + | + |
| 20 | PA0634 | PA0634 | Unknown | 10 | 50.4 | 5.06 | 12.4 | − | + | + | + |
| 21 | PA0624 | PA0624 | Unknown | 6 | 68 | 4.62 | 12 | + | + | + | + |
| 23 | PA4385 | GroEL | Chaperone GroEL | 22 | 41.5 | 5.04 | 57.1 | + | + | + | + |
| Stationary-phase proteins | |||||||||||
| 11 | PA0423 | PasP | Protease | 10 | 67.5 | 6.1 | 20.8 | + | + | + | + |
| 28 | PA2939 | PA2939 | Aminopeptidase | 6 | 9.9 | 5.1 | 57.5 | − | + | − | + |
| 57 | PA4922 | Azu | Azurin | 2 | 23.4 | 6.9 | 16.0 | + | − | − | + |
| 72 | PA1178 | OprH | N-terminal fragment (10 kDa) of porin OprH | 7 | 38.8 | 9.0 | 21.6 | + | + | + | + |
Proteins were separated by 2-DE, and their identities were determined by peptide mass mapping.
Spot numbers are shown in Fig. 1 (exponential-phase proteins) and Fig. 2 (stationary-phase proteins).
PA no. is the open reading frame number in the P. aeruginosa strain PAO1 genome (http://www.pseudomonas.com).
Protein functions were predicted through BLASTP and TBLASTN searches, with at least 50% protein sequence identity obtained in each case.
Values for pI and mass are theoretical values from deduced amino acid sequence of the identified gene and open reading frame, respectively.
The presence of proteins that are normally associated with the cytosol, periplasm, or outer membranes in the samples was most likely due to bacterial-cell lysis or membrane vesicle exocytosis during growth of the bacteria. Detection of these proteins by sera from the CF patients indicates that significant bacterial lysis occurs in the CF patient lung during chronic infection by P. aeruginosa.
Host interactions with proteins produced by P. aeruginosa during stationary phase.
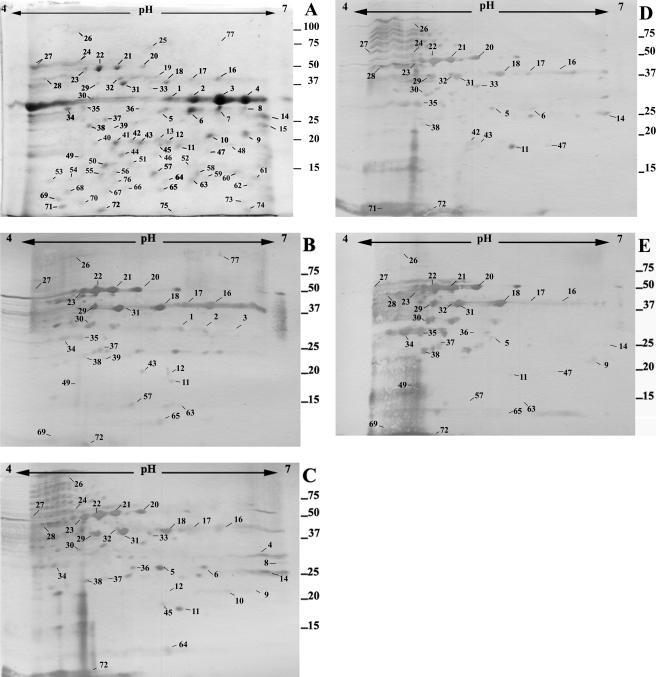
We next investigated antibody responses to proteins secreted by P. aeruginosa grown until an early stationary phase of growth. Extracellular proteins isolated from early stationary-phase cultures were separated by 2-DE and detected with Coomassie or analyzed using Western blotting (Fig. 2). Seventy-seven proteins were visualized, and patient antibodies detected 51 of them. Western blotting using pooled control sera weakly detected 22 proteins only after prolonged exposure (>15 min) (data not shown). The identities of four protein spots from stationary-phase cultures that were detected strongly by sera from at least two CF patients were determined (Table 1). These proteins were two secreted proteases, azurin, and a fragment of an outer membrane protein, OmpH1. Two other secreted proteins, LasB protease (spot numbers [SN] 1, 2, 3, and 4 in Fig. 2) and PrpL protease (SN 14 in Fig. 2) were presumptively identified using pI and molecular weight coordinates from a prior 2-DE map of the extracellular proteome (32).
