Abstract
Summary: Mycothiol (MSH; AcCys-GlcN-Ins) is the major thiol found in Actinobacteria and has many of the functions of glutathione, which is the dominant thiol in other bacteria and eukaryotes but is absent in Actinobacteria. MSH functions as a protected reserve of cysteine and in the detoxification of alkylating agents, reactive oxygen and nitrogen species, and antibiotics. MSH also acts as a thiol buffer which is important in maintaining the highly reducing environment within the cell and protecting against disulfide stress. The pathway of MSH biosynthesis involves production of GlcNAc-Ins-P by MSH glycosyltransferase (MshA), dephosphorylation by the MSH phosphatase MshA2 (not yet identified), deacetylation by MshB to produce GlcN-Ins, linkage to Cys by the MSH ligase MshC, and acetylation by MSH synthase (MshD), yielding MSH. Studies of MSH mutants have shown that the MSH glycosyltransferase MshA and the MSH ligase MshC are required for MSH production, whereas mutants in the MSH deacetylase MshB and the acetyltransferase (MSH synthase) MshD produce some MSH and/or a closely related thiol. Current evidence indicates that MSH biosynthesis is controlled by transcriptional regulation mediated by σB and σR in Streptomyces coelicolor. Identified enzymes of MSH metabolism include mycothione reductase (disulfide reductase; Mtr), the S-nitrosomycothiol reductase MscR, the MSH S-conjugate amidase Mca, and an MSH-dependent maleylpyruvate isomerase. Mca cleaves MSH S-conjugates to generate mercapturic acids (AcCySR), excreted from the cell, and GlcN-Ins, used for resynthesis of MSH. The phenotypes of MSH-deficient mutants indicate the occurrence of one or more MSH-dependent S-transferases, peroxidases, and mycoredoxins, which are important targets for future studies. Current evidence suggests that several MSH biosynthetic and metabolic enzymes are potential targets for drugs against tuberculosis. The functions of MSH in antibiotic-producing streptomycetes and in bioremediation are areas for future study.
INTRODUCTION
The class Actinobacteria is a very large and diverse group of gram-positive, high-G+C bacteria whose members produce a variety of morphologies, from small cocci to highly branched mycelia (145). Members of the Actinobacteria are encountered in a wide range of ecosystems, from soil and seawater to the skin, lungs, and gastrointestinal tract of humans, and are responsible for the production of commercial products, including amino acids, vitamins, and antibiotics (52, 155), as well as for causing important human diseases, such as leprosy, tuberculosis, and diphtheria. In addition, members of the Actinobacteria are used in the biodegradation of organic compounds during bioremediation (62). One feature which is common to most of the Actinobacteria is the production of mycothiol (MSH; also designated AcCys-GlcN-Ins), a small thiol that is often present in millimolar amounts and has analogous functions to glutathione (GSH), which is not found in Actinobacteria (84).
In this review, we discuss what is currently known about the biochemistry, biosynthesis, catabolism, and function of MSH. The majority of the early work on MSH was carried out with mycobacteria, and this has laid the basis for more recent studies of other Actinobacteria. Several recent reviews which cover aspects of MSH biochemistry have appeared. An excellent review by Bhave et al. (6) considers MSH metabolism in the larger general context of sulfur metabolism. Detailed coverage of MSH-dependent enzymes with bioinformatic analysis and comparison to analogous GSH-dependent enzymes is found in a review by Rawat and Av-Gay (114). Hand and Honek (41) consider MSH in the context of the spectrum of thiols found in microorganisms. In this review, we have sought to minimize coverage of those aspects that are well treated in these recent reviews and to emphasize relevant chemical details where these have been elucidated, but substantial overlap of coverage in some areas could not be avoided.
STRUCTURE, PREPARATION, PROPERTIES, AND DISTRIBUTION OF MSH
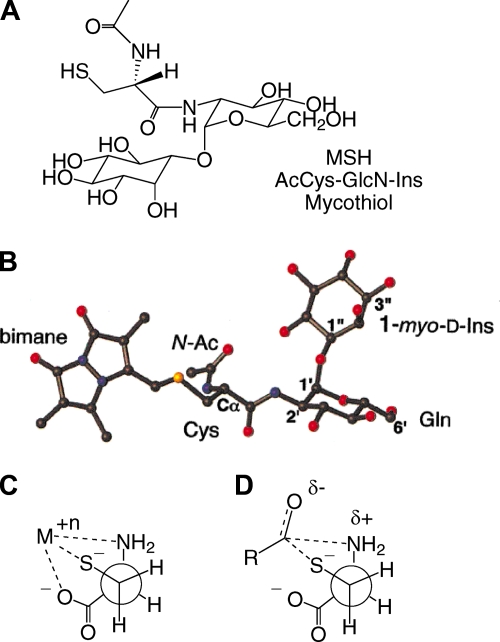
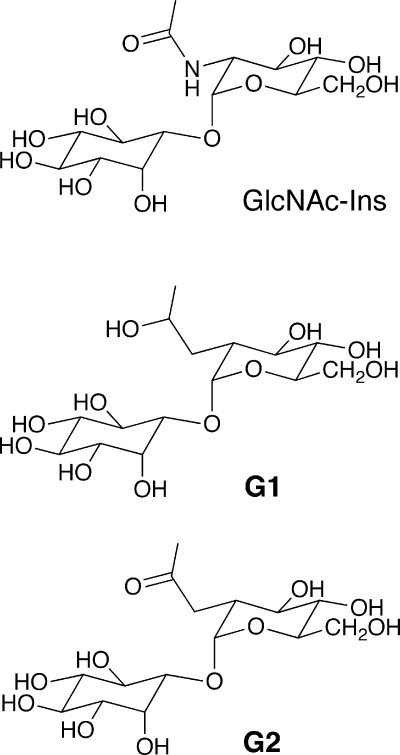
MSH, or 1-O-[2-[[(2R)-2-(acetylamino)-3-mercapto-1-oxopropyl]amino]-2-deoxy-α-d-glucopyranosyl]-d-myo-inositol, was initially isolated and its structure elucidated from Streptomyces strains (87, 123) and from Mycobacterium bovis (133). The common name mycothiol was proposed by Spies and Steenkamp (133). The structure (Fig. 1A) involves a cysteine residue in which the amino group is acetylated and the carboxyl group is amide linked to d-glucosamine, which is in turn α(1-1) linked to myo-inositol. The amide bond linking AcCys to GlcN-Ins in MSH is sufficiently unique that it is cleaved only by specialized enzymes, such as the MSH S-conjugate amidase (Mca) (see below).
FIG. 1.
(A) Structure of MSH. (B) Conformation of mycothiol-S-bimane determined by NMR. (Reprinted in part with permission from reference 69. Copyright 2003 American Chemical Society.) (C) Cysteine complexed to heavy metal. (D) Transition state for S-N-acyl migration of S-acylcysteine thioesters.
Bewley and coworkers (69) determined the conformation of mycothiol-S-bimane (MSmB) in aqueous solution by nuclear magnetic resonance (NMR), producing the mean structure shown in Fig. 1B. The conformation of the GlcN-Ins moiety was found to be essentially the same as that determined for GlcNAc-Ins, which is not surprising given the rigidity of the GlcN and Ins rings. Theoretical calculations of the favored conformation for MSH in an implicit water environment employed several different methods and generated several low-energy clusters (40). The global minimum cluster most closely matched the results determined by NMR on MSmB, and it was suggested that multiple conformations are in rapid equilibrium to produce the mean result determined by NMR.
A limiting factor in the study of MSH biochemistry has been the availability of MSH and its precursors. Once the structure was established, its chemical synthesis was undertaken, but generating GlcN-Ins with the correct stereochemistry is a challenging task. Jardine et al. (50) reported the production of a mixture of GlcN-Ins isomers and use of a cell-free enzyme preparation from Mycobacterium smegmatis to convert the correct stereoisomer into MSH. An acetylated derivative of the correct GlcN-Ins stereoisomer was synthesized by the Bewley group (99), coupled to AcCySmB, and deacetylated to generate the bimane derivative of MSH, MSmB. Lee and Rosazza (65) produced the acetylated GlcN-Ins by a different route and coupled it to N,S-diacetyl-l-cysteine to produce peracetylated MSH. Basic hydrolysis generated a mixture of MSH and MSH disulfide (MSSM), with an overall yield of ∼0.4%. A high yield of GlcN-Ins was obtained in a recently reported synthesis of this substrate for use in a kinetic study of MshC (28). The complexity and poor yields involved in chemical synthesis have thus far made isolation of MSH a more efficient means of its generation.
In our laboratory, we isolate MSH from M. smegmatis by using thiol affinity chromatography on activated thiopropyl agarose, followed by elution with dithiothreitol and preparative high-performance liquid chromatography (HPLC) of the eluent (142). Steenkamp and Vogt (135) described an alternative approach to isolation of MSH involving reaction of the thiol with 2-S-(2′-thiopyridyl)-6-hydroxynaphthyldisulfide to generate 2-S-(mycothiolyl)-6-hydroxynaphthyldisulfide, followed by solid-phase extraction on a C18 cartridge and preparative HPLC purification of the disulfide in the concentrated eluent. The disulfide was reduced with dithiothreitol, and MSH was purified by preparative HPLC. GlcN-Ins can be generated from the bimane derivative MSmB or other S-conjugates of MSH (MSR) by treatment with the S-conjugate amidase Mca (see below) and purification by solid-phase extraction (86).
A key property of MSH is its resistance to autoxidation. Cysteine undergoes heavy metal-catalyzed autoxidation much more rapidly than GSH does (45, 138, 140), a property attributable to the ability of cysteine to provide three metal ligands in a favorable geometric arrangement (Fig. 1C). The affinities of the substituents for heavy metals decrease in the order -S− > -NH2 > -COO−. Since autoxidation produces hydrogen peroxide, which is lethal to cells (45), cysteine is a liability to aerobic cells and is generally maintained at a low concentration. Cysteine derivatives found at high levels in cells have the amino and carboxyl groups blocked in order to slow autoxidation. In GSH, the γ-glutamyl and glycine residues reduce the Cu-catalyzed autoxidation rate 8- to 26-fold (45, 138, 140). In MSH, the acetyl and GlcN-Ins residues make Cu-catalyzed autoxidation of MSH some 30-fold slower than that of cysteine and 7-fold slower than that of GSH (87). Cys-GlcN-Ins autoxidizes ∼11-fold faster than MSH, which shows that blocking the amino group of Cys is of major importance (94). Why MSH is more resistant than GSH to autoxidation is not apparent but may be associated with the absence of any metal-chelating residues in the molecule other than the thiol group. The metal ion autoxidation rate is influenced by the thiol pKa and possibly by the redox potential, but these have not been reported for MSH.
Another problem for Cys in the cell comes from the presence of coenzyme A (CoA) thioesters. S-Acyl transfer reactions between thiols and thioesters are facile reactions (36, 66), and with the presence of acetyl-CoA and other acyl-CoAs at significant levels, the equilibrium generation of cysteine thioester species is highly likely. The difficulty is that cysteine thioester species are able to undergo a facile intramolecular acyl transfer reaction from sulfur to nitrogen via a favorable cyclic transition state (Fig. 1D) to generate the more stable amides, RCONH-Cys. With the amino group blocked, as in GSH or MSH, such rearrangements do not occur, and the thioester can revert to the acyl-CoA if the concentration of CoA is favorable. This aspect of thiol biochemistry has received little attention, in part because of the difficulty in measuring intracellular levels of specific acyl thioesters. The Cys acyl transfer problem and the Cys autoxidation problem are key reasons that cellular levels of Cys are kept low.
MSH has been found to occur only in Actinobacteria, as summarized in Table 1 (64, 84, 89). However, not all Actinobacteria produce MSH. One Arthrobacter species and one each of Actinomyces and Agromyces species gave negative analyses from determinations for samples obtained at a single growth time. However, MSH levels can vary markedly with the growth phase, and it would be imprudent to conclude that MSH is not produced by a species from analyses at a single growth time. Thus, for the marine streptomycete CNQ703, the values varied 40-fold over the full range of growth and secondary metabolite production (Table 1). MSH has not been found in representative samples of low-GC gram-positive bacteria, gram-negative bacteria, plants, fungi, or animals (84). Thus, we conclude that MSH biosynthesis is restricted to the Actinobacteria.
TABLE 1.
MSH content of Actinobacteriaa
| Strain(s) | MSH contentb |
|---|---|
| Actinomadura hibisca ATCC 53557 | L |
| Actinomyces israelii ATCC 10049 | N |
| Agromyces ramosus ATCC 25173 | N |
| Arthrobacter globiformis ATCC 8010 | N |
| Corynebacterium diphtheriae UCSD 21 | L |
| Kocurea rosea ATCC 144 | M |
| Marine Streptomycetaceaec | |
| CNQ525, CNQ687, and CNQ695 | L |
| CNR252, CNQ698, CNQ719, and CNQ766 | M |
| CNR530 and CNQ857 | H |
| CNQ703 | L-M |
| Marine Thermomonosporaceaec | |
| CNR363 | L |
| CNR431 | H |
| Micrococcus agilis ATCC 966 | L |
| Micrococcus luteus UCSD 22 | M |
| Micrococcus luteus ATCC 4698 | M |
| Micrococcus kristinae ATCC 27570 | L |
| Micromonospora carbonacea ATCC 27144 | L |
| Micromonospora floridensis JCM 3265 | H |
| Micromonospora fulvopurpurea JCM 5696 | M |
| Mycobacterium avium NJH9151 and NJH1854-4 | M |
| Mycobacterium chelonae UCSD 102 | M |
| Mycobacterium fortuitum UCSD 101 | M |
| Mycobacterium smegmatis mc2155 and mc26 | H |
| Mycobacterium tuberculosis ATCC 25618 | H |
| Mycobacterium tuberculosis PZAR UCSD 100 | M |
| Nocardia sp. strain NRRL 5646d | M |
| Nocardia asteroides UCSD 1 and UCSD 3 | M |
| Nocardiopsis mutabilis ATCC 31520 | L |
| Salinispora arenicola CNR107, CNR003, CNR643, | |
| CNH725, and CNQ976c | M |
| Streptomyces clavuligerus ATCC 270674 | M |
| Streptomyces coelicolor A3(2) | M |
| Streptomyces jumonjinensis ATCC 29864 | M |
| Streptomyces lactamdurans ATCC 27382 | M |
| Streptomyces lividans 1326 | M |
MSH BIOSYNTHESIS
The Biosynthetic Pathway
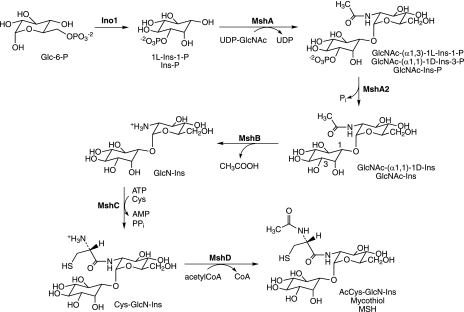
MSH biosynthesis was first elaborated in mycobacteria and is accomplished in five steps, starting from 1l-myo-inositol-1-phosphate (1l-Ins-1-P or Ins-P), which is produced from glucose-6-phosphate (Glc-6-P) by inositol phosphate synthase (Ino1) (Fig. 2). The first dedicated intermediate in MSH biosynthesis is 1-O-(2-acetamido-2-deoxy-α-d-glucopyranosyl)-d-myo-inositol-3-phosphate (GlcNAc-Ins-P), produced from UDP-GlcNAc and Ins-P by the glycosyltransferase MshA (91, 92). The second step involves the dephosphorylation of GlcNAc-Ins-P, catalyzed by MshA2, which is encoded by a gene that is not yet identified. The third step is the deacetylation of 1-O-(2-acetamido-2-deoxy-α-d-glucopyranosyl)-d-myo-inositol (GlcNAc-Ins) by the metalloprotein MshB (85) to yield 1-O-(2-amino-2-deoxy-α-d-glucopyranosyl)-d-myo-inositol (GlcN-Ins), the major intermediate found in extracts of mycobacteria (14, 93, 118). The substrate and products involved in the fourth and fifth steps of MSH biosynthesis were first identified by Steenkamp and coworkers (9) and confirmed by Anderberg et al. (1). The fourth step in the pathway is the ATP-dependent ligation of cysteine with GlcN-Ins by MshC, a homolog of Cys-tRNA synthetase, to produce 1-O-[2-[[(2R)-2-amino-3-mercapto-1-oxopropyl]amino]-2-deoxy-α-d-glucopyranosyl]-d-myo-inositol (Cys-GlcN-Ins) (125). The final step of MSH biosynthesis is acetylation of the amino group of cysteine in Cys-GlcN-Ins by acetyl-CoA under catalysis by a GCN5 acetyltransferase, MshD, yielding MSH (59).
FIG. 2.
Biosynthesis of MSH. myo-Inositol-1-phosphate synthase (Ino1) generates Ins-P, MSH glycosyltransferase (MshA) links Ins-P to GlcNAc, MSH phosphatase (MshA2) generates GlcNAc-Ins, MSH deacetylase (MshB) produces GlcN-Ins, MSH ligase (MshC) links Cys with GlcN-Ins, and MSH synthase (MSH acetyltransferase; MshD) acetylates Cys-GlcN-Ins to produce MSH. Note that myo-inositol has a plane of symmetry through C-2 and C-5, making C-1 and C-3 equivalent so GlcN-Ins-P can be named as a derivative of 1l-Ins-1-P or 1d-Ins-3-P, and that both systems are used.
The MSH biosynthesis genes for Mycobacterium tuberculosis, Mycobacterium smegmatis, Streptomyces coelicolor, Corynebacterium glutamicum, and Rhodococcus jostii RHA-1 are listed in Table 2. All gene assignments for these organisms have been confirmed experimentally, with the exception of those for R. jostii RHA-1. In each organism, the genes are not arranged in operons and are found throughout the respective genome.
TABLE 2.
MSH biosynthesis genes from selected Actinobacteria with sequenced genomes
| Gene | Gene name in:
|
||||
|---|---|---|---|---|---|
| M. tuberculosis | M. smegmatisa | S. coelicolorb | C. glutamicumc | R. jostiid | |
| mshA | Rv0486 | MSMEG0933 | SCO4204 | NCgl0389 | ro02073 |
| mshB | Rv1170 | MSMEG5129 | SCO5126 | NCgl1055 | ro05935 |
| mshC | Rv2130c | MSMEG4189 | SCO1663 | NCgl1457 | ro00877 |
| mshD | Rv0819 | MSMEG5854 | SCO4151 | NCgl2487 | ro04861 |
Mycobacterium smegmatis mc2155 MSH biosynthesis predicted proteins are 56% to 76% identical to the M. tuberculosis H37Rv orthologs.
Streptomyces coelicolor A3(2) MSH biosynthesis predicted proteins are 42 to 55% identical to the M. tuberculosis orthologs.
Corynebacterium glutamicum (ATCC 13032) MSH biosynthesis predicted proteins are 35 to 56% identical to the M. tuberculosis orthologs.
Rhodococcus jostii RHA-1 MSH biosynthesis predicted proteins are 47 to 66% identical to the M. tuberculosis orthologs.
MshA, the MSH Glycosyltransferase, and MshA2, the MSH Phosphatase
MSH biosynthesis begins with the formation of the pseudodisaccharide phosphate GlcNAc-Ins-P by MshA, a glycosyltransferase identified by Tn5 mutagenesis of M. smegmatis (91). MshA orthologs are found in the complete genomes of many Actinobacteria, and representative examples are given in Table 2. MshA is the only identified member of the GT-B superfamily (CAZy [http://www.cazy.org]) in M. tuberculosis that is not directly involved with cell wall biosynthesis (5). It was initially found that M. smegmatis mshA mutants produce no GlcN-Ins or GlcNAc-Ins (91, 96), but the actual substrates and biochemistry of MshA were only recently elucidated (92). This was done by showing that extracts of M. smegmatis produced GlcN-Ins from 1d,l-Ins-1-P and UDP-GlcNAc but not from myo-inositol or 1-d-Ins-1-P and UDP-GlcNAc, indicating that 1l-Ins-1-P was the inositol donor for MshA. This was verified with authentic 1l-Ins-1-P produced by Archaeoglobus fulgidus inositol phosphate synthase (18). With 1l-Ins-1-P as the substrate of MshA, the product is predicted to be GlcNAc-Ins-P (Fig. 2), and this was verified by HPLC-mass spectrometry analysis (92).
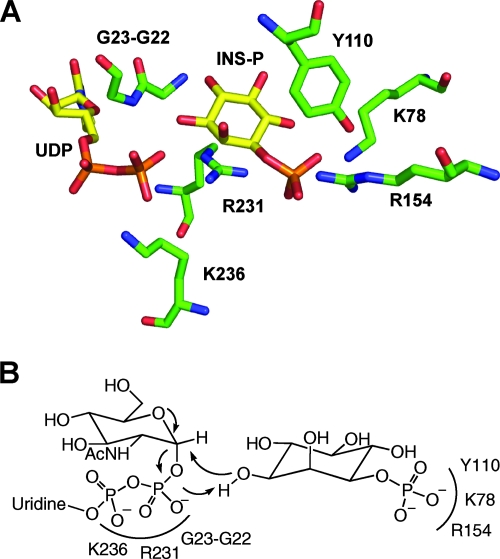
The MshA protein proved difficult to clone and express in active form as a His6-tagged protein from M. tuberculosis, Streptomyces coelicolor, and Nocardia farcinica but was successfully produced by the Blanchard group from Corynebacterium glutamicum (146). The enzyme was found to exhibit Michaelis-Menten kinetics with both 1l-Ins-1-P and UDP-GlcNAc, producing Km values of 240 ± 10 and 210 ± 20 μM, respectively, and a kcat value of 12.5 ± 0.2 s−1. The rate expression with variation of both substrates corresponded to that expected for a sequential mechanism. Crystal structures were obtained for the apo-enzyme, the binary complex with UDP, and the ternary complex with UDP and 1l-Ins-1-P (146). The results demonstrate that a substantial conformational change occurs upon UDP binding, which generates the binding site for 1l-Ins-1-P. Numerous residues involved in the binding of UDP and 1l-Ins-1-P could be identified, and a few of these are shown in Fig. 3. The phosphate residue of inositol is stabilized by electrostatic interactions with K78 and R154 and by multiple hydrogen bonds, likely explaining why the enzyme is inactive with myo-inositol (92). UDP is oriented and stabilized by interaction with R231 and K236 as well as with other residues. The highly conserved G22-G23 sequence is also part of the active site, with G23 hydrogen bonding to the β-phosphate of UDP. This explains why a G32D mutant of M. smegmatis MshA (G32 is equivalent to G22 of C. glutamicum) is totally inactive (91), since replacement of the hydrogen side chain of glycine with the carboxymethyl side chain of aspartic acid would be expected to greatly disrupt the active site (Fig. 3).
FIG. 3.
(A) PyMOL structure of selected active-site residues of MshA from Corynebacterium glutamicum in complex with UDP and Ins-P (146). (B) Summary of proposed catalytic mechanism for production of GlcNAc-Ins-P by MshA (146).
The crystal structure data provided the basis for a molecular model of the ternary complex of MshA with UDP-GlcNAc and 1l-Ins-1-P and for a proposed mechanism of catalysis (146). The slow step of the reaction is considered to involve a nucleophilic internal substitution in which the UDP-GlcNAc bond ionizes to generate a C-1 cation stabilized by the neighboring oxygen lone pair (Fig. 3B). Simultaneously, the β-phosphate residue functions as a general base, with the hydroxyl group of inositol allowing the resulting oxygen anion to attack the cationic site, generating a product with a retained configuration.
Dephosphorylation of GlcNAc-Ins-P generates GlcNAc-Ins, the substrate of MshB. This phosphatase, termed MshA2, is currently unidentified. There are four inositol monophosphatase (IMP) homologs to human IMP in the M. tuberculosis genome, including Rv2701c (suhB), Rv3137, Rv1604 (impA), and Rv2131c (cysQ,) which are 28, 25, 23, and 19% identical, respectively, to the human enzyme. M. tuberculosis SuhB has been cloned and expressed and was found to have activity with 1d-Ins-P, Ins-2-P, glucitol-6-P, and 2′-AMP, similar to the human, plant, and Escherichia coli IMP substrate specificity (102), but SuhB has not been assayed for MshA2 activity. Since no phosphatase mutants were identified in the screening of chemical (96, 118) or transposon (59, 91, 118) M. smegmatis mutant libraries for MSH-deficient mutants, it seems likely that more than one phosphatase in mycobacteria has MshA2 activity. Even alkaline phosphatase has been shown to dephosphorylate GlcNAc-Ins-P (146).
Inositol is utilized in two known pathways in mycobacteria, namely, the production of phosphatidylinositol for phosphatidylinositol mannosides and the biosynthesis of MSH. In M. tuberculosis, inositol is supplied as 1l-Ins-1-P from glucose-6-P (Fig. 2) by inositol phosphate synthase (Ino1) (81). The essential ino-1 gene was knocked out in M. tuberculosis, and mutants were inositol auxotrophs, had normal levels of phosphatidylinositol mannosides, lipomannan, and lipoarabinomannan but low levels of MSH (Y. Av-Gay, personal communication), and were attenuated for growth in SCID mice (81). The finding of any MSH in this mutant is surprising. The immediate product of the reaction of ino-1 is 1l-Ins-1-P, and the knockout mutant requires myo-inositol to produce phosphatidylinositol for cell wall synthesis. There are only two known types of enzymes that utilize inositol-1-phosphate in mycobacteria, i.e., inositol monophosphatase (SuhB [101, 102] and CysQ [37, 43]) and MshA. MshA cannot utilize myo-inositol or 1d-Ins-1-P, produced from the action of phospholipase C activity on phosphatidyinositol, to produce MSH (92). It is possible that a myo-inositol kinase activity exists in M. tuberculosis and produces the 1l-Ins-1-P required for MSH biosynthesis, as postulated for M. smegmatis (92).
MshB, the MSH Deacetylase
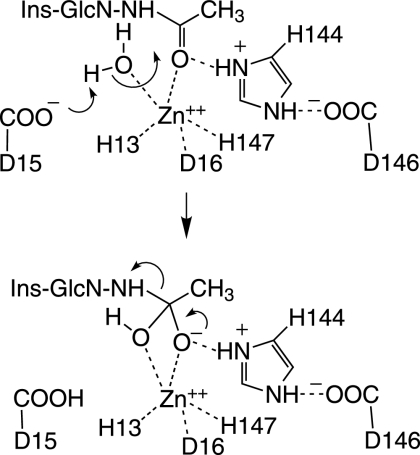
The third MSH biosynthesis enzyme, MshB, was discovered as a homolog of the MSH S-conjugate amidase (Mca) (see below), an MSH-dependent detoxification protein (85). MshB is a divalent metalloprotein which deacetylates GlcNAc-Ins and also has overlapping amidase activity with Mca (90). MshB is completely inactivated by the chelator 1,10-phenathroline, and the apoenzyme is activated by Zn2+, Ni2+, Mn2+, and Co2+ but not by Ca2+ or Mg2+. The native form of MshB was expected to be a Zn2+ metalloprotein, like its homolog Mca (see below), and this was confirmed by X-ray fluorescence scanning and the coordination geometry determined by X-ray crystallography (71). A second structure of MshB was determined with bound mercury and octylglycoside in the active site (73). In both cases, the metal binding ligands were identified as His13, Asp16, and His147, and Asp15 was identified as a plausible general base for deprotonation of water in the catalytic step.
A proposed catalytic mechanism for MshB, based on the crystal structure (71), is shown in Fig. 4. The conserved Asp15 residue serves as a general base for removal of a proton from water, serving as a nucleophile in the attack on the amide carbonyl which is polarized by the zinc. The developing negative charge on the carbonyl oxygen is stabilized by hydrogen bonding to the protonated His144 connected in a charge-relay system to Asp146. Decomposition of the tetrahedral intermediate requires transfer of a proton to the amine of the leaving GlcN-Ins. The general acid serving this function could be either protonated Asp15 (71) or protonated His144 (46).
FIG. 4.
Proposed mechanism for the M. tuberculosis deacetylase MshB (71). The active site is formed with Asp15, His13, and His147 coodinating a Zn ion required for catalysis and protein stability. The active-site Zn also polarizes the acetyl carbon-oxygen bond of bound GlcNAc-Ins for attack by a hydroxyl ion generated from water by protonation of Asp15. The forming acetate is hydrogen bonded to His144 or Asp15 to complete the hydrolysis.
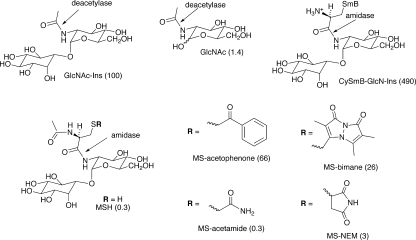
A study of the substrate specificity of M. tuberculosis MshB produced some curious results, as shown in Fig. 5 (90). Removal of the Ins residue from the natural substrate, GlcNAc-Ins, results in a 70-fold decrease in rate, whereas replacement of the acetyl group by AcCys to generate MSH produces a 330-fold drop in rate. MshB, like Mca (see below), has amidase activity with MSR that in some cases (R = CH2COC6H5) approaches the activity with GlcNAc-Ins but is significantly lower than the corresponding activity of Mca for most substrates. An exception is CySmB-GlcN-Ins, which is 60-fold more reactive with MshB than with Mca and nearly 5-fold more reactive with MshB than with its native substrate, GlcNAc-Ins. Since Cys-GlcN-Ins is present at only very low levels in mycobacteria, it is unlikely that significant levels of S-conjugates of Cys-GlcN-Ins are formed under native conditions. The overlapping activities of MshB and Mca are examined further in subsequent sections.
FIG. 5.
Relative rates of M. tuberculosis MSH deacetylase (MshB) cleavage of amide bonds in acyl glucosamine derivatives. The substrate specificity of MshB overlaps that of MSH S-conjugate amidase (Mca) (see Fig. 11). The natural substrate of MshB is GlcNAc-Ins, whose rate is set at 100 as a reference.
MshC, the MSH Ligase
The fourth MSH biosynthesis gene product, MshC, forms an amide bond between the amine of GlcN-Ins and the carboxylate of l-cysteine. MshC was identified by purification of the ligase activity from crude extracts of M. smegmatis, followed by N-terminal sequencing (125). MshC is a homolog of Cys-tRNA synthetase, which is unusual among tRNA synthetases because it is a Zn2+-containing protein (83).
Cloning, expression, and purification of MshC from M. tuberculosis proved difficult in E. coli by use of conventional His affinity tags. The native enzyme has been cloned and expressed in an M. smegmatis MshC-deficient mutant, but its purification is rather tedious (95). Expression and affinity purification of M. tuberculosis MshC have been achieved in an MshC M. smegmatis mutant by use of the N-terminal fusion tags glutathione S-transferase and maltose binding protein or the B1 domain of streptococcal protein G (38).
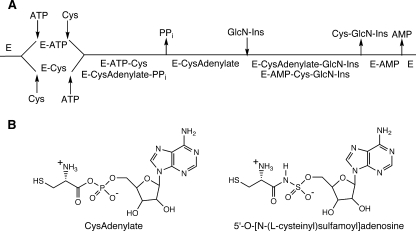
MshC from M. smegmatis proved less fastidious, was expressed in E. coli as an N-terminally His-tagged protein, and was purified with a high yield (28). The M. smegmatis enzyme was used for a detailed study of the reaction mechanism by the Blanchard group (28). In order to study the mechanism of the MshC-catalyzed reaction, the group carried out a chemical synthesis of GlcN-Ins, needed as a substrate, and conducted initial velocity and inhibition studies of the steady-state mechanism. The data were consistent with a bi uni uni bi ping pong mechanism, as shown in Fig. 6A, although the order of product release of AMP versus Cys-GlcN-Ins was not defined by the results. The rate-determining turnover of E-ATP-Cys to E-CysAdenylate-PPi competes with that of E-CysAdenylate-GlcN-Ins to E-AMP-Cys-GlcN-Ins, with the rates being ∼9.4 and ∼5.2 s−1, respectively, yielding a calculated steady-state kcat of 3.3 s−1, in good accord with the experimental value (3.15 s−1). Steady-state Km values were determined for ATP (1.84 ± 0.06 mM), Cys (0.10 ± 0.01 mM), and GlcN-Ins (0.16 ± 0.05 mM).
FIG. 6.
(A) Bi uni uni bi ping pong kinetic mechanism for M. smegmatis MshC (28). (B) Structures of the Cys adenylate intermediate and its Cys sulfamoyl adenylate inhibitor analog.
In a subsequent study, the initial steps of the MshC-catalyzed reaction were examined by positional isotope exchange using [βγ-18O6]ATP (157). In the presence of cysteine, formation of E-CysAdenylate-PPi occurs with cleavage of the α-P-16O bond to release PPi with a single 16O and six 18O atoms. The reaction is reversible and produces isotopic mixing of the 16O among the β- and γ-phosphate oxygens of ATP, as measured by 31P NMR. The rate is dependent on the cysteine concentration, showing that the formation of E-CysAdenylate-PPi is reversible. However, no inhibition of exchange was observed even at cysteine concentrations of 100 times the Km value, consistent with random binding of Cys and ATP (Fig. 6A) or ordered binding with Cys preceding ATP (Fig. 6A, lower route). The rate of isotopic exchange was eliminated in the presence of pyrophosphatase, showing that the PPi dissociates from the E-CysAdenylate complex and rebinds to produce the positional isotopic exchange. In the presence of high concentrations of GlcN-Ins, the exchange activity was eliminated. This result indicates that dissociation of PPi from the enzyme complex is faster than reformation of ATP and Cys from the E-CysAdenylate-PPi complex. Alternatively, if GlcN-Ins can bind to this complex prior to dissociation of PPi (an alternative pathway to that shown in Fig. 6A), then this result shows that reaction of the complex with GlcN-Ins is faster than the overall rate for regeneration and dissociation of ATP from the enzyme.
The CysAdenylate analog 5′-O-[N-(l-cysteinyl) sulfamoyl]adenosine (Fig. 5B) is a known nanomolar inhibitor of Cys- and prolyl tRNA synthetases (53) and was shown to be a potent inhibitor of His-tagged M. smegmatis MshC, having an ATP competitive inhibition constant of 304 ± 40 nM (28). Cys sulfamoyl adenylate was also found to be a 50 nM inhibitor of maltose binding protein-linked M. tuberculosis MshC, whereas the aspartyl and seryl sulfamoyl adenosine analogs were less potent inhibitors of M. tuberculosis MshC (38, 38a), in accord with their expected lower affinities for the active-site zinc.