FIG. 2.
Extracellular proteins from stationary-phase P. aeruginosa PAO1. Extracellular proteins from stationary-phase cells were separated on 2-DE gels (pH gradient for isoelectric focusing, pH 4 to 7). (A) Proteins detected by Coomassie staining. (B to E) Proteins detected by sera from CF patients (1 to 4) following Western blotting. The numbers represent the protein spots listed in Table 1 and the text. The approximate positions of molecular mass markers (kDa) are indicated.
Azurin is a copper-containing redox protein that is located in the periplasm and is las and rhl quorum-sensing system regulated (32, 44). Secretion of azurin occurs when P. aeruginosa cells are grown in a medium containing eukaryotic proteins, such as peptone in LB medium (58). The Zn-dependent aminopeptidase (PA2939) is a secreted enzyme (11) and is regulated by the las quorum-sensing system (32, 49). Microarray analysis had previously indicated that expression of this gene is induced during early stationary phase (40, 49). PasP is also a secreted protease (29), and its recognition by patient sera shows that it is made during infections in CF patients, as well as in corneal infections, where it was first implicated. The last identified protein was the N-terminal fragment of an outer membrane protein, OmpH1. This fragment may have been present due to cell lysis or may have been secreted in membrane vesicles that were released by P. aeruginosa (4); PA2939 has also been reported to be released in membrane vesicles (4).
The influence of iron-deficient conditions on the extracellular proteome.
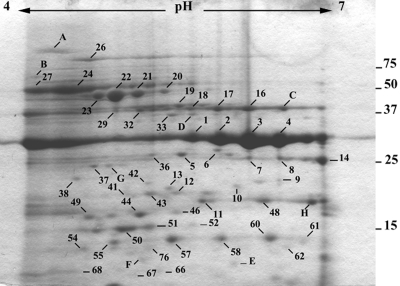
The growth medium (LB broth) used in this study had a free-iron (Fe3+) content of approximately 6.1 μM (13). In vivo, Fe3+ ions are bound by host proteins, and the concentration of free iron is approximately 10−18 M (9). The effect of a lowered iron concentration on the extracellular proteins produced by P. aeruginosa was examined by growing the bacteria in LB broth supplemented with the iron chelator EDDHA. The presence of EDDHA in LB broth had no apparent effect on the rate of growth of the bacteria (data not shown). Only proteins above 10 kDa were analyzed for comparing extracellular proteins produced in either an iron-limited or more iron-replete environment. Iron limitation resulted in a reduction in the number of extracellular proteins (Fig. 3), but the amounts of several proteins (for example, SN 14, 33, and 48 and protein spots corresponding to LasB) were apparently increased. Iron limitation also revealed eight proteins visualized with Coomassie (A to H in Fig. 3) that were not detected when EDDHA was omitted from the growth medium. Five of these (B, C, D, E, and F) were detected by one or more patient sera (data not shown).
FIG. 3.
Effects of EDDHA on extracellular proteins from stationary-phase P. aeruginosa PAO1. Extracellular proteins were prepared from bacteria grown in LB broth containing EDDHA and separated on 2-DE gels (pH gradient for isoelectric focusing, pH 4 to 7). Shown are proteins detected by Coomassie staining. The numbers represent the protein spots listed in the text. The letters represent proteins that were observed only following the addition of EDDHA to the growth medium. The approximate positions of molecular mass markers (kDa) are indicated.
Comparison of the extracellular proteomes of strains PAO1 and Pa4.