Currently, no crystal structure is available for MshC, but a crystal structure for the homologous E. coli Cys-tRNA synthase has been reported (83).
MshD, the MSH Synthase (MSH Acetyltransferase)
MSH synthase (MshD) is the fifth and final enzyme in the MSH biosynthesis pathway and catalyzes the acetylation of the amino group of Cys-GlcN-Ins by acetyl-CoA (59). MshD is a member of a large family of GCN5-related N-acetyltransferases (GNATs) (23). It appears to have two separate regions with similarity to the pfam00583 domain for acetyltransferase and is approximately twice the size of homologous proteins, indicating that MshD is the result of a gene duplication (59). Blanchard and coworkers (147) have determined a crystal structure of M. tuberculosis MshD (Rv0819) showing that the C-terminal domain binds acetyl-CoA and that the N-terminal acetyl-CoA binding site is not functional. Initial kinetic studies indicated that acetyl-CoA is the preferred CoA thioester but that both acetyl-CoA and propionyl-CoA are MshD substrates (148). A crystal structure for the complex of MshD and CoA mixed disulfide with Cys-GlcN-Ins indicates a large conformational change upon substrate binding (148). No inhibitors of MshD have been reported.
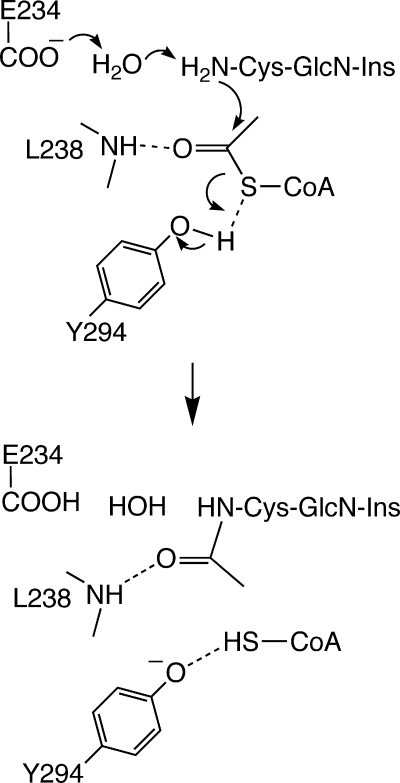
Kinetic studies indicated a ternary complex mechanism common to the GNAT superfamily of N-acetyltransferases in which both substrates must be present for the reaction to occur (148). Based upon the crystal structure of the mixed disulfide of Cys-GlcN-Ins and CoA and the pH dependence of enzyme activity, the catalytic mechanism shown in Fig. 7 was proposed. Glu234 is proposed to act as general base via an intervening water molecule, the Leu238 backbone NH stabilizes the negative charge developed in the transition state on the carbonyl of the acetyl residue, and Tyr294 provides a proton to generate CoA in the thiol form (CoASH). The Km values for acetyl-CoA and Cys-GlcN-Ins were determined to be 40 ± 5 μM and 82 ± 22 μM, respectively.
FIG. 7.
Proposed catalytic mechanism for M. tuberculosis MshD (148). The acetyl group from acetyl-CoA is transferred to the Cys amine of Cys-GlcN-Ins in a bound ternary complex, with subsequent release of CoASH, in a mechanism similar to that of other GCN5 acetyltransferases.
Regulation of MSH Biosynthesis
There is a regulator preceding MshA in M. tuberculosis (Rv0485) annotated as a transcriptional regulator similar to an N-acetylglucosamine repressor from Vibrio cholerae. Similarly, a transcriptional regulator precedes MshD in M. tuberculosis (Rv0818), but no functional studies of the genes have been reported. Indirect evidence suggests that mRNA levels of the MSH biosynthetic genes are relatively constant in M. tuberculosis, since none of these genes have been identified in various studies examining changes in mRNA levels under various conditions, such as stationary phase and low oxygen (151), exposure to hydrogen peroxide and palmitic acid, growth within macrophages (128), and exposure to nitric oxide (150). MSH biosynthetic enzymes are influenced by additional DNA sequences, since M. smegmatis mutants lacking all MshC activity have been isolated containing lesions outside their structural gene (118).
Sigma factors are involved in the regulation of MSH metabolism in Streptomyces coelicolor A3(2). A sigma factor responsive to osmotic and peroxide stresses, σB, has been linked to upregulation of mshA, mshC, and mshD in S. coelicolor (63). S1 nuclease mapping showed that expression of mshA, mshC, and mshD was induced by osmotic stress and that the induction of mshA and mshC did not occur in a σB-deficient mutant. Levels of MSH were only slightly lower (<2-fold) in the σB null mutant, suggesting that σB may not be a major regulator of MSH metabolism under these conditions.
In S. coelicolor, the response to the production of disulfide bonds within the cell (disulfide stress) is regulated by the sigma factor σR. The activity of σR is controlled by the anti-sigma factor RsrA, which responds to changes in the intracellular thiol-disulfide redox balance (54). Using a consensus sequence of σR target promoters, Paget et al. (104) predicted that σR regulates the expression of more than 30 genes, including genes involved in thiol metabolism. When MSH levels were examined in a σR-deficient mutant, they were fourfold lower than parental levels during all phases of growth. More recently, the role of σR in MSH metabolism was elaborated by Park and Roe (106). A complex is formed between σR and the reduced form of the σR regulator. Exposure of the oxidized disulfide form of RsrA to thiols generates RsrAred, and thiols generated allow it to bind Zn and form RsrA(Zn)red (161). RsrA(Zn)red binds σR, preventing it from activating its target genes. σR activates direct expression of a host of genes, including mshA and mca, and indirect expression of mshB, mshC, and mshD (106). Treatment of cells with reagents that deplete MSH, including the thiol oxidant diamide and the thiol alkylating agents monobromobimane (mBBr) and N-ethylmaleimide, leads to oxidation of RsrA and the release of σR, with subsequent expression of the σR regulon. The consensus promoter sequence for mshA and mca was identified as GGAAT-N18-GTT, differing in only 1 nucleotide of spacer length from the sequence determined by Paget et al. (104). Other genes that undergo increased expression with σR include rsrA and sigR, producing amplification of the redox sensing system, and trxAB (encoding thioredoxin and thioredoxin reductase), which also provides reducing capacity. The sigR gene is absent in mycobacteria, but a related gene product, σH (MT3320 in M. tuberculosis CDC1551), regulates responses to both heat shock and redox stress (55). Several components of the thioredoxin system were found to be regulated by σH in M. tuberculosis, but none of the MSH biosynthetic genes were noted.
MUTANTS IN MSH BIOSYNTHESIS
Mycobacterium smegmatis
Studies using mutants defective in MSH production have been invaluable in identifying the genes encoding each of the MSH biosynthetic enzymes, in determining which of the genes is essential for the biosynthesis of MSH, and in defining the various functions within the cell that require MSH. The initial studies on MSH biosynthesis were carried out with M. smegmatis, and viable mutants with reduced activity of each of the MSH biosynthetic enzymes, except for MshA2, have been recovered. One or more of these M. smegmatis MSH mutants has exhibited an increased sensitivity to reactive oxygen and nitrogen species, to alkylating agents, and to antibiotics, suggesting that MSH is involved in mutiple detoxification mechanisms.
A summary of the thiol content of the M. smegmatis MSH mutants is given in Table 3. Data for M. smegmatis mc2155 were taken from multiple analyses performed in our laboratory (1, 59, 94). Mutants in the mshA gene produce undetectable levels of MSH and none of the MSH intermediates. The first MshA-deficient mutant isolated was a chemical mutant (96) that contained a single base change within mshA, producing a G32D mutation, but contributions to the phenotype by additional changes in the chemical mutant were not excluded (91). Using site-directed mutagenesis, we reconstructed this mutant and confirmed that the single base change generating the G32D mutant results in a complete loss in synthesis of GlcNAc-Ins, GlcN-Ins, and MSH. In addition, an mshA::Tn5 mutant was identified from a Tn5 transposon library after selection for resistance to isoniazid (100 μg/ml), a characteristic which had previously been established as a phenotype for MSH-deficient M. smegmatis (96, 118). The mshA::Tn5 mutant was devoid of GlcNAc-Ins, GlcN-Ins, and MSH.
TABLE 3.
Production of MSH and MSH intermediates by M. smegmatis mutants
| Enzyme | Strain | Concn (μmol/g dry weight) of MSH or intermediate
|
|||
|---|---|---|---|---|---|
| GlcNAc-Ins | GlcN-Ins | Cys-GlcN-Ins | MSH | ||
| Wild typea | mc2155 | <0.1 | 0.2-1.0 | ∼0.008 | 10 ± 3 |
| MshAb | mshA::Tn5 | <0.01 | <0.01 | <0.01 | <0.01 |
| MshBc | Myco504 | 2.6 ± 0.2 | <0.01 | <0.02 | 1.0 ± 0.2 |
| MshCd | Tn1 | NDe | 2.6 | <0.002 | <0.004 |
| MshDf | mshD::Tn5 | ND | 0.35 ± 0.05 | 0.6-2 | 0.12 ± 0.01g |
The MshB mutant Myco504 contains a kanamycin resistance cassette at base 207 within mshB (117). This mutant accumulates elevated levels of the intermediate GlcNAc-Ins (Table 3), confirming that the major deacetylase activity within the MSH biosynthetic pathway has been inactivated. The mshB mutant also produces about 10% of normal levels of MSH, indicating that the cell contains an additional activity that is capable of deacetylating GlcNAc-Ins. A possible candidate for this deacetylase is the related protein Mca, which has weak GlcNAc-Ins deacetylase activity (136). However, complementation of Myco504 with an expression plasmid expressing Mca did not increase the level of MSH in the complemented mshB mutant (117), so involvement of another deacetylase is possible.
The Tn1 transposon mutant is completely defective in MshC activity, has undectable levels of Cys-GlcN-Ins and MSH, and has elevated levels of GlcN-Ins (118). Interestingly, the transposon insertion in this mutant and a companion MshC-deficient mutant, the Tn2 mutant, is not within the mshC gene but at an unknown location within the chromosome.
The mshD::Tn5 mutant is a transposon mutant which was also initially identified by its resistance to isoniazid. The MshD mutant produces small amounts of MSH as well as high levels of the MshD substrate Cys-GlcN-Ins, a novel thiol (formyl-Cys-GlcN-Ins), and moderate amounts of another novel thiol, succinyl-Cys-GlcN-Ins (59, 94). Formyl-Cys-GlcN-Ins is closely related to MSH in structure, with the CH3CO- residue replaced by HCO- on the nitrogen of Cys. Experimentally, formyl-Cys-GlcN-Ins has been shown to replace MSH in reactions of two MSH-dependent enzymes, Mca and Mtr (mycothione reductase), with reduced but significant rates (94). This suggests that high levels of formyl-Cys-GlcN-Ins may partially substitute for MSH in the MshD-deficient mutant. Chemical transacylation of Cys-GlcN-Ins by acetyl-CoA and succinyl-CoA is the most likely source of the small amounts of MSH and succinyl-Cys-GlcN-Ins produced in this mutant.
The following is a summary of phenotypic characteristics observed among the various M. smegmatis MSH mutants.
Poor growth during initial culture.
We have observed that MshA- and MshC-deficient mutants are slower to grow from frozen stocks during the initial stages of culture, and this may reflect their lack of MSH. This suggests that MSH may provide some protection during adaptation to growth in an oxygen-rich environment and during the initiation of cellular metabolism.
Sensitivity to reactive oxygen and nitrogen species and redox cycling agents.
The MSH mutants have been reported to exhibit some degree of sensitivity to reactive oxygen species and redox cycling agents, with the results dependent upon the type and amount of compound used and the manner in which the assay was performed. Compounds which produced greater inhibition of growth in a disk diffusion assay included hydrogen peroxide, menadione, plumbagin, and t-butyl hydrogen peroxide (with MshA, MshB, MshC, and MshD mutants) (116-118). The MshA chemical mutant and MshC-deficient mutants (Tn1 and Tn2) were at least 10-fold more sensitive to killing by hydrogen peroxide in a 2-h broth exposure (96, 118). The MshA mutant was also reported to be more sensitive to killing by gaseous nitric oxide (79). When bacteria were continuously exposed to gaseous nitric oxide, toxicity to the mutant began after 4 h of exposure to the gas, compared to 7 h for wild-type M. smegmatis.
Sensitivity to alkylating agents.
The MSH S-conjugate amidase (Mca) takes part in the detoxification of alkylating agents, suggesting that MSH-deficient mutants should be more sensitive to these agents (86). The MshC mutants were two- to fourfold more sensitive to killing by the alkylating agents mBBr, iodoacetamide, and 1-chloro-2,4-dinitrobenzene (118). In a separate study using a disk diffusion assay, the MshA-, MshB-, MshC-, and MshD-deficient mutants were all sensitive to 1-chloro-2,4-dinitrobenzene at 0.2 μmol, and all except the MshB mutant were sensitive to iodoacetamide at 0.05 μmol (116).
Resistance to isoniazid and to ethionamide.
One of the most pronounced characteristics observed in the first MSH mutant isolated was its >25-fold increase in resistance to isoniazid (96). In the subsequent isolation of additional MSH mutants, resistance to 100 μg/ml isoniazid was used in the initial mutant screen. In this manner, the mshA::Tn5 and mshD::Tn5 mutants were isolated (59, 91). The MshB-deficient mutant, which makes 5 to 10% of normal MSH levels, has been reported to be only slightly more resistant to isoniazid than the wild type in a disk assay (116, 117), suggesting that moderate levels of MSH are sufficient to confer normal sensitivity to isoniazid. Enhanced resistance to ethionamide is another characteristic of the MshA-, MshB-, and MshD-deficient mutants, with complete resistance at 50 μg/ml in a disk assay (116). Using an agar plate assay, the MIC99 of ethionamide for the mshB mutant Myco504 was 150 μg/ml, which was sixfold higher than that for the parental strain (117). Both isoniazid and ethionamide are prodrugs which need to be activated intracellularly and whose reactive groups must be unmasked before inhibiting their targets (4). The high levels of resistance of the MSH mutants to isoniazid and to ethionamide suggest that MSH is involved in the activation of these drugs.
Sensitivity to other antibiotics.
In contrast to the enhanced resistance to isoniazid and to ethionamide, the MSH mutants have displayed increased sensitivities to a variety of antibiotics, including rifamycin, streptomycin, erythromycin, and azithromycin (96, 116-118). For example, the MshA mutant was 10-fold more sensitive to rifampin and ∼20-fold more sensitive to erythromycin than the wild type by use of Etest strips (116).
Production of novel thiols.
The mshD mutant produces two novel thiols, formyl-Cys-GlcN-Ins and succinyl-Cys-GlcN-Ins (94). The blockage at the MshD step results in an accumulation of high levels of Cys-GlcN-Ins within the cell. It seems likely that cellular succinyl-CoA undergoes S-transacylation with Cys-GlcN-Ins, followed by S-N acyl transfer, to form succinyl-Cys-GlcN-Ins (Fig. 1D), a process shown to occur chemically in solution (94). This is similar to the nonenzymatic formation of MSH via acetyl-CoA in the mutant. The origin of formyl-Cys-GlcN-Ins in the mshD mutant is likely to be enzymatic.
Mycobacterium tuberculosis
Using a temperature-sensitive shuttle phasmid to introduce targeted disruptions within the chromosome of M. tuberculosis Erdman, we have been able to generate viable mutants in the mshB (14) and mshD (13) genes but not in the mshA (12) and mshC (124) genes (Table 4). The mshB mutant, which carries a disruption in gene Rv1170, produced ∼20% of wild-type levels of MSH during exponential growth. With prolonged culture, MSH levels increased over 20-fold, to levels significantly higher than those in wild-type M. tuberculosis (14). GlcNAc-Ins levels were dramatically elevated and GlcN-Ins levels were reduced in the mutant, establishing that the Rv1170 gene encodes the major MshB activity in the MSH biosynthetic pathway of M. tuberculosis but that another deacetylase activity can partially substitute for Rv1170 activity during MSH biosynthesis. The mshD mutant produced ∼1% of normal MSH levels and high levels of the MshD substrate Cys-GlcN-Ins (13). The dominant thiol in the MshD mutant was formyl-Cys-GlcN-Ins, similar to the situation in M. smegmatis.
TABLE 4.
Production of MSH and MSH intermediates by M. tuberculosis mutants
Because the MshB- and MshD-deficient mutants produce significant levels of either MSH or the MSH-like thiol formyl-Cys-GlcN-Ins, they are not ideal organisms for characterizing a MSH-free state in M. tuberculosis. However, certain characteristics which suggest functions for MSH in M. tuberculosis during its growth and response to stress were apparent in both of these mutants. Both the MshB- and MshD-deficient mutants grew poorly on agar media lacking Middlebrook OADC supplement (oleic acid, NaCl, albumin, dextrose, and catalase), resulting in a reduced plating efficiency and smaller colony size. The defect was most pronounced for the MshD-deficient mutant, with a >3-log drop in plating efficiency on plates lacking OADC versus ADS supplement (albumin, dextrose, NaCl) (13). Log-phase cultures of the MshB-deficient mutant were ∼2.5 times more sensitive to the toxic oxidant cumene hydroperoxide after 7 h of exposure and 2 to 4 times more sensitive to rifampin, depending upon the concentration of drug. The MshD-deficient mutant was moderately more sensitive to killing by hydrogen peroxide and more restricted in growth in moderately acidic (pH 5.5) medium. In a genome-wide screen of nonessential M. tuberculosis genes required for growth in macrophages, an mshD mutant was one of the most impaired (120). The mshD mutant failed to grow in cultures of primary murine macrophages that modeled all stages of the host immune response, including inactivated macrophages, gamma interferon-preactivated macrophages, and gamma interferon-postactivated macrophages. The mshD mutant had not previously been identified as being defective for growth in the spleens of mice after intravenous infection of mice (127), indicating that the mshD gene is critical for survival under distinct conditions. These results indicate that growth of M. tuberculosis in environments where antimicrobial factors, including reactive oxygen and reactive nitrogen intermediates, are formed, such as within macrophages, relies upon MSH-dependent systems.