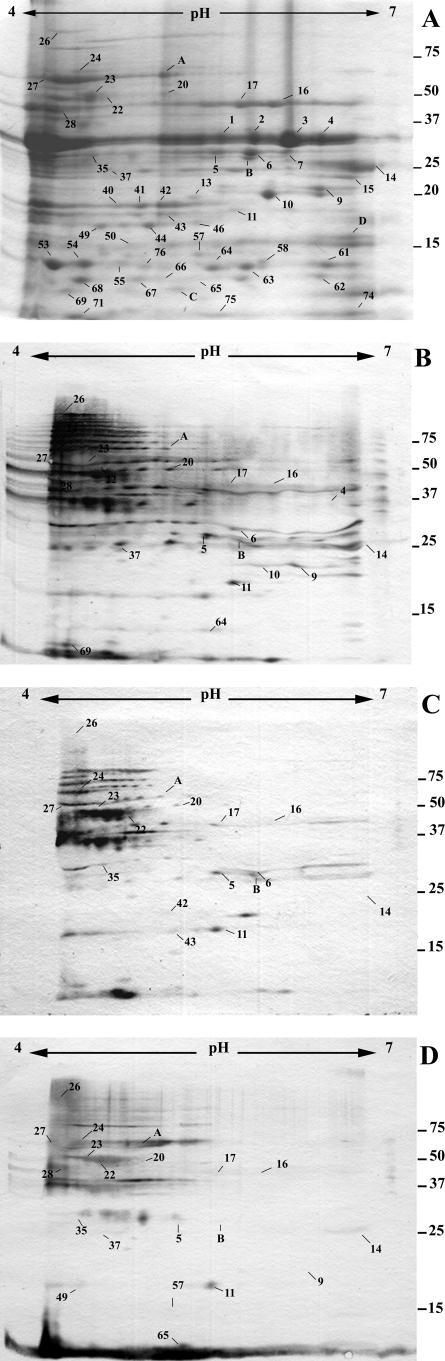
The cellular proteomes of different strains of P. aeruginosa are very similar, whereas the extracellular proteomes can vary greatly between strains (33, 53). In order to investigate the interaction of CF sera with proteins from a second strain of P. aeruginosa, we prepared extracellular proteins from Pa4, an isolate originally obtained from a urinary tract infection (30). This strain was chosen in order to focus on proteins that are common to multiple strains of P. aeruginosa and not specific to CF strains. The two P. aeruginosa strains had equivalent rates of growth in LB broth (data not shown). The Pa4 extracellular proteome (Fig. 4) comprised at least 51 proteins clearly visible following Coomassie staining. Comparison of the extracellular proteomes revealed significant conservation, with 47 spots having the same pI/molecular weight coordinates as proteins in the PAO1 extracellular proteome, and these are likely to be the same proteins. Furthermore, PA4 proteins detected in the Western analysis gave results in concordance with the proteins detected for PAO1 (Fig. 4). The antibody titer to proteins, as represented by intensity on immunoblots, varied considerably between the two strains (Fig. 2 and 4). Interestingly, some proteins not detected using Coomassie staining had strong signals on Western blots, indicating high antigenicity to proteins made in small amounts in laboratory culture. These results suggest that a subset of secreted proteins and of proteins resulting from cell lysis are common to strains in this study and to strains infecting the CF patients.
FIG. 4.
Extracellular proteins from P. aeruginosa strain Pa4. Extracellular proteins were prepared from bacteria grown in LB broth until stationary phase and separated on 2-DE gels (pH gradient for isoelectric focusing, pH 4 to 7). (A) Proteins detected by Coomassie staining. (B to D) Proteins detected by sera from CF patients (2, 3, and 4) following Western blotting. The numbers represent the protein spots listed in the text. The letters represent proteins produced by strain PA4 but not observed for PAO1. The approximate positions of molecular mass markers (kDa) are indicated. Western blots with patient 1 serum were excluded due to high background.
DISCUSSION
A major challenge in medical microbiology is to determine how the growth of bacteria in the laboratory is related to the growth of the same microbes during infection. The overall aim of this research was to draw correlations between proteins made by P. aeruginosa in vitro and those that are made in vivo in the CF lung. To do this, we took advantage of the known IgG antibody response of CF patients to P. aeruginosa infection (37, 54). Immunoblotting showed that all four sera used had significant titers of anti-Pseudomonas antibodies, as expected, although there were also striking differences between different patients (Fig. 1 to 4). The differences may be because of differences between the proteomes of the infecting strains and strains PAO1 and Pa4 and also because of differences in anti-Pseudomonas antibody titers between patients. Such differences have been noticed previously (24, 36, 37) and are correlated with the infectious load of Pseudomonas within the patient and the length of time that the patient has been infected.