MSH Mutants in Other Actinobacteria
Mutants in MSH biosynthesis have been isolated in several other Actinobacteria, and those mutants which have been characterized have phenotypes that are quite similar to those observed with the M. smegmatis mutants. In Streptomyces coelicolor, disruption of the mshA, mshC, or mshD gene abolished all detectable MSH, while a MshB mutant produced 10% of wild-type levels (107). This indicates that the deacetylase activity of the MshB protein can be provided by other enzymes, while MshA, MshC, and MshD are essential for MSH biosynthesis in S. coelicolor, which is identical to the situation in M. smegmatis. The MshA and MshC mutants were reported to differentiate more slowly, but all of the mutants grew as well as the wild type in yeast extract-malt extract liquid medium. Corynebacterium glutamicum is an MSH-containing soil organism that is commercially used to produce amino acids and vitamins and that can metabolize aromatic compounds. Mutations in mshC and in mshD resulted in no MSH, while a mutation in mshB reduced MSH levels (29). The mshC and mshD mutants were unable to grow in media with gentisate or 3-hydroxybenzoate as a carbon source, and this observation led to the discovery of an MSH-dependent maleylpyruvate isomerase. A MshD mutant has been constructed in Amycolatopsis mediterranei, a streptomycete strain used to produce rifamycin SV (17). Although MSH was not quantitated in this mutant, the mutant was reported to grow slower than the wild type on plates and was more sensitive to hydrogen peroxide and the antibiotics apramycin and erythromycin.
MSH-DEPENDENT ENZYMES
Overview
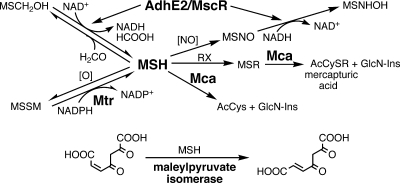
Enzymes which require MSH are involved in a number of cellular processes, including detoxification of electrophilic compounds, inactivation of reactive oxygen and nitrogen species, reductions, and isomerizations. The activities of some of the more well-characterized enzymes are summarized in Fig. 8. MSH reacts spontaneously with formaldehyde to produce an adduct, H2C(OH)SM, which is a substrate for the formaldehyde dehydrogenase AdhE2 (103). AdhE2-catalyzed oxidation of formaldehyde to formate detoxifies formaldehyde originating from metabolic or environmental sources. It was later shown that the mycobacterial enzyme exhibits S-nitrosomycothiol (MSNO) reductase activity (MscR) and that the MscR-specific activity is markedly higher than the AdhE2-specific activity (149). A key enzyme of MSH metabolism is MSH disulfide reductase (Mtr), which maintains MSH in its reduced state. MSSM is generated by MSH autoxidation but probably also by as yet unidentified MSH-dependent peroxidases. Reaction of MSH with a wide range of alkylating agents produces adducts that are cleaved by MSH S-conjugate amidase (Mca) to generate a mercapturic acid (AcCySR) and GlcN-Ins, with the latter being used by the cell to regenerate MSH (86). All of these reactions involve detoxification functions by MSH.
FIG. 8.
Summary of well-characterized MSH-dependent reactions where MSH is either a substrate or cofactor. MSH is the redox buffer for Actinobacteria, is oxidized by cellular oxidants, [O], and is maintained in a highly reduced status by MSH disulfide reductase (Mtr). Formaldehyde (HCOOH) and MSNO are converted to formate and MSH sulfinamide (MSONH2), respectively, by the alcohol dehydrogenase (AdhE2)/MSNO reductase (MscR). Thiol-reactive drugs form MSR that are cleaved by MSH S-conjugate amidase (Mca) to produce the N-acetyl-CyS-conjugate (AcCySR), which is excreted. The pseudodisaccharide GlcN-Ins is retained in the cell and reenters the MSH biosynthesis pathway. Another important function of Mca is the degradation of MSH to provide cysteine (from acetylcysteine [AcCys]) and GlcN-Ins as needed for other metabolic pathways. MSH is also required for the metabolism of 3-hydroxybenzoate; the MSH-dependent step is the isomerization of maleyl-pyruvate to fumaryl-pyruvate by maleylpyruvate isomerase (see Fig. 16 for more details).
An initially suprising finding was that Mca has low activity with MSH itself to produce AcCys and GlcN-Ins (86, 136). Subsequent studies, described below, revealed that this is an important step in the catabolism of MSH, allowing it to serve as a reserve resource of cysteine, glucosamine, and myo-inositol. MSH was also shown to function as a coenzyme in the cis-trans isomerization catalyzed by maleylpyruvate isomerase (29).
Mtr, the MSH Disulfide Reductase (Mycothione Reductase)
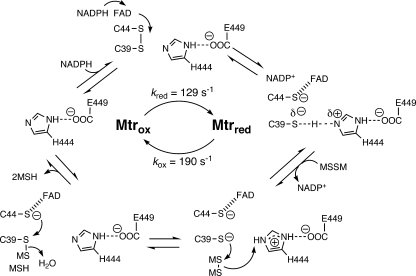
The hallmark of the thiol redox buffer of any cell is the presence of a specific disulfide reductase that allows a highly reduced status to be maintained within the cell. Due to the substantial CoA content in mycobacteria, it was not initially clear whether MSH or CoA was the redox buffer until the M. tuberculosis genome sequence was published (19). The genome did not show single-active-site cysteine-type CoA disulfide reductases homologous to Staphylococcus aureus CoA disulfide reductase (21) but did indicate a gene, annotated as gorA (GSH reductase), later shown to be the MSH disulfide reductase by Patel and Blanchard (108). MSH disulfide reductase, named mycothione reductase by analogy with GSH reductase and trypanothione reductase, was one of the first enzymes of MSH metabolism to be characterized, and it plays a key role in maintaining MSH in a reduced state. The Rv2855 gene in M. tuberculosis was cloned and overexpressed in M. smegmatis as the native protein, was purified to homogeneity, and was shown to reduce MSSM and the disulfide of des-myo-inositol MSH (108, 109). Subsequent sequencing of genomes allowed the identification of genes encoding Mtr in M. smegmatis (MSMEG2611), Corynebacterium glutamicum (NCg1832), and Rhodococcus jostii RHA1 (ro006610), with identities of >50%. The closest homolog in Streptomyces coelicolor (SCO2180) has only 27% identity and 44% similarity, and there are several other homologs with nearly comparable scores, so its assignment requires experimental confirmation.
Kinetic studies on M. tuberculosis Mtr by Patel and Blanchard (108-110) have provided a detailed mechanism for the reaction. The deduced mechanism is shown in Fig. 9 and is similar to the mechanism proposed for GSH reductase (121, 158). For the reductive reaction, binding of NADPH to the oxidized enzyme is very rapid; subsequent transfer of hydride to flavin adenine dinucleotide and reduction of the active-site disulfide occur at a rate of 129 ± 2 s−1 at 25°C, comparable to the corresponding value of 110 s−1 for E. coli GSH reductase (121). Mtr will also utilize NADH, albeit less efficiently, with the maximal rate for reduction of the active-site disulfide being 20 ± 1 s−1. In the oxidative reaction, loss of NADP+ and binding of MSSM lead to cleavage of the substrate disulfide and formation of an MSS enzyme intermediate, followed by reduction of this disulfide and loss of the MSH products. The rate of the oxidative reaction for Mtr is 190 s−1 at 25°C, whereas the corresponding step for E. coli GSH reductase is significantly faster, at a rate of 490 s−1 at 4°C (121).
FIG. 9.
Proposed catalytic mechanism for reduction of MSSM by MSSM reductase, Mtr (110). The oxidized enzyme (Mtrox) binds NADPH, and the active-site disulfide (C-44-S-S-C-39) is reduced by electrons from NADPH mediated by flavin adenine dinucleotide to generate reduced enzyme (Mtrred). Mtrred binds the substrate disulfide, MSSM, and releases the oxidized pyridine nucleotide, NADP+. Cleavage of bound MSSM by active-site C-39S⊖ to generate bound M-S-S-C-39 is followed by cleavage of M-S-S-C-39 by C-44S⊖ and release of two MSH molecules to regenerate Mtrox. ⊖, negative charge.
Mtr appears to have a substantial degree of specificity with respect to disulfide substrates (Table 5). For residues attached to the carboxyl group of cysteine, no activity is seen without the GlcN residue. The Ins residue can be replaced by a benzyl residue with only a minor loss of activity, and this derivative represents a synthetically accessible alternative substrate potentially useful in screening for Mtr inhibitors (137). All substitutions at the amino residue of cysteine, including simple removal of the acetyl moiety or its replacement by a formyl residue, produce significant losses of activity. The thiol titrating agent 5,5′-dithiobis(2-nitrobenzoic acid) was >105-fold less reactive than MSSM (109).
TABLE 5.
Relative reactivities of various disulfides with mycothione reductase, using NADPH as a cofactor, normalized to MSSM rate of 100
Although Mtr was one of the first MSH-dependent enzymes studied (108, 109), the Mtr crystal structure and the identification of Mtr-specific inhibitors have not been reported and represent important areas for future study. Several quinones function as “subversive substrates” of Mtr, through promotion of redox cycling, and are toxic to mycobacteria (see below).
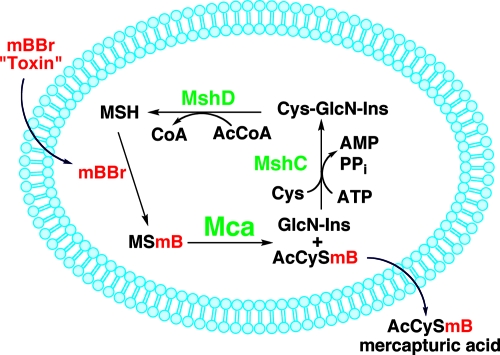
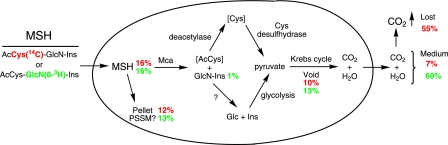
Mca, the MSH S-Conjugate Amidase
MSH S-conjugate amidase was accidentally discovered in our laboratory during an attempt to label MSH in M. smegmatis cells with mBBr prior to pelleting and extraction in order to ascertain whether the pelleting of cells influenced the MSH level. The cellular content of the labeled MSH, MSmB, proved to be much lower than expected, and analysis of the medium revealed the presence of a high level of AcCySmB. This is one of a class of compounds, known as mercapturic acids, having the general structure AcCySR and long known as the final product excreted in urine of a GSH-dependent, multiorgan detoxification pathway in animals (16). The process in mycobacteria is summarized in Fig. 10, with mBBr as the thiol-reactive toxin (86). Rapid diffusion of mBBr into cells is followed by reaction with MSH to generate MSmB. Mca catalyzes the cleavage of MSmB. The mercapturic acid, AcCySmB, is rapidly lost from the cell, while GlcN-Ins is retained intracellularly and utilized for the resynthesis of MSH.
FIG. 10.
MSH-dependent detoxification of thiol-reactive drugs and metabolites, as reported for M. smegmatis and S. coelicolor (86, 106, 136). The thiol-reactive reagent mBBr reacts with MSH, and the MSR is hydrolyzed by MSH S-conjugate amidase (Mca) to GlcN-Ins and the excreted mercapturic acid AcCySmB. GlcN-Ins is retained in the cell and recycled for MSH synthesis, using MshC and MshD.
Mca was purified from M. smegmatis and sequenced to identify the corresponding protein, encoded by Rv1082, in M. tuberculosis (86). Genes encoding the closest homolog to Mca and their percentages of identity to Rv1082 in representative Actinobacteria are as follows: M. smegmatis MSMEG5261, 78%; Rhodococcus jostii RHA1 ro05852, 69%; C. glutamicum NCg10948, 59%; and S. coelicolor SCO4967, 52%. The uniform occurrence of mca in MSH-producing bacteria indicates that Mca plays an important role in MSH function.
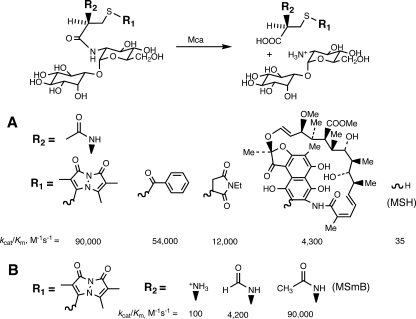
The M. tuberculosis Mca, cloned and expressed in E. coli, was purified to homogeneity and shown to be a zinc-containing enzyme that is inactivated by the metal chelator 1,10-phenanthroline (136). The cleavage of MSR by Mca follows simple Michaelis-Menten kinetics, and the enzyme is active with a wide range of substrates, including MSH itself (136). Reaction rates for a spectrum of substrates are given in Fig. 11. With respect to the residue attached to the sulfur of MSH, R1, aryl-CH2 residues are the most reactive, but a range of structures give substantial reactivity, including the highly complex conjugate of rifamycin S (Fig. 11A). MSH itself has a much lower reactivity, but one that is significant at the millimolar intracellular levels of MSH, even though MSH inhibits Mca at these levels (136). As discussed below, this activity of Mca is important in the turnover of MSH. Mca activity is substantially dependent upon the integrity of the MSH moiety itself. Removal of the inosityl residue from MSmB to generate AcCySmB-GlcN reduces the value of kcat/Km 900-fold, to 100 M−1 s−1 (136). Removal of the acetyl residue also reduces the rate 900-fold, whereas replacement of acetyl by a formyl residue results in an ∼20-fold drop in rate (Fig. 11B). Thus, Mca is quite sensitive to the residues present at both ends of the MSH moiety but has substantial activity with a wide range of groups linked to the sulfur atom.
FIG. 11.
Substrate specificity of M. tuberculosis MSH S-conjugate amidase (Mca) for MSR (136), where the MSH moiety is held constant and the S-conjugate moiety (R1) is varied (A) or the S-conjugate moiety (bimane) is held constant and the MSH moiety is varied, from Cys-GlcN-Ins to formyl Cys-GlcN-Ins to MSH (B).
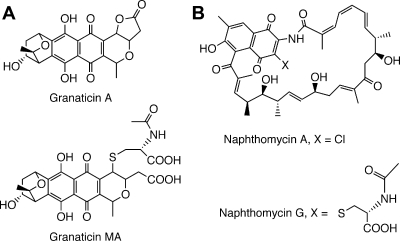
A variety of mercapturic acid derivatives have been isolated from cultures of various Actinobacteria and are logical products of Mca-catalyzed cleavage of MSH-derived adducts (88). Kormann et al. (60) showed that the antibiotic granaticin A is produced by Streptomyces violaceoruber Tü 7 during the initial days of culture but disappears after 9 days, coinciding with the appearance of the corresponding mercapturic acid, granaticin MA (Fig. 12A). The antibiotic activity of granaticin MA was at least an order of magnitude less than that of granaticin A. When 14C-labeled granaticin A was added to the culture, a large fraction of the label was recovered in the broth as granaticin MA. This provides strong support for the operation of the MSH-dependent detoxification pathway generating the mercapturic acid via the action of Mca.
FIG. 12.
Antibiotics produced in Actinobacteria fermentation broths from Streptomyces violaceruber Tü (granaticin A and its mercapturic acid, granaticin MA) (A) and Streptomyces sp. strain E/784 (naphthomycin A and its mercapturic acid, naphthomycin G) (B).
Another example is the antibiotic naphthomycin A (Fig. 12B), an active inhibitor of a number of gram-positive and gram-negative bacteria (82), and the corresponding mercapturic acid, naphthomycin G, which lacks biological activity (78). This constitutes an example where MSH and Mca likely function in a detoxification mode.
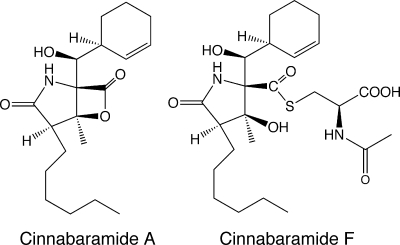
Some mercapturic acids isolated from cultures of Streptomyces spp. have been found to have biological activities similar to those of their precursors in assays against mammalian targets. A recent example involves the cinnabaramides A and F (Fig. 13), isolated from Streptomyces strain JS360 (134). Cinnabaramide F is the mercapturic acid derived from cinnabaramide A, and both have comparable activities in the low nanomolar range in a human 20S proteasome inhibitory assay.
FIG. 13.
Human 20S proteasome inhibitors produced by Streptomyces JS360 cinnabaramide A and its mercapturic acid, cinnabaramide F (134).