Many of the extracellular proteins made by cells in stationary phase in LB broth were recognized by serum from at least one patient, indicating that they are also made in vivo; however, some proteins that were present in culture supernatants were not recognized by the sera. The most likely explanation for the second class of proteins is that the genes encoding them are not expressed in vivo, at least not in sufficient quantities to invoke a detectable antibody response. A second explanation is that the P. aeruginosa strains infecting the patients do not have the genes required to encode these proteins. However the genomes of different strains of P. aeruginosa are highly conserved, with 89 to 97% of the PAO1 open reading frames being detected in 38 other strains that were analyzed (15, 56). Consistent with this, almost all of the extracellular proteins made by strain Pa4 corresponded to PAO1 proteins with the same pI and molecular weight. There is also very high sequence identity between corresponding genes from different strains (43), so it is unlikely that an antibody against a protein from a P. aeruginosa strain in CF would fail to detect the corresponding protein from PAO1 because of sequence differences. This issue could be resolved by using a larger number of patient sera and by preparing extracellular proteins from the infecting strains grown in culture, rather than from PAO1. It should be noted that the differences in the extracellular proteomes of strains Pa4 and PAO1 are less marked than those between PAO1 and a cytotoxic strain, 6206 (33), and comparable to those observed for CF strain C (44). The extracellular proteome of a strain of P. aeruginosa that is widespread in patients with CF in Australia is also very similar to that of strain PAO1 (data not shown).
The extracellular proteome was strikingly affected by the growth phase of the bacteria, and others have also found that the number and concentration of secreted proteins increase as P. aeruginosa enters stationary phase (53). There were fewer proteins in the supernatants of cultures in the exponential growth phase, and most of those identified are unlikely to be secreted proteins; instead, they are more likely to be surface associated or to result from cell lysis or the formation of membrane vesicles. However, many of these proteins were recognized by patient sera, indicating that they are visible to the immune system during infection. The recognition of cytoplasmic and periplasmic proteins by patient sera suggests that there is significant bacterial-cell lysis or formation of membrane vesicles during infection in CF.
Patient sera interacted with significantly more proteins from stationary-phase than logarithmic-phase cells, consistent with the notion that in vivo the bacteria are in a physiological state that more closely resembles stationary phase. Sputa from patients with CF contain compounds secreted by P. aeruginosa for quorum sensing (41), implying that quorum sensing is a regulatory mechanism employed by the bacteria during chronic CF infection. Production of at least four of the proteins that were detected by patient sera (azurin and the three proteases elastase, PrpL, and PasP) is regulated by quorum sensing (3, 32), showing that not only quorum-sensing signals, but also the protein end products of quorum-sensing regulatory pathways, are active in vivo.
The addition of sputa from CF patients to growth medium results in the upregulation of genes involved in iron acquisition (35), and there is also evidence that P. aeruginosa in the CF lung is growing under iron-restricted conditions (23). Fewer extracellular proteins were produced by cells grown in the presence of the iron chelator EDDHA, but the amounts of several extracellular proteins were markedly increased (Fig. 3). Iron limitation also resulted in the production of at least eight proteins that were not otherwise detected under more iron-replete conditions. Five of these were detected by sera from at least one patient, consistent with the hypothesis that P. aeruginosa experiences conditions of iron starvation in the CF lung. These data demonstrate that the concentration of iron in the growth medium affects the extracellular proteome, including proteins that are detected by patient sera. Synthesis of at least two iron-regulated proteins, PrpL and exotoxin A, is under the direction of the iron-regulated alternative sigma factor PvdS (34, 55). Analysis of proteins secreted by PAO1 carrying a mutation in PvdS identified only two proteins, SN 43 and PrpL (SN 14), that were absent compared to wild-type PAO1, indicating that under the laboratory conditions used few proteins are PvdS regulated (data not shown). Both of these proteins were detected by two of the patient sera, consistent with earlier findings that the pvdS gene is transcribed in vivo (23).
In conclusion, our results show that immunoproteomics provides a powerful tool to investigate the physiology of P. aeruginosa in CF infections and to identify uncharacterized proteins that are made and secreted by the bacteria during infection. Further work will be required to determine the identities of the remaining proteins recognized by patient sera and to understand the differences in the immune responses of different patients.
Acknowledgments
We are very grateful to Christopher Hewitt and four anonymous CF patients for providing the sera used in this study.
Funds to cover costs associated with protein identification were provided by the University of Otago Functional Genomics, Gene Expression and Proteomics Research Theme.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 28 July 2008.