Homologs of mca with low levels of identity to the M. tuberculosis enzyme are found in the biosynthetic operons of various antibiotic-producing Actinobacteria, suggesting that the encoded proteins may be involved in MSH-dependent antibiotic detoxification (88). Alternatively, such mca homologs may be involved in tailoring GlcNAc residues in the biosynthesis of aminoglycoside antibiotics (139). An updated list is provided by Rawat and Av-Gay in their recent review (114). Experimental tests of the functions of these mca homologs in antibiotic biosynthesis operons are important. Yu et al. (160) tested the effect of inactivation of most genes in the rifamycin biosynthesis operon of Amycolatopsis mediterranei S699 upon rifamycin B production. Although knockout of the rifO gene, a homolog of mca, was not reported in this communication, this mutant has been created and found to produce normal levels of rifamycin B (Tin-Wein Yu and Rolf Müller, personal communication). Thus, the product of rifO is apparently not required to protect the cells against rifamycin B toxicity.
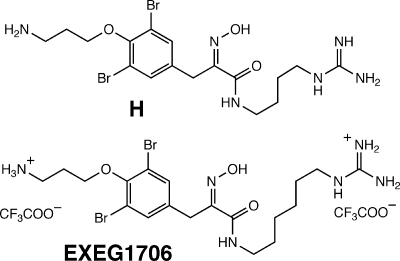
A search for natural product inhibitors of Mca by the Bewley group (97, 100) identified a spectrum of compounds, with one of the most potent being hydroxamate H (Fig. 14), isolated from the marine sponge Oceanapia sp. and having 50% inhibitory concentrations (IC50s) for Mca in the low micromolar range. Subsequent synthesis of a series of related compounds identified the closely related compound EXEG1706 (Fig. 14) as a potent inhibitor of a spectrum of gram-positive bacteria, including species that do not produce MSH (113).
FIG. 14.
Hydroxamate inhibitors of MSH S-conjugate amidase. H, a bromotyrosine alkyloid from the marine sponge Oceanapia sp. (100), and the closely related synthetic analog EXEG1706 (113) are shown.
MscR, MSNO Reductase/Formaldehyde Dehydrogenase
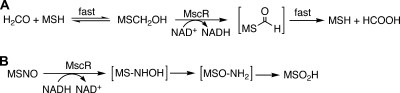
The first MSH-dependent enzyme activity to be purified was that of a factor-dependent formaldehyde dehydrogenase from Amycolatopsis methanolica (144) and Rhodococcus erythropolis (24), for which the factor proved to be MSH (80). Sequence analysis showed that the enzyme is a divergent medium-chain dehydrogenase/reductase (103). The cloned His6-tagged M. tuberculosis enzyme proved difficult to purify in active form when expressed in E. coli or M. smegmatis (149). The native M. smegmatis enzyme was purified by Vogt et al. (149) and shown to also exhibit much higher MSNO reductase activity, leading to its designation as MscR. They undertook detailed kinetic studies of both the formaldehyde dehydrogenase and the MSNO reductase reactions. The reactions catalyzed are shown in Fig. 15.
FIG. 15.
Two known activities of the alcohol dehydrogenase previously annotated AdhE2 in the M. tuberculosis genome. (A) MSH-dependent formaldehyde dehydrogenase reaction with MscR, generating formate in Amycolatopsis methanolica and Rhodococcus erythropolis (80), and with M. smegmatis MscR (149). (B) MSNO reaction with MscR, generating MSH sulfinic acid (MSO2H) in M. smegmatis (149).
Thiols, including MSH, react rapidly with formaldehyde to produce a hemithioacetal (Fig. 15A). This was shown to be the substrate of MscR in a reaction studied with excess formaldehyde (149). The rate was followed by the reduction of NAD+ and shown to be dependent upon MSH and NAD+ concentrations, in accord with a rapid-equilibrium ordered reaction mechanism with Km values of 354 ± 116 and 17.3 ± 0.8 μM for NAD+ and MSH, respectively, and a turnover number of 24.7 s−1. MSH was not consumed, indicating that the expected product, the thioester of formate, was rapidly hydrolyzed under the reaction conditions to regenerate MSH.
The MSNO reductase reaction of MscR (Fig. 15B) was studied using MSNO synthesized from MSH and sodium nitrite (149). The kinetic dependence upon MSNO and NADH was consistent with an ordered bireactant system with a Km for MSNO of ∼1 mM and a turnover number of ∼2,000 s−1, some 80-fold greater than that for the formaldehyde dehydrogenase reaction. The stoichiometry of the reaction, 1 equivalent of NADH per equivalent of MSNO, indicated that the initial product is MSNHOH, an unstable species that readily rearranges to the sulfinamide, MSONH2. The latter was presumed to hydrolyze under the conditions used to produce the sulfinic acid MSO2H, which was the characterized product of the in vitro reaction.
Treatment of M. smegmatis with S-nitrosoglutathione led to the production of 0.5 equivalent of glutathione disulfide and 1.0 equivalent of nitrate per equivalent of S-nitrosoglutathione consumed (149), indicating that the NO was oxidized in vivo to nitrate rather than reduced to ammonia as the in vitro studies indicate. Attempts to mimic the in vivo production of nitrate in the in vitro system by incorporating hemoglobin N in the in vitro system to facilitate the generation of nitrate had only limited success. Thus, the details of the pathway of NO metabolism in mycobacterial cells require further clarification, but the high specific activity of MscR toward MSNO leaves little doubt that this enzyme plays a key initial role in the detoxification of nitric oxide. This notion is supported by recent studies by the Av-Gay group (79), who showed that the MSH-producing organism M. smegmatis was more resistant to NO than a broad range of bacteria that do not produce MSH, including E. coli and other GSH-producing bacteria. They showed that an MshA-deficient M. smegmatis mutant that does not produce MSH was more sensitive to NO than the parental strain.
Homologs of M. tuberculosis MscR (Rv2259) in other Actinobacteria and their percentages of identity include the following: M. smegmatis MSMEG4340, 84%; Rhodococcus jostii RHA1 ro02587, 74%; S. coelicolor SCO0741, 74%; and C. glutamicum NCg10313, 65%. This demonstrates that this enzyme is broadly associated with MSH metabolism. In the transposon site hybridization (TraSH) analysis of genes required for growth of M. tuberculosis, MscR was classified as an essential gene (126).
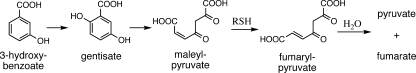
Maleylpyruvate Isomerase
The conversion of maleylpyruvate to fumarylpyruvate is an important reaction in the metabolism of aromatic compounds via the gentisate pathway (Fig. 16). In GSH-producing organisms, it is catalyzed by glutathione S-transferase zeta (8), also known as maleylpyruvate isomerase. Maleylpyruvate isomerase activity was reported (39) for extracts of 11 different strains of gram-positive bacteria, including species of Nocardia, Corynebacterium, Arthrobacter, and Rhodococcus, genera subsequently shown to produce MSH. A GSH-independent maleylpyruvate isomerase (NCgl2918) was subsequently identified in Corynebacterium glutamicum (130). It was cloned and expressed in E. coli as a C-terminal His6 construct and affinity purified to produce an active enzyme with a specific activity of ∼19 μmol min−1 mg−1 that was not stimulated by GSH. It was subsequently shown that the activity of this enzyme is markedly stimulated by 20 μM MSH (29). At this level of MSH, the enzyme was determined to have an apparent Km for maleylpyruvate of ∼150 μM and a Vmax of ∼1,500 μmol min−1 mg−1, but the apparent Km for MSH was not determined. Deletion mutants of C. glutamicum in mshC and mshD were shown not to produce significant levels of MSH and to be unable to grow on 3-hydroxybenzoate or gentisate (29). It appears that the dominant activity is that of an MSH-dependent maleylpyruvate isomerase but that a lower-level, thiol-independent activity may also be present. The MSH-dependent maleylpyruvate isomerase from C. glutamicum has been crystallized and, unlike the human GSH-dependent enzyme, was found to be a zinc metalloprotein with a novel fold (153). The MSH-dependent maleylpyruvate isomerase also lacked the dehalogenation activity found for the human enzyme. Homologs of the C. glutamicum enzyme are found in Rhodococcus jostii RHA1 (ro01864; 68% identity) and Streptomyces coelicolor (SCO1959; 44% identity), but no significant homologs are found in the mycobacteria.
FIG. 16.
Thiol-dependent maleylpyruvate isomerase function in the gentisate pathway.
Other Enzymes
There is indirect evidence for additional MSH-dependent enzymes based upon the phenotypic characterization of MSH-deficient mutants. Enhanced sensitivity to peroxides in MSH-deficient M. smegmatis (96, 116, 118), M. tuberculosis (13, 14), and Amycolatopsis mediterranei (17) suggests the presence of an MSH-dependent peroxidase. Increased killing of MSH-deficient M. smegmatis by alkylating agents and certain antibiotics (96, 116, 118) may be associated with MSH-dependent S-transferase activity. A protein identified in M. smegmatis (MSMEG0608) as associated with resistance to diamide is a possible S-transferase (115).
MSH has been associated with the detoxification of methylglyoxal by a glyoxylase I-type activity. This was evidenced by production of S-lactoylmycothiol in extracts of M. smegmatis (http://nobelprize.org/nobel_prizes/chemistry/laureates/2004/rose-autobio.html). This is similar to the situation in GSH-containing bacteria, where glyoxylase I produces S-lactoylglutathione from GSH and methylglyoxal, the toxic side product of several metabolic pathways, including glycerol catabolism (112).
CELLULAR THIOL CONTENT AND REDOX STATUS
MSH is the dominant thiol in most Actinobacteria, with coenzyme A (CoASH) and cysteine occurring at significant but generally lower levels (84). MSH functioning as a thiol buffer has a key role, in addition to the thioredoxin system, in maintaining a highly reducing environment where protein cysteines are retained in their thiol form (22). As with the thiol/disulfide status of GSH in GSH-producing organisms (35), the thiol/disulfide status of MSH (ratio of MSH to low-molecular-weight MSS forms) in Actinobacteria may impact the activity of the cell. For example, the Dos regulon in M. tuberculosis plays a key role in transition to the persistent state (105), and it was recently shown that DosS, which is a heme-containing protein, functions as a redox sensor (61). In GSH-producing organisms, the thiol/disulfide status is important in regulating protein function, with thioredoxin and glutaredoxin playing centrol roles (11, 47). Glutathionylation of proteins, which is the production of protein-SSG disulfides, was recently explored as a regulatory process under both oxidative stress and constitutive conditions (20, 31, 34, 49, 67) and was earlier shown to be important in the dormancy of fungal spores (10, 26). Analogous mycothiolation of proteins could also be important, and there is some indication that mycothiolation of proteins occurs in mycobacteria (15).
MSH is highly reduced in growing M. smegmatis (94) and M. tuberculosis (13) cells and becomes more oxidized as the cells enter stationary phase. The level of total thiols in M. tuberculosis was found to be related to the growth phase and increases almost fourfold from early log to late stationary phase (13). During a 60-day culture in Middlebrook 7H9 plus ADS, the dominant thiol in M. tuberculosis Erdman was MSH (Table 6). The MSH level rose from 18 nmol/109 cells (∼5 mM) to over 70 nmol/109 cells (∼20 mM), with the greatest increase occuring in the transition from log to stationary phase. The oxidized form of MSH (MSS), which is produced in the cell by autoxidation as well as by peroxide reactions, increased to an even greater extent (almost 10-fold) than MSH did, and this is reflected in the 3-fold decrease in the thiol/disulfide ratio of MSH at the late culture times. The second most prominent thiol in M. tuberculosis was CoA, with small amounts of the MSH intermediate Cys-GlcN-Ins detected in the stationary cultures. The thiol form of Cys was not detected in wild-type M. tuberculosis during growth in 7H9 medium, but significant levels of Cys disulfide (0.15 to 0.22 nmol/109 cells) were detected from 17 days onward (13). An MSH level similar to that of M. tuberculosis was reported for M. bovis BCG (17 to 25 nmol/109 cells) grown in 7H9 plus ADS to stationary phase, with an MSH/MSS ratio of 50:1 (141). For comparison with a GSH-containing bacterium, the GSH/glutathione disulfide ratio in E. coli was reported to be 100 (132). In the M. tuberculosis MshD-deficient mutant, the ratio of thiols to disulfides was greatly reduced, with major increases in Cys disulfide and Cys-GlcN-Ins disulfide (13). This resulted in a total thiol-to-total disulfide ratio of 13 during exponential phase in the MshD mutant, which declined to 6 after 60 days of culture. This constitutes a markedly more oxidized redox state than that for a total thiol-to-disulfide ratio of ∼130 for wild-type M. tuberculosis during log phase and of ∼45 after 60 days of culture.
TABLE 6.
Thiol levels in M. tuberculosis Erdman (13)
| Thiol | Level (nmol/109 cells) on day:
|
Level (nmol/109 cells) in M. smegmatis (range) | ||||
|---|---|---|---|---|---|---|
| 3 | 7 | 17 | 33 | 60 | ||
| MSH | 18 ± 1 | 22 ± 2 | 64 ± 5 | 75 ± 5 | 65 ± 11 | 20-28 |
| Cys-GlcN-Ins | ≤0.02 | ≤0.01 | 0.038 ± 0.003 | 0.06 ± 0.03 | 0.09 ± 0.03 | <0.02 |
| Cys | ≤0.06 | ≤0.04 | ≤0.03 | ≤0.03 | ≤0.03 | 0.14-0.46 |
| CoA | ≤0.8 | ≤0.8 | 2.1 ± 0.3 | 4.1 ± 0.2 | 5.1 ± 0.6 | 0.7-5 |
| MSH/MSS | 180 ± 50 | 160 ± 50 | 133 ± 22 | 96 ± 16 | 50 ± 14 | 200-1,300 |
The MSH and CoA levels in M. smegmatis are quite similar to those detected in M. tuberculosis, though the marked increase in MSH during stationary phase and the drop in thiol/disulfide ratio were not observed in M. smegmatis during prolonged culture (94). Another difference is that measurable amounts of Cys thiol have consistently been detected in M. smegmatis.
Thiol levels and, more specifically, MSH levels are generally lower in Actinobacteria other than mycobacteria. For example, Streptomyces coelicolor MSH levels were measured to be 3 to 6 μmol/g dry weight, the Nocardia asteroides level was 3.5 μmol/g dry weight, the Micrococcus luteus level was 3.6 μmol/g dry weight, and the Corynebacterium diphtheriae level was 0.18, compared to a range of 3 to 19 μmol/g dry weight for seven strains of mycobacteria (84, 107). The current exceptions are the high levels of thiols recently observed in some of the marine Actinobacteria (Table 1 (89), with one of the Streptomycetaceae (strain CNQ530) producing a total thiol level of ∼50 μmol/g dry weight.
A novel thiol related to MSH was observed in 13 of 17 marine Actinomycetes strains examined and was the major thiol in two of the “Mar2” species (89). This thiol was purified from strain CNQ703 and was shown to be N-propionyl-des-N-acetyl MSH (PdAMSH). It appeared in stationary-phase cultures at the time of secondary metabolite production. In strain CNQ703, MSH was the major thiol upon entry into stationary phase on day 2, but its level fell 50-fold from days 3 to 10. From days 2 to 3 of culture, a prominent green-black secondary metabolite was produced, and PdAMSH increased 40-fold to become the dominant thiol from days 3 to 10. M. tuberculosis MshD has been reported to utilize propionyl-CoA as well as the preferred substrate acetyl-CoA (148), so PdAMSH is likely also generated by MshD when propionyl-CoA is abundant and acetyl-CoA is limiting. In the production of polyketide secondary metabolites, propionyl-CoA can be utilized as an initiating subunit and in the production of methylmalonyl-CoA extender subunits (152). It is not presently known what function PdAMSH production serves.
In addition to growth phase, MSH levels and thiol/disulfide ratios may also be influenced by environmental conditions, such as oxidative or osmotic stress or nutrient starvation, and can be regulated, at least in Streptomyces, by sigma factors. Exposure of M. bovis BCG to the thiol oxidant diamide caused a reduction in MSH levels and a lowering of the MSH/MSS ratio that was more pronounced when the cells were incubated in 0.9% saline (141). In Streptomyces coelicolor, levels of MSH can be altered during stress and are influenced by multiple sigma factors, as discussed above (63, 104, 106). Thus, a variety of internal as well as environmental factors appear able to influence the amount and redox status of thiols within Actinobacteria.
MSH TURNOVER
The unexpected discoveries that M. smegmatis mutants devoid of MSH are able to actively import MSH and to accumulate MSH to levels at or above the wild-type content opened the way to examine the turnover of MSH (15). When stationary-phase cells of the MshA- and MshC-deficient mutants were loaded with MSH and resuspended in conditioned stationary-phase medium and the MSH content followed with time, it was found that their half-lives for loss of MSH were ∼50 h and ∼16 h, respectively. The degradation of MSH by Mca generates GlcN-Ins, which can be utilized by MshC to regenerate MSH in the MshA-deficient mutant but not in the MshC-deficient mutant, explaining why the lifetime of MSH is longer in the MshA-deficient mutant.