REFERENCES
- 1.Anderson, T. R., T. C. Montie, M. D. Murphy, and V. P. McCarthy. 1989. Pseudomonas aeruginosa flagellar antibodies in patients with cystic fibrosis. J. Clin. Microbiol. 272789-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arevalo-Ferro, C., J. Buschmann, G. Reil, A. Gorg, L. Wiehlmann, B. Tummler, L. Eberl, and K. Riedel. 2004. Proteome analysis of intraclonal diversity of two Pseudomonas aeruginosa TB clone isolates. Proteomics 41241-1246. [DOI] [PubMed] [Google Scholar]
- 3.Arevalo-Ferro, C., M. Hentzer, G. Reil, A. Gorg, S. Kjelleberg, M. Givskov, K. Riedel, and L. Eberl. 2003. Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 51350-1369. [DOI] [PubMed] [Google Scholar]
- 4.Bauman, S. J., and M. J. Kuehn. 2006. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect. 82400-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckmann, C., M. Brittnacher, R. Ernst, N. Mayer-Hamblett, S. I. Miller, and J. L. Burns. 2005. Use of phage display to identify potential Pseudomonas aeruginosa gene products relevant to early cystic fibrosis airway infections. Infect. Immun. 73444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bragonzi, A., D. Worlitzsch, G. B. Pier, P. Timpert, M. Ulrich, M. Hentzer, J. B. Andersen, M. Givskov, M. Conese, and G. Doring. 2005. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J. Infect. Dis. 192410-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brett, M. M., A. T. Ghoneim, and J. M. Littlewood. 1988. Prediction and diagnosis of early Pseudomonas aeruginosa infection in cystic fibrosis: a follow-up study. J. Clin. Microbiol. 261565-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brett, M. M., A. T. Ghoneim, and J. M. Littlewood. 1987. Serum IgG antibodies in patients with cystic fibrosis with early Pseudomonas aeruginosa infection. Arch. Dis. Child. 62357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullen, J. J., H. J. Rogers, and E. Griffiths. 1978. Role of iron in bacterial infection. Curr. Top. Microbiol. Immunol. 801-35. [DOI] [PubMed] [Google Scholar]
- 10.Burns, J. L., R. L. Gibson, S. McNamara, D. Yim, J. Emerson, M. Rosenfeld, P. Hiatt, K. McCoy, R. Castile, A. L. Smith, and B. W. Ramsey. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183444-452. [DOI] [PubMed] [Google Scholar]
- 11.Cahan, R., I. Axelrad, M. Safrin, D. E. Ohman, and E. Kessler. 2001. A secreted aminopeptidase of Pseudomonas aeruginosa. Identification, primary structure, and relationship to other aminopeptidases. J. Biol. Chem. 27643645-43652. [DOI] [PubMed] [Google Scholar]
- 12.Chmiel, J. F., and P. B. Davis. 2003. State of the art: why do the lungs of patients with cystic fibrosis become infected and why can't they clear the infection? Respir. Res. 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diggle, S. P., S. Matthijs, V. J. Wright, M. P. Fletcher, S. R. Chhabra, I. L. Lamont, X. Kong, R. C. Hider, P. Cornelis, M. Camara, and P. Williams. 2007. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 1487-96. [DOI] [PubMed] [Google Scholar]
- 14.Driscoll, J. A., S. L. Brody, and M. H. Kollef. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67351-368. [DOI] [PubMed] [Google Scholar]
- 15.Ernst, R. K., D. A. D'Argenio, J. K. Ichikawa, M. G. Bangera, S. Selgrade, J. L. Burns, P. Hiatt, K. McCoy, M. Brittnacher, A. Kas, D. H. Spencer, M. V. Olson, B. W. Ramsey, S. Lory, and S. I. Miller. 2003. Genome mosaicism is conserved but not unique in Pseudomonas aeruginosa isolates from the airways of young children with cystic fibrosis. Environ. Microbiol. 51341-1349. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, R. L., J. L. Burns, and B. W. Ramsey. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168918-951. [DOI] [PubMed] [Google Scholar]