To examine the fate of MSH during turnover, MSH containing either [U-14C]cysteine or [6-3H]glucosamine was enzymatically synthesized, and the radiolabeled MSHs were loaded separately into the MshC-deficient mutant of M. smegmatis in stationary phase. The fate of each radiolabel was followed during 24-h incubations, and the results are summarized in Fig. 17. Immediately after MSH loading, ∼10% of the label was found associated with the pellet fraction, and this increased only slightly over the course of the incubation. Since labels from both Cys and GlcN were found in similar amounts, this suggested that MSH itself had become associated with protein (protein mycothiolation), in a process analogous to protein glutathionylation that has received substantial recent attention (20, 32, 34, 49).
FIG. 17.
Fate of [U-14C]cysteine-labeled (red) and [6-3H]glucosamine-labeled (green) MSH loaded for 3 h into stationary-phase cells of MshC-deficient M. smegmatis, resuspended after being washed in conditioned stationary-phase medium, and incubated for 24 h with sampling. Data presented represent the percent distribution of label after 24 h (15). Pellet, amount of label in resuspended pellet after extraction in hot 50% acetonitrile; void, amount of label eluted in void volume of mBBr-labeled extract separated by reversed-phase C18 HPLC (nonthiol glycolysis and Krebs cycle intermediates); medium, total amount of label in medium; lost, amount of label not recovered in cells or medium.
An identical small portion (16%) of the label from Cys and GlcN remained in cellular MSH, and 1% of the 3H from GlcN was found in GlcN-Ins (Fig. 17). For AcCys-GlcN(6-3H)-Ins, ∼100% of the counts was recovered, with 60% recovered from the culture medium. This is consistent with cleavage of MSH by Mca and hydrolysis of GlcN-Ins to generate glucose and myo-inositol. myo-Inositol is readily metabolized by M. smegmatis and can serve as a sole carbon source. Glucose was proposed to be metabolized through pyruvate and the Krebs cycle, with the 3H label lost to the medium as water. AcCys, produced from MSH by Mca, was deacetylated by an active deacetylase detected in cell extracts (15). The resulting [U-14C]cysteine was converted to pyruvate by Cys desulfhydrase (15, 156), recently identified as Rv3025c in M. tuberculosis (131). Pyruvate was metabolized through the Krebs cycle, with the loss of label as carbon dioxide. The complete metabolism of MSH occurred in stationary-phase cells in stationary-phase conditioned medium, which represents starvation conditions.
These studies show that turnover of MSH in stationary-phase M. smegmatis occurs with a half-life of less than a day and that MSH can serve as a resource for Cys and for other metabolites generated in or from glycolytic or Krebs cycle intermediates. Many enzymes involved in these processes, including the active transporter of MSH, the AcCys deacetylase, and the GlcN-Ins hydrolase, remain to be identified.
MSH DRUG TARGETS IN MYCOBACTERIUM TUBERCULOSIS
Initial interest in MSH as a drug target stemmed from its novel structure and its presence as the dominant thiol in mycobacteria. In evaluating which of the enzymes of MSH metabolism are suitable as potential drug targets, several criteria are important (111). The target must exhibit “drugability,” the ability of the active site to accommodate small-molecule inhibitors (68, 129). Essentiality for growth, as demonstrated in gene knockout experiments, is also a key criterion. However, many drugs that are lethal to growing M. tuberculosis cells fail to kill the bacterium in the latent or dormant state (159). Effectiveness against latent M. tuberculosis is now considered a prime criterion for new tuberculosis drugs (3) but is not readily established prior to drug development. To the extent that MSH is necessary to protect cells against reactive oxygen species and reactive nitrogen intermediates, we might expect it to be important in the dormant state. If the turnover of MSH in dormant M. tuberculosis is comparable to that in the stationary phase of M. smegmatis (half-life of ∼16 to 50 h), then the enzymes of MSH biosynthesis and metabolism are active in the dormant state. Which enzymes are likely to be the most effective targets for drug development?
Inhibition of MSH production simultaneously inactivates all MSH-dependent metabolic processes, so the enzymes of MSH biosynthesis would appear to be the prime targets. The first enzyme of the pathway, MshA, is essential for MSH production in M. smegmatis and is likely to be a drugable target in M. tuberculosis. MshA is a member of the GT-B-fold (5; http://www.cazy.org/fam/acc_GT.html) glycosyltransferases, which include MurG. A screening assay has been reported for E. coli MurG, which also utilizes UDP-GlcNAc, and low-micromolar small-molecule inhibitors were identified, which suggests that MshA may also be a drugable target. The hits were relatively specific for MurG and did not inhibit other UDP-GlcNAc-utilizing glycosyltransferases tested (48).
The essentiality of MshA in M. tuberculosis is unclear. The TraSH method (127) gave conflicting results, indicating that mshA is apparently essential for M. tuberculosis but not for the control organism M. bovis BCG (126; C. Sassetti, personal communication). We were not successful in making a mshA gene knockout without the presence of mshA elsewhere in the genome, which is consistent with the essentiality of MshA (12). Recently, the isolation of an M. tuberculosis mutant in mshA was mentioned in the analysis of M. smegmatis mutants by Rawat et al. (116), but the manuscript reporting details has not yet appeared. Thus, the essentiality of MshA for M. tuberculosis during growth and dormancy is still uncertain.
An oxidation product of UDP-GlcNAc, 2′,3′-dialdehydo-UDP-N-acetylglucosamine (7), is the only reported inhibitor of MshA. This compound inhibited MshA in M. smegmatis extracts with an IC50 of ∼0.2 mM, similar to that with UDP-N-acetylglucosamine 2-epimerase (7, 93). This supports the view that MshA is a drugable target. A limiting factor in the search for drugs directed against MshA has been the inability to readily generate active purified M. tuberculosis MshA.
Little is known about the MSH phosphatase, MshA2. Since no MSH-deficient mutants in a phosphatase were identified during screening of mutant libraries for MSH-deficient mutants, it seems likely that there are multiple phosphatases with overlapping MshA2 activity, making this a poor drug target.
MSH deacetylase, MshB, is a close homolog of Mca, as previously discussed, and is thought to be a poor drug target because mutants in M. smegmatis (117) and M. tuberculosis (14) produce significant amounts of MSH. The compensating deacetylase activity for MshB mutants in M. tuberculosis is likely to derive from the homologous Mca or Rv0323c protein. The Bewley group recently reported synthetic small-molecule inhibitors of MshB developed around the cyclohexyl thioglycoside analog of GlcN-Ins (Fig. 18) (76), originally introduced by Knapp et al. (58). With Mca, the substrate analog of MSmB (Fig. 18, R1) exhibits half the activity of MSmB (the most active substrate of Mca). Of some 23 analogs with various R groups based upon natural product inhibitors of Mca, the most active inhibitor was the R2 analog (Fig. 18), which has IC50s of 7 μM and 33 μM for MshB and Mca, respectively (76). The effects of this inhibitor on MSH levels and viability in mycobacteria have not been reported.
FIG. 18.
The cyclohexyl thioglycoside substrate analog of MSmB (R1) is an efficient substrate of Mca, and the nonsubstrate analog (R2) is an inhibitor of Mca and MshB.
MshC is expected to be the most promising drug target to block MSH biosynthesis. MshC is required in the synthesis of MSH from Ins-P and also in the recycling pathway that regenerates MSH from GlcN-Ins generated by the action of Mca (Fig. 10). The ample binding sites required for ATP, Cys, and GlcN-Ins suggest that MshC should be a drugable target. Also, MshC is a close homolog of Cys-tRNA synthase, which along with the other tRNA synthases is a validated drug target for Staphylococcus aureus (111). In that study, drug target validation consisted of essential genes coding for drugable proteins where the bacterial enzyme was significantly different in structure from the human ortholog (111). Several lines of evidence indicate that mshC is essential in M. tuberculosis, including results from TraSH analysis (126) and those from directed gene knockout studies (124). If the turnover time of MSH in the stationary-phase M. smegmatis MshC mutant (half-life of ∼16 h) is indicative of that in dormant M. tuberculosis, then inhibition of MSH biosynthesis and MshC in particular could be on a time scale consistent with shortening tuberculosis chemotherapy.
Initial efforts to identify inhibitors of MshC used a high-throughput screen with a small library selected for ATP binding proteins (95). One hit, designated NTF1836 (Fig. 19), was found to have an IC50 of ∼100 μM in the MshC assay and produced a 60% decrease in the MSH content of M. smegmatis at 30 μM (27, 95). However, it also inhibited M. smegmatis growth, indicating that it has targets other than MshC, since M. smegmatis does not require MSH for growth. A likely alternative target is cysteinyl-tRNA synthetase. This could be an advantage, since development of resistance to a drug that has two essential targets is highly unlikely (51). However, it necessitates that the inhibition of human cysteinyl-tRNA synthetase be sufficiently less to provide a usable therapeutic window.
FIG. 19.
Structure of MshC inhibitor NTF1836.
MshD is also likely to be a drugable target considering the substantial surfaces involved in the binding of the substrate moieties (148). The mshD mutant produces a small amount of MSH as well as a substantial level of formyl-Cys-GlcN-Ins, and the mshD gene is not essential for growth of M. tuberculosis (13). This indicates that MshD, like MshB, is not a good target to eliminate MSH biosynthesis. However, in a survey of an M. tuberculosis transposon mutant library for survival in unactivated and activated mouse macrophages, the mshD mutant was found to be one of the most sensitive of the more than 2,000 mutants characterized (120). This further demonstrates that under conditions of oxidative challenge, the MSH protective systems play an important role. A drug inhibiting MshD would be useful in the further evaluation of this target, but none have been reported.
Turning now to the enzymes of MSH metabolism, we first consider the MSH disulfide reductase Mtr. Like the related enzymes GSH reductase (2) and trypanothione reductase (75), for which inhibitors are known, Mtr is expected to be a drugable target. However, whether Mtr is essential for growth of M. tuberculosis is unclear. The TraSH method of Sassetti et al. (126) identified mtr as essential for growth of M. tuberculosis, whereas a transposon library generated by McAdam et al. (72) produced a single viable M. tuberculosis mutant in mtr. Treatment of M. tuberculosis H37Rv with 10 μM antisense oligonucleotide to Mtr RNA produced no inhibition of growth at 4 days but a two-thirds inhibition at 6 days of growth (44). An effort to produce a specific gene knockout of mtr is needed to provide a clear indication of the essentiality of this gene.
Benzo- and naphthoquinone derivatives are reduced by Mtr and can serve as “subversive substrates” that promote redox cycling (109). A series of natural and synthetic naphthoquinone derivatives related to 7-methyljuglone (Fig. 20, S1) was prepared and examined for substrate activity with Mtr and for toxicity to M. tuberculosis by Mahapatra et al. (70). Several homologs of S1 (S2 to S4) exhibited comparable toxicities toward M. tuberculosis, but the correlation with inhibition of Mtr was weak, suggesting that the compounds may also target other enzymes. No specific inhibitors of Mtr have been reported as yet.
FIG. 20.
Quinone-subversive substrates of M. tuberculosis MSSM reductase Mtr (70).
MSH S-conjugate amidase, or Mca, plays an important role in MSH-dependent detoxification. As one of the earliest enzymes of MSH metabolism identified and readily assayed, it has received considerable attention as a target for inhibitor development. It was not identified as essential for in vitro growth of M. tuberculosis in the TraSH survey (126), but the mca gene is upregulated in the enduring hypoxic response believed to be important in M. tuberculosis latency (122). A specific gene knockout of mca in M. smegmatis exhibited increased sensitivity to a variety of toxins and antibiotics (119), suggesting that inhibition of Mca might enhance the actions of other antitubercular drugs. Novel bromotyrosine alkaloids from an Australian sponge, Oceanapia sp., were found to be low-micromolar inhibitors of Mca (97, 98, 100). One scaffold is particularly accessible for chemical synthesis (30, 56), and synthetic analogs of the natural product, especially EXEG1706 (Fig. 14), show good activity against mycobacteria as well as methicillin- and vancomycin-resistant Staphylococcus aureus (113). This was somewhat surprising, because low-GC bacteria, including Staphylococcus, do not have MSH metabolism. However, MSH S-conjugate amidase and the other M. tuberculosis paralogs, MshB and the uncharacterized protein Rv0323c, belong to a large family of zinc metalloproteinases with homologs in eukaryotes (143, 154), low-GC gram-positive bacteria, such as the staphylococci and the bacilli (25, 139), and Deinococcus-Thermus (42). Since the bromotyrosine natural products (100) and analogs contain the zinc-binding hydroxamate moiety, it is not difficult to envision inhibition of this large family of proteins by inhibitors such as EXEG1706 (Fig. 14) (113).
The other identified MSH-dependent metabolic enzyme, MscR (MSNO reductase/formaldehyde dehydrogenase; Rv2259), is expected to be a drugable target based on the sizes of its substrates (MSNO and NADH). The TraSH survey indicated that this enzyme is essential in M. tuberculosis, but this has not yet been confirmed by attempted construction of a specific gene knockout. It is not apparent why an NO detoxifying enzyme should be essential under conditions where there is not any NO stress. Perhaps the formaldehyde reductase activity of AdhE2/MscR (Fig. 14) is more important during in vitro growth than the MSNO reductase activity. MscR is a potentially important target but requires further evaluation.
MSH structural analogs can serve as subversive substrates or as inhibitors for MSH biosynthetic or metabolism enzymes. Inhibitors of MshB were synthesized as the carbo-analogs G1 and G2 of GlcNAc-Ins, with changes to the C-2 nitrogen (Fig. 21) (33). Both analogs inhibited MSH biosynthesis by ∼50% at 200 μg/ml in M. smegmatis. It has been reported that M. smegmatis will take up GlcNAc-Ins (96) and MSH (15) from the medium, so it is not surprising that the analogs G1 and G2 are effective at inhibiting MSH biosynthesis.
FIG. 21.
Synthetic nonreacting analogs of the substrate (GlcNAc-Ins) for the MSH deacetylase MshB in which the NH residue is replaced by CH2 (G2) and the corresponding alcohol (G1); both G1 and G2 inhibit MSH biosynthesis in M. smegmatis, about twofold, at 200 μg/ml (33).
Substrate analogs for Mca have been synthesized based on the incorporation of quinic acid in place of myo-d-inositol, which has given an efficient parallel synthesis of several analogs (77). Most of the analogs have activity against Mca at 50 μM, but no data were provided on mycobacteria. Other Mca inhibitors have been produced with the S-cyclohexyl group substituted for myo-d-inositol, as illustrated by the bimane derivative of an MSH S-cyclohexyl thioglycoside, which was half as active as the bimane derivative of MSH with Mca (58). A series of analogs based on the aminotriol scaffold (Fig. 18) (R = variable), an analog of GlcN-Ins, were reported by Knapp et al. (57) and included four compounds with bactericidal activity with M. tuberculosis H37Rv at 6.25 μg/ml. No information is provided as to which of the MSH biosynthesis or metabolism genes were inhibited by these MSH analogs.
Validation of MSH as a drug target is provided by inhibitors like the ones produced by Knapp and coworkers (57) and like NTF1836, where MSH enzymes are specifically targeted and bactericidal lead compounds emerge.
CONCLUSIONS
MSH is the major low-molecular-weight thiol and redox buffer in the Actinobacteria, a very large and geographically diverse phylogenetic clade in the eubacteria. Substantial progress has been made in elucidating its biochemistry and microbiology. MSH, like GSH, is a stabilized form of cysteine, which is required by all organisms for protein and CoA synthesis. MSH has a unique amide bond (Cys-GlcN), as does GSH (γ-Glu-Cys), not subject to cleavage by normal proteases but selectively cleaved by specialized enzymes, namely, MSH S-conjugate amidase for MSH and γ-glutamyl transpeptidase for GSH. Cysteine is problematic in cells due to its rapid oxidation and its reversible S-acylation in the presence of acyl-CoA coupled with rearrangement to stable N-acyl derivatives. Blocking of the amino group by the γ-glutamyl and acetyl groups in GSH and MSH, respectively, limits heavy metal binding and accompanying autoxidation as well as rearrangement of S-acyl derivatives. The γ-glutamyl moiety of GSH and the GlcN-Ins moiety of MSH make these thiols hydrophilic and prevent the loss of these cysteine reserves from the cell.
The pathway of MSH biosynthesis has been established, and the enzymes involved include a glycosyltransferase (MshA), a phosphatase (MshA2), a deacetylase (MshB), an ATP-dependent ligase (MshC), and an acetyltransferase (MshD). The genes encoding all of the enzymes have been identified, with the exception of mshA2. MshA and MshC are essential for MSH biosynthesis. Mutants in MSH biosynthesis genes have been produced in the soil-dwelling Actinobacteria, which demonstrates that MSH is not essential in these organisms that generally have large genomes. Mutants in mshA and mshC have not been reported for M. tuberculosis Erdman, indicating that MSH is essential for M. tuberculosis. A difference in these two types of Actinobacteria is the unique adaptation to life in the human host, whereas soil-dwelling Actinobacteria grow in diverse environments which require a more varied genome. One advantage of the generally larger genome size of soil Actinobacteria is the antioxidant activities driven by the large network of sigma factors, including sigR and sigB, which can compensate for the absence of MSH.