- 17.Haas, G., G. Karaali, K. Ebermayer, W. G. Metzger, S. Lamer, U. Zimny-Arndt, S. Diescher, U. B. Goebel, K. Vogt, A. B. Roznowski, B. J. Wiedenmann, T. F. Meyer, T. Aebischer, and P. R. Jungblut. 2002. Immunoproteomics of Helicobacter pylori infection and relation to gastric disease. Proteomics 2313-324. [DOI] [PubMed] [Google Scholar]
- 18.Handfield, M., D. E. Lehoux, F. Sanschagrin, M. J. Mahan, D. E. Woods, and R. C. Levesque. 2000. In vivo-induced genes in Pseudomonas aeruginosa. Infect. Immun. 682359-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Hirose, I., K. Sano, I. Shioda, M. Kumano, K. Nakamura, and K. Yamane. 2000. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 14665-75. [DOI] [PubMed] [Google Scholar]
- 21.Hollsing, A. E., M. Granstrom, M. L. Vasil, B. Wretlind, and B. Strandvik. 1987. Prospective study of serum antibodies to Pseudomonas aeruginosa exoproteins in cystic fibrosis. J. Clin. Microbiol. 251868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoshino, T., K. Kose-Terai, and K. Sato. 1992. Solubilization and reconstitution of the Pseudomonas aeruginosa high affinity branched-chain amino acid transport system. J. Biol. Chem. 26721313-21318. [PubMed] [Google Scholar]
- 23.Hunt, T. A., W. T. Peng, I. Loubens, and D. G. Storey. 2002. The Pseudomonas aeruginosa alternative sigma factor PvdS controls exotoxin A expression and is expressed in lung infections associated with cystic fibrosis. Microbiology 1483183-3193. [DOI] [PubMed] [Google Scholar]
- 24.Kappler, M., A. Kraxner, D. Reinhardt, B. Ganster, M. Griese, and T. Lang. 2006. Diagnostic and prognostic value of serum antibodies against Pseudomonas aeruginosa in cystic fibrosis. Thorax 61684-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurien, B. T., and R. H. Scofield. 2003. Protein blotting: a review. J. Immunol. Methods 2741-15. [DOI] [PubMed] [Google Scholar]
- 26.Lewenza, S., J. L. Gardy, F. S. Brinkman, and R. E. Hancock. 2005. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 15321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 21051-1060. [DOI] [PubMed] [Google Scholar]
- 28.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marquart, M. E., A. R. Caballero, M. Chomnawang, B. A. Thibodeaux, S. S. Twining, and R. J. O'Callaghan. 2005. Identification of a novel secreted protease from Pseudomonas aeruginosa that causes corneal erosions. Investig. Ophthalmol. Vis. Sci. 463761-3768. [DOI] [PubMed] [Google Scholar]
- 30.Meyer, J.-M., A. Stintzi, D. D. Vos, P. Cornelis, R. Tappe, K. Taraz, and H. Budzikiewicz. 1997. Use of siderophores to type pseudomonads: the three Pseudomonas aeruginosa pyoverdine systems. Microbiology 14335-43. [DOI] [PubMed] [Google Scholar]
- 31.Neuhoff, V., N. Arold, D. Taube, and W. Ehrhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9255-262. [DOI] [PubMed] [Google Scholar]
- 32.Nouwens, A. S., S. A. Beatson, C. B. Whitchurch, B. J. Walsh, H. P. Schweizer, J. S. Mattick, and S. J. Cordwell. 2003. Proteome analysis of extracellular proteins regulated by the las and rhl quorum sensing systems in Pseudomonas aeruginosa PAO1. Microbiology 1491311-1322. [DOI] [PubMed] [Google Scholar]
- 33.Nouwens, A. S., M. D. Willcox, B. J. Walsh, and S. J. Cordwell. 2002. Proteomic comparison of membrane and extracellular proteins from invasive (PAO1) and cytotoxic (6206) strains of Pseudomonas aeruginosa. Proteomics 21325-1346. [DOI] [PubMed] [Google Scholar]
- 34.Ochsner, U. A., Z. Johnson, I. L. Lamont, H. E. Cunliffe, and M. L. Vasil. 1996. Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol. Microbiol. 211019-1028. [DOI] [PubMed] [Google Scholar]
- 35.Palmer, K. L., L. M. Mashburn, P. K. Singh, and M. Whiteley. 2005. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 1875267-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pressler, T., B. Frederiksen, M. Skov, P. Garred, C. Koch, and N. Hoiby. 2006. Early rise of anti-pseudomonas antibodies and a mucoid phenotype of Pseudomonas aeruginosa are risk factors for development of chronic lung infection—a case control study. J. Cyst. Fibros. 59-15. [DOI] [PubMed] [Google Scholar]
- 37.Pressler, T., S. S. Pedersen, F. Espersen, N. Hoiby, and C. Koch. 1990. IgG subclass antibodies to Pseudomonas aeruginosa in sera from patients with chronic P. aeruginosa infection investigated by ELISA. Clin. Exp. Immunol. 81428-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramsey, D. M., and D. J. Wozniak. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56309-322. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., D. W. Russell, and N. Irwin. 2000. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 40.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 1852066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407762-764. [DOI] [PubMed] [Google Scholar]
- 42.Smith, R. S., and B. H. Iglewski. 2003. Pseudomonas aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 656-60. [DOI] [PubMed] [Google Scholar]
- 43.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 1851316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sriramulu, D. D., M. Nimtz, and U. Romling. 2005. Proteome analysis reveals adaptation of Pseudomonas aeruginosa to the cystic fibrosis lung environment. Proteomics 53712-3721. [DOI] [PubMed] [Google Scholar]
- 45.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406959-964. [DOI] [PubMed] [Google Scholar]
- 46.Ulanova, M., T. D. Petersen, O. Ciofu, P. Jensen, M. Hahn-Zoric, L. A. Hanson, and N. Hoiby. 1997. The clonal antibody response to Pseudomonas aeruginosa heat shock protein is highly diverse in cystic fibrosis patients. APMIS 105449-456. [PubMed] [Google Scholar]
- 47.Vasil, M. L. 2007. How we learnt about iron acquisition in Pseudomonas aeruginosa: a series of very fortunate events. Biometals 20587-601. [DOI] [PubMed] [Google Scholar]
- 48.Vasil, M. L., and U. A. Ochsner. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34399-413. [DOI] [PubMed] [Google Scholar]
- 49.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 1852080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, V. E., J. G. Frelinger, R. K. Barth, and B. H. Iglewski. 2006. Quorum sensing: dynamic response of Pseudomonas aeruginosa to external signals. Trends Microbiol. 1455-58. [DOI] [PubMed] [Google Scholar]
- 51.Walduck, A., T. Rudel, and T. F. Meyer. 2004. Proteomic and gene profiling approaches to study host responses to bacterial infection. Curr. Opin. Microbiol. 733-38. [DOI] [PubMed] [Google Scholar]
- 52.Wang, J., S. Lory, R. Ramphal, and S. Jin. 1996. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol. Microbiol. 221005-1012. [DOI] [PubMed] [Google Scholar]
- 53.Wehmhoner, D., S. Haussler, B. Tummler, L. Jansch, F. Bredenbruch, J. Wehland, and I. Steinmetz. 2003. Inter- and intraclonal diversity of the Pseudomonas aeruginosa proteome manifests within the secretome. J. Bacteriol. 1855807-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.West, S. E., L. Zeng, B. L. Lee, M. R. Kosorok, A. Laxova, M. J. Rock, M. J. Splaingard, and P. M. Farrell. 2002. Respiratory infections with Pseudomonas aeruginosa in children with cystic fibrosis: early detection by serology and assessment of risk factors. JAMA 2872958-2967. [DOI] [PubMed] [Google Scholar]
- 55.Wilderman, P. J., A. I. Vasil, Z. Johnson, M. J. Wilson, H. E. Cunliffe, I. L. Lamont, and M. I. Vasil. 2001. Characterization of an endoprotease (PrpL) encoded by a PvdS-regulated gene Pseudomonas aeruginosa. Infect. Immun. 695385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1008484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaborina, O., N. Dhiman, M. Ling Chen, J. Kostal, I. A. Holder, and A. M. Chakrabarty. 2000. Secreted products of a nonmucoid Pseudomonas aeruginosa strain induce two modes of macrophage killing: external-ATP-dependent, P2Z-receptor-mediated necrosis and ATP-independent, caspase-mediated apoptosis. Microbiology 1462521-2530. [DOI] [PubMed] [Google Scholar]