Mutants deficient in MSH exhibit increased sensitivity to various toxins and antibiotics, indicating that MSH is involved in their detoxification. Several key enzymes of MSH-dependent metabolism have been identified and have a wide distribution in Actinobacteria. These include mycothione reductase (Mtr), which maintains MSH in a highly reduced state; MSH S-conjugate amidase (Mca), which cleaves MSH and its S-conjugates; and the formaldehyde dehydrogenase/MSNO reductase MscR, which detoxifies H2CO and NO. An MSH-dependent maleylpyruvate isomerase appears to have a more limited distribution. A variety of additional MSH-dependent enzymes associated with MSH-dependent detoxification are likely to be identified, including one or more MSH S-transferases, MSH peroxidases, and mycoredoxins, all of which would be analogs of corresponding enzymes in GSH metabolism. Given the genomic size and metabolic versatility of many Actinobacteria, it is quite possible that additional unique MSH-dependent enzymatic processes will be uncovered.
A driving force behind much of the early research on MSH has been to identify targets for potential drugs to treat tuberculosis. Promising targets include MshA, MshC, Mtr, and MscR. MshD and MshB/Mca may also have some potential. Initial results with an inhibitor of MshC suggest that it can be effective against dormant tuberculosis.
Despite the rapid growth in MSH research in the last 5 years, many of its functions and metabolic enzymes have yet to be discovered. An exciting new area of MSH metabolism is the detoxification of aromatic contaminants of soil by bioremediation organisms, as illustrated by the MSH-dependent degradation of hydroxybenzoate by C. glutamicum (29). Of particular interest are the larger-genome bioremediation Actinobacteria, such as the recently sequenced organism R. jostii RHA-1, which is a “catabolic powerhouse” well known for biodegradation of xenobiotics (74). This organism has over 1,000 oxidoreductases and may utilize MSH as a cofactor in many of its bioconversions. Research on MSH-dependent detoxification and bioremediation will easily inspire biochemists for years before a full understanding of the scope of MSH metabolism is known in detail.
Acknowledgments
We are indebted to the National Institutes of Health for research funding under grants AI49174 and AI72133 and to the National Science Foundation for support under grant MCB-0235705.
We thank Chris Hamilton for critical comments on the manuscript and for helpful discussions. We thank Christopher Sassetti for permission to use unpublished data. We thank Jossef Av-Gay, Mamta Rawat, and John Blanchard for many informative discussions on MSH biochemistry.
REFERENCES
- 1.Anderberg, S. J., G. L. Newton, and R. C. Fahey. 1998. Mycothiol biosynthesis and metabolism: cellular levels of potential intermediates in the biosynthesis and degradation of mycothiol. J. Biol. Chem. 27330391-30397. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. E. 1998. Glutathione: an overview of biosynthesis and modulation. Chem. Biol. Interact. 111-1121-14. [DOI] [PubMed] [Google Scholar]
- 3.Barry, C. E., III, and K. Duncan. 2004. Tuberculosis—strategies towards anti-infectives for a chronic disease. Drug Discov. Today 1491-496. [Google Scholar]
- 4.Baulard, A. R., J. C. Betts, J. Engohang-Ndong, S. Quan, R. A. McAdam, P. J. Brennan, C. Locht, and G. S. Besra. 2000. Activation of the pro-drug ethionamide is regulated in mycobacteria. J. Biol. Chem. 27528326-28331. [DOI] [PubMed] [Google Scholar]
- 5.Berg, S., D. Kaur, M. Jackson, and P. J. Brennan. 2007. The glycosyltransferases of Mycobacterium tuberculosis—roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology 1735R-56R. [DOI] [PubMed] [Google Scholar]
- 6.Bhave, D. P., W. B. Muse III, and K. S. Carroll. 2007. Drug targets in mycobacterial sulfur metabolism. Infect. Disord. Drug Targets 7140-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blume, A., H. Chen, W. Reutter, R. R. Schmidt, and S. Hinderlich. 2002. 2′,3′-Dialdehydo-UDP-N-acetylglucosamine inhibits UDP-N-acetylglucosamine 2-epimerase, the key enzyme of sialic acid biosynthesis. FEBS Lett. 521127-132. [DOI] [PubMed] [Google Scholar]
- 8.Board, P. G., and M. W. Anders. 2005. Human glutathione transferase zeta. Methods Enzymol. 40161-77. [DOI] [PubMed] [Google Scholar]
- 9.Bornemann, C., M. A. Jardine, H. S. C. Spies, and D. J. Steenkamp. 1997. Biosynthesis of mycothiol: elucidation of the sequence of steps in Mycobacterium smegmatis. Biochem. J. 325623-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brody, S., S. D. Mikolajczyk, and R. C. Fahey. 1983. Levels of sulfhydryls and disulfides in proteins from Neurospora crassa conidia and mycelia. J. Bacteriol. 156703-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchanan, B. B., and Y. Balmer. 2005. Redox regulation: a broadening horizon. Annu. Rev. Plant Biol. 56187-220. [DOI] [PubMed] [Google Scholar]
- 12.Buchmeier, N., and R. C. Fahey. 2006. The mshA gene encoding the glycosyltransferase of mycothiol biosynthesis is essential in Mycobacterium tuberculosis Erdman. FEMS Microbiol. Lett. 26474-79. [DOI] [PubMed] [Google Scholar]
- 13.Buchmeier, N. A., G. L. Newton, and R. C. Fahey. 2006. A mycothiol synthase mutant of Mycobacterium tuberculosis has an altered thiol-disulfide content and limited tolerance to stress. J. Bacteriol. 1886245-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchmeier, N. A., G. L. Newton, T. Koledin, and R. C. Fahey. 2003. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol. Microbiol. 471723-1732. [DOI] [PubMed] [Google Scholar]
- 15.Bzymek, K. P., G. L. Newton, P. Ta, and R. C. Fahey. 2007. Mycothiol import by Mycobacterium smegmatis and function as a resource for metabolic precursors and energy production. J. Bacteriol. 1896796-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chasseaud, L. F. 1976. Conjugation with glutathione and mercapturic acid excretion, p. 77-114. In I. M. Arias and W. B. Jakoby (ed.), Glutathione: metabolism and function. Raven Press, New York, NY.
- 17.Chen, L., H. Yu, Y. Lu, and W. Jiang. 2005. Cloning and preliminary characterization of lh3 gene encoding a putative acetyltransferase from a rifamycin SV-producing strain Amycolatopsis mediterranei. Biotechnol. Lett. 271129-1134. [DOI] [PubMed] [Google Scholar]
- 18.Chen, L., C. Zhou, H. Yang, and M. F. Roberts. 2000. Inositol-1-phosphate synthase from Archaeoglobus fulgidus is a class II aldolase. Biochemistry 3912415-12423. [DOI] [PubMed] [Google Scholar]
- 19.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 20.Dalle-Donne, I., R. Rossi, D. Giustarini, R. Colombo, and A. Milzani. 2007. S-Glutathionylation in protein redox regulation. Free Radic. Biol. Med. 43883-898. [DOI] [PubMed] [Google Scholar]
- 21.delCardayré, S. B., K. P. Stock, G. L. Newton, R. C. Fahey, and J. E. Davies. 1998. Coenzyme A disulfide reductase: purification and characterization of the enzyme component of the primary thiol/disulfide redox system in Staphylococcus aureus. J. Biol. Chem. 2735744-5751. [DOI] [PubMed] [Google Scholar]
- 22.den Hengst, C. D., and M. J. Buttner. Redox control in actinobacteria. Biochim. Biophys. Acta, in press. [DOI] [PubMed]
- 23.Dyda, F., D. C. Klein, and A. B. Hickman. 2000. GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 2981-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eggeling, L., and H. Sahm. 1985. The formaldehyde dehydrogenase of Rhodococcus erythropolis, a trimeric enzyme requiring a cofactor and active with alcohols. Eur. J. Biochem. 150129-134. [DOI] [PubMed] [Google Scholar]
- 25.Fadouloglou, V. E., A. Deli, N. M. Glykos, E. Psylinakis, V. Bouriotis, and M. Kokkinidis. 2007. Crystal structure of the BcZBP, a zinc-binding protein from Bacillus cereus. FEBS J. 2743044-3054. [DOI] [PubMed] [Google Scholar]
- 26.Fahey, R. C., S. Brody, and S. D. Mikolajczyk. 1975. Changes in the glutathione thiol-disulfide status of Neurospora crassa conidia during germination and aging. J. Bacteriol. 121144-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahey, R. C., G. L. Newton, P. Ta, and N. Buchmeier. March 2008. Dibenzothiazepinones and oxides thereof as inhibitors of bacterial MshC biosynthesis enzymes, homologs thereof, and inhibitor identification methods. U.S. patent WO 2008036139.
- 28.Fan, F., A. Luxenburger, G. F. Painter, and J. S. Blanchard. 2007. Steady-state and pre-steady-state kinetic analysis of Mycobacterium smegmatis cysteine ligase (MshC). Biochemistry 4611421-11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng, J., Y. Che, J. Milse, Y. J. Yin, L. Liu, C. Ruckert, X. H. Shen, S. W. Qi, J. Kalinowski, and S. J. Liu. 2006. The gene ncgl2918 encodes a novel maleylpyruvate isomerase that needs mycothiol as cofactor and links mycothiol biosynthesis and gentisate assimilation in Corynebacterium glutamicum. J. Biol. Chem. 28110778-10785. [DOI] [PubMed] [Google Scholar]
- 30.Fetterolf, B., and C. A. Bewley. 2004. Synthesis of a bromotyrosine-derived natural product inhibitor of mycothiol-S-conjugate amidase. Bioorg. Med. Chem. Lett. 143785-3788. [DOI] [PubMed] [Google Scholar]
- 31.Fratelli, M., L. O. Goodwin, U. A. Orom, S. Lombardi, R. Tonelli, M. Mengozzi, and P. Ghezzi. 2005. Gene expression profiling reveals a signaling role of glutathione in redox regulation. Proc. Natl. Acad. Sci. USA 10213998-14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallogly, M. M., and J. J. Mieyal. 2007. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 7381-391. [DOI] [PubMed] [Google Scholar]
- 33.Gammon, D. W., R. Hunter, D. J. Steenkamp, and T. T. Mudzunga. 2003. Synthesis of 2-deoxy-2-C-alkylglucosides of myo-inositol as possible inhibitors of a N-deacetylase enzyme in the biosynthesis of mycothiol. Bioorg. Med. Chem. Lett. 132045-2049. [DOI] [PubMed] [Google Scholar]
- 34.Ghezzi, P. 2005. Regulation of protein function by glutathionylation. Free Radic. Res. 39573-580. [DOI] [PubMed] [Google Scholar]
- 35.Gilbert, H. F. 1995. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 2518-28. [DOI] [PubMed] [Google Scholar]
- 36.Grillo, M. P., and L. Z. Benet. 2002. Studies on the reactivity of clofibryl-S-acyl-CoA thioester with glutathione in vitro. Drug Metab. Dispos. 3055-62. [DOI] [PubMed] [Google Scholar]
- 37.Gu, X., M. Chen, H. Shen, X. Jiang, Y. Huang, and H. Wang. 2006. Rv2131c gene product: an unconventional enzyme that is both inositol monophosphatase and fructose-1,6-bisphosphatase. Biochem. Biophys. Res. Commun. 339897-904. [DOI] [PubMed] [Google Scholar]
- 38.Gutierrez-Lugo, M. T., G. L. Newton, R. C. Fahey, and C. A. Bewley. 2006. Cloning, expression and rapid purification of active recombinant mycothiol ligase as B1 immunoglobulin binding domain of streptococcal protein G, glutathione-S-transferase and maltose binding protein fusion proteins in Mycobacterium smegmatis. Protein Expr. Purif. 50128-136. [DOI] [PubMed] [Google Scholar]
- 38a.Gutierrez-Lugo, M. T., and C. A. Bewley. 2006. Abstr. 47th Annu. Meet. Am. Soc. Pharmacognosy, abstr. P-135.
- 39.Hagedorn, S. R., G. Bradley, and P. J. Chapman. 1985. Glutathione-independent isomerization of maleylpyruvate by Bacillus megaterium and other gram-positive bacteria. J. Bacteriol. 163640-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hand, C. E., F. I. Auzanneau, and J. F. Honek. 2006. Conformational analyses of mycothiol, a critical intracellular glycothiol in Mycobacteria. Carbohydr. Res. 3411164-1173. [DOI] [PubMed] [Google Scholar]
- 41.Hand, C. E., and J. F. Honek. 2005. Biological chemistry of naturally occurring thiols of microbial and marine origin. J. Nat. Prod. 68293-308. [DOI] [PubMed] [Google Scholar]
- 42.Handa, N., T. Terada, Y. Kamewari, H. Hamana, J. R. Tame, S. Y. Park, K. Kinoshita, M. Ota, H. Nakamura, S. Kuramitsu, M. Shirouzu, and S. Yokoyama. 2003. Crystal structure of the conserved protein TT1542 from Thermus thermophilus HB8. Protein Sci. 121621-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatzios, S. K., A. T. Iavarone, and C. R. Bertozzi. 2008. Rv2131c from Mycobacterium tuberculosis is a CysQ 3′-phosphoadenosine-5′-phosphatase. Biochemistry 475823-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayward, D., I. Wiid, and P. van Helden. 2004. Differential expression of mycothiol pathway genes: are they affected by antituberculosis drugs? IUBMB Life 56131-138. [DOI] [PubMed] [Google Scholar]
- 45.Held, K. D., and J. E. Biaglow. 1994. Mechanisms for the oxygen radical-mediated toxicity of various thiol-containing compounds in cultured mammalian cells. Radiat. Res. 13915-23. [PubMed] [Google Scholar]
- 46.Hernick, M., and C. A. Fierke. 2005. Zinc hydrolases: the mechanisms of zinc-dependent deacetylases. Arch. Biochem. Biophys. 43371-84. [DOI] [PubMed] [Google Scholar]
- 47.Holmgren, A., C. Johansson, C. Berndt, M. E. Lonn, C. Hudemann, and C. H. Lillig. 2005. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem. Soc. Trans. 331375-1377. [DOI] [PubMed] [Google Scholar]
- 48.Hu, Y., J. S. Helm, L. Chen, C. Ginsberg, B. Gross, B. Kraybill, K. Tiyanont, X. Fang, T. Wu, and S. Walker. 2004. Identification of selective inhibitors for the glycosyltransferase MurG via high-throughput screening. Chem. Biol. 11703-711. [DOI] [PubMed] [Google Scholar]
- 49.Hurd, T. R., N. J. Costa, C. C. Dahm, S. M. Beer, S. E. Brown, A. Filipovska, and M. P. Murphy. 2005. Glutathionylation of mitochondrial proteins. Antioxid. Redox Signal. 7999-1010. [DOI] [PubMed] [Google Scholar]
- 50.Jardine, M. A., H. S. Spies, C. M. Nkambule, D. W. Gammon, and D. J. Steenkamp. 2002. Synthesis of mycothiol, 1d-1-O-(2-[N-acetyl-l-cysteinyl]amino-2-deoxy-alpha-d-glucopyranosyl)-myo-inositol, principal low molecular mass thiol in the actinomycetes. Bioorg. Med. Chem. 10875-881. [DOI] [PubMed] [Google Scholar]
- 51.Jenwitheesuk, E., J. A. Horst, K. L. Rivas, W. C. Van Voorhis, and R. Samudrala. 2008. Novel paradigms for drug discovery: computational multitarget screening. Trends Pharmacol. Sci. 2962-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Kramer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Puhler, D. A. Rey, C. Ruckert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 1045-25. [DOI] [PubMed] [Google Scholar]
- 53.Kamtekar, S., W. D. Kennedy, J. Wang, C. Stathopoulos, D. Soll, and T. A. Steitz. 2003. The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases. Proc. Natl. Acad. Sci. USA 1001673-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang, J. G., M. S. Paget, Y. J. Seok, M. Y. Hahn, J. B. Bae, J. S. Hahn, C. Kleanthous, M. J. Buttner, and J. H. Roe. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 184292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 998330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kende, A. S., J. Lan, and J. Fan. 2004. Total synthesis of a dibromotyrosine alkaloid inhibitor of mycothiol S-conjugate amidase. Tetrahedron Lett. 45133-135. [Google Scholar]
- 57.Knapp, S., B. Amorelli, E. Darout, C. C. Ventocilla, L. M. Goldman, R. A. Huhn, and E. C. Minnihan. 2005. A family of mycothiol analogues. J. Carbohydr. Chem. 24103-130. [Google Scholar]
- 58.Knapp, S., S. Gonzalez, D. S. Myers, L. L. Eckman, and C. A. Bewley. 2002. Shortcut to mycothiol analogues. Org. Lett. 44337-4339. [DOI] [PubMed] [Google Scholar]
- 59.Koledin, T., G. L. Newton, and R. C. Fahey. 2002. Identification of the mycothiol synthase gene (mshD) encoding the acetyltransferase producing mycothiol in actinomycetes. Arch. Microbiol. 178331-337. [DOI] [PubMed] [Google Scholar]
- 60.Kormann, E., H. Pape, and D. Münster. 1984. Modification of the antibiotic granaticin A by the producing organism, Streptomyces violaceoruber Tü, p. 105-109. In 3rd European Congress of Biotechnology, vol. 1. Verlag Chemie, Weinheim, Germany. [Google Scholar]
- 61.Kumar, A., J. C. Toledo, R. P. Patel, J. R. Lancaster, Jr., and A. J. Steyn. 2007. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc. Natl. Acad. Sci. USA 10411568-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Larkin, M. J., L. A. Kulakov, and C. C. Allen. 2005. Biodegradation and Rhodococcus—masters of catabolic versatility. Curr. Opin. Biotechnol. 16282-290. [DOI] [PubMed] [Google Scholar]
- 63.Lee, E. J., N. Karoonuthaisiri, H. S. Kim, J. H. Park, C. J. Cha, C. M. Kao, and J. H. Roe. 2005. A master regulator sigma B governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol. Microbiol. 571252-1264. [DOI] [PubMed] [Google Scholar]
- 64.Lee, S., H. Bergeron, P. C. Lau, and J. P. Rosazza. 2007. Thiols in nitric oxide synthase-containing Nocardia sp. strain NRRL 5646. Appl. Environ. Microbiol. 733095-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee, S., and J. P. Rosazza. 2004. First total synthesis of mycothiol and mycothiol disulfide. Org. Lett. 6365-368. [DOI] [PubMed] [Google Scholar]
- 66.Li, C., M. P. Grillo, and L. Z. Benet. 2003. In vitro studies on the chemical reactivity of 2,4-dichlorophenoxyacetyl-S-acyl-CoA thioester. Toxicol. Appl. Pharmacol. 187101-109. [DOI] [PubMed] [Google Scholar]
- 67.Lind, C., R. Gerdes, Y. Hamnell, I. Schuppe-Koistinen, H. B. von Lowenhielm, A. Holmgren, and I. A. Cotgreave. 2002. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Arch. Biochem. Biophys. 406229-240. [DOI] [PubMed] [Google Scholar]
- 68.Lipinski, C. A. 2000. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 44235-249. [DOI] [PubMed] [Google Scholar]
- 69.Mahadevan, J., G. M. Nicholas, and C. A. Bewley. 2003. Solution conformations of mycothiol bimane, 1-d-GlcNAc-alpha-(1→1)-d-myo-Ins and 1-d-GlcNAc-alpha-(1→1)-l-myo-Ins. J. Org. Chem. 683380-3386. [DOI] [PubMed] [Google Scholar]
- 70.Mahapatra, A., S. P. Mativandlela, B. Binneman, P. B. Fourie, C. J. Hamilton, J. J. Meyer, F. van der Kooy, P. Houghton, and N. Lall. 2007. Activity of 7-methyljuglone derivatives against Mycobacterium tuberculosis and as subversive substrates for mycothiol disulfide reductase. Bioorg. Med. Chem. 157638-7646. [DOI] [PubMed] [Google Scholar]
- 71.Maynes, J. T., C. Garen, M. M. Cherney, G. L. Newton, D. Arad, Y. Av-Gay, R. C. Fahey, and M. N. James. 2003. The crystal structure of 1-d-myo-inosityl 2-acetamido-2-deoxy-alpha-d-glucopyranoside deacetylase (MshB) from Mycobacterium tuberculosis reveals a zinc hydrolase with a lactate dehydrogenase fold. J. Biol. Chem. 27847166-47170. [DOI] [PubMed] [Google Scholar]
- 72.McAdam, R. A., S. Quan, D. A. Smith, S. Bardarov, J. C. Betts, F. C. Cook, E. U. Hooker, A. P. Lewis, P. Woollard, M. J. Everett, P. T. Lukey, G. J. Bancroft, W. R. Jacobs, Jr., and K. Duncan. 2002. Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology 1482975-2986. [DOI] [PubMed] [Google Scholar]
- 73.McCarthy, A. A., N. A. Peterson, R. Knijff, and E. N. Baker. 2004. Crystal structure of MshB from Mycobacterium tuberculosis, a deacetylase involved in mycothiol biosynthesis. J. Mol. Biol. 3351131-1141. [DOI] [PubMed] [Google Scholar]
- 74.McLeod, M. P., R. L. Warren, W. W. Hsiao, N. Araki, M. Myhre, C. Fernandes, D. Miyazawa, W. Wong, A. L. Lillquist, D. Wang, M. Dosanjh, H. Hara, A. Petrescu, R. D. Morin, G. Yang, J. M. Stott, J. E. Schein, H. Shin, D. Smailus, A. S. Siddiqui, M. A. Marra, S. J. Jones, R. Holt, F. S. Brinkman, K. Miyauchi, M. Fukuda, J. E. Davies, W. W. Mohn, and L. D. Eltis. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 10315582-15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meiering, S., O. Inhoff, J. Mies, A. Vincek, G. Garcia, B. Kramer, M. Dormeyer, and R. L. Krauth-Siegel. 2005. Inhibitors of Trypanosoma cruzi trypanothione reductase revealed by virtual screening and parallel synthesis. J. Med. Chem. 484793-4802. [DOI] [PubMed] [Google Scholar]
- 76.Metaferia, B. B., B. J. Fetterolf, S. Shazad-Ul-Hussan, M. Moravec, J. A. Smith, S. Ray, M. T. Gutierrez-Lugo, and C. A. Bewley. 2007. Synthesis of natural product-inspired inhibitors of Mycobacterium tuberculosis mycothiol-associated enzymes: the first inhibitors of GlcNAc-Ins deacetylase. J. Med. Chem. 506326-6336. [DOI] [PubMed] [Google Scholar]
- 77.Metaferia, B. B., S. Ray, J. A. Smith, and C. A. Bewley. 2007. Design and synthesis of substrate-mimic inhibitors of mycothiol-S-conjugate amidase from Mycobacterium tuberculosis. Bioorg. Med. Chem. Lett. 17444-447. [DOI] [PubMed] [Google Scholar]
- 78.Meyer, M., W. Keller-Schierlein, S. Megahed, and A. Segre. 1986. Stoffwechselprodukte van Mikroorganismen. Schwefelhaltige ana-Verbindungen des Naphthomycin-Typus. Helf. Chim. Acta 691356-1364. [Google Scholar]
- 79.Miller, C. C., M. Rawat, T. Johnson, and Y. Av-Gay. 2007. Innate protection of Mycobacterium smegmatis against the antimicrobial activity of nitric oxide is provided by mycothiol. Antimicrob. Agents Chemother. 513364-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Misset-Smits, M., P. W. van Ophem, S. Sakuda, and J. A. Duine. 1997. Mycothiol, 1-O-(2′-[N-acetyl-l-cysteinyl]amido-2′-deoxy-α-d-glucopyranosyl)-d-myo-inositol, is the factor of NAD/factor-dependent formaldehyde dehydrogenase. FEBS Lett. 409221-222. [DOI] [PubMed] [Google Scholar]
- 81.Movahedzadeh, F., D. A. Smith, R. A. Norman, P. Dinadayala, J. Murray-Rust, D. G. Russell, S. L. Kendall, S. C. Rison, M. S. McAlister, G. J. Bancroft, N. Q. McDonald, M. Daffe, Y. Av-Gay, and N. G. Stoker. 2004. The Mycobacterium tuberculosis ino1 gene is essential for growth and virulence. Mol. Microbiol. 511003-1014. [DOI] [PubMed] [Google Scholar]
- 82.Mukhopadhyay, T., C. M. Franco, G. C. Reddy, B. N. Ganguli, and H. W. Fehlhaber. 1985. A new ansamycin antibiotic, naphthomycin H from a Streptomyces species Y-83,40369. J. Antibiot. (Tokyo) 38948-951. [DOI] [PubMed] [Google Scholar]
- 83.Newberry, K. J., Y.-M. Hou, and J. J. Perona. 2002. Structural origins of amino acid selection without editing by cysteinyl-tRNA synthetase. EMBO J. 212778-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. delCardayré, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 1781990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newton, G. L., Y. Av-Gay, and R. C. Fahey. 2000. N-Acetyl-1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside deacetylase (MshB) is a key enzyme in mycothiol biosynthesis. J. Bacteriol. 1826958-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Newton, G. L., Y. Av-Gay, and R. C. Fahey. 2000. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry 3910739-10746. [DOI] [PubMed] [Google Scholar]
- 87.Newton, G. L., C. A. Bewley, T. J. Dwyer, R. Horn, Y. Aharonowitz, G. Cohen, J. Davies, D. J. Faulkner, and R. C. Fahey. 1995. The structure of U17 isolated from Streptomyces clavuligerus and its properties as an antioxidant thiol. Eur. J. Biochem. 230821-825. [DOI] [PubMed] [Google Scholar]
- 88.Newton, G. L., and R. C. Fahey. 2002. Mycothiol biochemistry. Arch. Microbiol. 178388-394. [DOI] [PubMed] [Google Scholar]
- 89.Newton, G. L., P. R. Jensen, J. B. MacMillan, W. Fenical, and R. C. Fahey. An N-acyl homolog of mycothiol is produced in marine actinomycetes. Arch. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 90.Newton, G. L., M. Ko, P. Ta, Y. Av-Gay, and R. C. Fahey. 2006. Purification and characterization of Mycobacterium tuberculosis 1d-myo-inosityl-2-acetamido-2-deoxy-alpha-d-glucopyranoside deacetylase, MshB, a mycothiol biosynthetic enzyme. Protein Expr. Purif. 47542-550. [DOI] [PubMed] [Google Scholar]
- 91.Newton, G. L., T. Koledin, B. Gorovitz, M. Rawat, R. C. Fahey, and Y. Av-Gay. 2003. The glycosyltransferase gene encoding the enzyme catalyzing the first step of mycothiol biosynthesis (mshA). J. Bacteriol. 1853476-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Newton, G. L., P. Ta, K. P. Bzymek, and R. C. Fahey. 2006. Biochemistry of the initial steps of mycothiol biosynthesis. J. Biol. Chem. 28133910-33920. [DOI] [PubMed] [Google Scholar]
- 93.Newton, G. L., P. Ta, and R. C. Fahey. 2007. Tuberculosis: from lab research to field trials, abstr. 347. Abstr. Keystone Symp.
- 94.Newton, G. L., P. Ta, and R. C. Fahey. 2005. A mycothiol synthase mutant of Mycobacterium smegmatis produces novel thiols and has an altered thiol redox status. J. Bacteriol. 1877309-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Newton, G. L., P. Ta, D. Sareen, and R. C. Fahey. 2006. A coupled spectrophotometric assay for l-cysteine:1-d-myo-inosityl 2-amino-2-deoxy-alpha-d-glucopyranoside ligase and its application for inhibitor screening. Anal. Biochem. 353167-173. [DOI] [PubMed] [Google Scholar]
- 96.Newton, G. L., M. D. Unson, S. J. Anderberg, J. A. Aguilera, N. N. Oh, S. B. delCardayré, J. Davies, Y. Av-Gay, and R. C. Fahey. 1999. Characterization of a Mycobacterium smegmatis mutant defective in 1-d-myo-inosityl-2-amino-2-deoxy-alpha-d-glucopyranoside and mycothiol biosynthesis. Biochem. Biophys. Res. Commun. 255239-244. [DOI] [PubMed] [Google Scholar]
- 97.Nicholas, G. M., L. L. Eckman, G. L. Newton, R. C. Fahey, S. Ray, and C. A. Bewley. 2003. Inhibition and kinetics of Mycobacterium tuberculosis and Mycobacterium smegmatis mycothiol-S-conjugate amidase by natural product inhibitors. Bioorg. Med. Chem. Lett. 11601-608. [DOI] [PubMed] [Google Scholar]
- 98.Nicholas, G. M., L. L. Eckman, S. Ray, R. O. Hughes, J. A. Pfefferkorn, S. Barluenga, K. C. Nicolaou, and C. A. Bewley. 2002. Bromotyrosine-derived natural and synthetic products as inhibitors of mycothiol-S-conjugate amidase. Bioorg. Med. Chem. Lett. 122487-2490. [DOI] [PubMed] [Google Scholar]
- 99.Nicholas, G. M., P. Kovac, and C. A. Bewley. 2002. Total synthesis and proof of structure of mycothiol bimane. J. Am. Chem. Soc. 1243492-3493. [DOI] [PubMed] [Google Scholar]
- 100.Nicholas, G. M., G. L. Newton, R. C. Fahey, and C. A. Bewley. 2001. Novel bromotyrosine alkaloids: inhibitors of mycothiol S-conjugate amidase. Org. Lett. 31543-1545. [DOI] [PubMed] [Google Scholar]
- 101.Nigou, J., and G. S. Besra. 2002. Characterization and regulation of inositol monophosphatase activity in Mycobacterium smegmatis. Biochem. J. 361385-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nigou, J., L. G. Dover, and G. S. Besra. 2002. Purification and biochemical characterization of Mycobacterium tuberculosis SuhB, an inositol monophosphatase involved in inositol biosynthesis. Biochemistry 414392-4398. [DOI] [PubMed] [Google Scholar]
- 103.Norin, A., P. W. Van Ophem, S. R. Piersma, B. Persson, J. A. Duine, and H. Jornvall. 1997. Mycothiol-dependent formaldehyde dehydrogenase, a prokaryotic medium-chain dehydrogenase/reductase, phylogenetically links different eukaroytic alcohol dehydrogenases—primary structure, conformational modelling and functional correlations. Eur. J. Biochem. 248282-289. [DOI] [PubMed] [Google Scholar]
- 104.Paget, M. S., V. Molle, G. Cohen, Y. Aharonowitz, and M. J. Buttner. 2001. Defining the disulphide stress response in Streptomyces coelicolor A3(2): identification of the sigmaR regulon. Mol. Microbiol. 421007-1020. [DOI] [PubMed] [Google Scholar]
- 105.Park, H. D., K. M. Guinn, M. I. Harrell, R. Liao, M. I. Voskuil, M. Tompa, G. K. Schoolnik, and D. R. Sherman. 2003. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Park, J.-H., and J.-H. Roe. 2008. Mycothiol regulates and is regulated by a thiol-specific antisigma factor RsrA and sigma(R) in Streptomyces coelicolor. Mol. Microbiol. 68861-870. [DOI] [PubMed] [Google Scholar]
- 107.Park, J. H., C. J. Cha, and J. H. Roe. 2006. Identification of genes for mycothiol biosynthesis in Streptomyces coelicolor A3(2). J. Microbiol. 44121-125. [PubMed] [Google Scholar]
- 108.Patel, M., and J. Blanchard. 1998. Synthesis of des-myo-inositol mycothiol and demonstration of a mycobacterial specific reductase activity. J. Am. Chem. Soc. 12011538-11539. [Google Scholar]
- 109.Patel, M. P., and J. S. Blanchard. 1999. Expression, purification, and characterization of Mycobacterium tuberculosis mycothione reductase. Biochemistry 3811827-11833. [DOI] [PubMed] [Google Scholar]
- 110.Patel, M. P., and J. S. Blanchard. 2001. Mycobacterium tuberculosis mycothione reductase: pH dependence of the kinetic parameters and kinetic isotope effects. Biochemistry 403119-3126. [DOI] [PubMed] [Google Scholar]
- 111.Payne, D. J., M. N. Gwynn, D. J. Holmes, and D. L. Pompliano. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 629-40. [DOI] [PubMed] [Google Scholar]
- 112.Penninckx, M. J., C. J. Jaspers, and M. J. Legrain. 1983. The glutathione-dependent glyoxalase pathway in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2586030-6036. [PubMed] [Google Scholar]
- 113.Pick, N., M. Rawat, D. Arad, J. Lan, J. Fan, A. S. Kende, and Y. Av-Gay. 2006. In vitro properties of antimicrobial bromotyrosine alkaloids. J. Med. Microbiol. 55407-415. [DOI] [PubMed] [Google Scholar]
- 114.Rawat, M., and Y. Av-Gay. 2007. Mycothiol-dependent proteins in actinomycetes. FEMS Microbiol. Rev. 31278-292. [DOI] [PubMed] [Google Scholar]
- 115.Rawat, M., J. Heys, and Y. Av-Gay. 2003. Identification and characterization of a diamide sensitive mutant of Mycobacterium smegmatis. FEMS Microbiol. Lett. 220161-169. [DOI] [PubMed] [Google Scholar]
- 116.Rawat, M., C. Johnson, V. Cadiz, and Y. Av-Gay. 2007. Comparative analysis of mutants in the mycothiol biosynthesis pathway in Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 36371-76. [DOI] [PubMed] [Google Scholar]
- 117.Rawat, M., S. Kovacevic, H. Billman-Jacobe, and Y. Av-Gay. 2003. Inactivation of mshB, a key gene in the mycothiol biosynthesis pathway in Mycobacterium smegmatis. Microbiology 1491341-1349. [DOI] [PubMed] [Google Scholar]
- 118.Rawat, M., G. L. Newton, M. Ko, G. J. Martinez, R. C. Fahey, and Y. Av-Gay. 2002. Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals and antibiotics. Antimicrob. Agents Chemother. 463348-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rawat, M., M. Uppal, G. Newton, M. Steffek, R. C. Fahey, and Y. Av-Gay. 2004. Targeted mutagenesis of the Mycobacterium smegmatis mca gene, encoding a mycothiol-dependent detoxification protein. J. Bacteriol. 1866050-6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rengarajan, J., B. R. Bloom, and E. J. Rubin. 2005. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. USA 1028327-8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rietveld, P., L. D. Arscott, A. Berry, N. S. Scrutton, M. P. Deonarain, R. N. Perham, and C. H. Williams, Jr. 1994. Reductive and oxidative half-reactions of glutathione reductase from Escherichia coli. Biochemistry 3313888-13895. [DOI] [PubMed] [Google Scholar]
- 122.Rustad, T. R., M. I. Harrell, R. Liao, and D. R. Sherman. 2008. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS ONE 3e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sakuda, S., Z.-Y. Zhou, and Y. Yamada. 1994. Structure of a novel disulfide of 2-(N-acetylcysteinyl)amido-2-deoxy-α-d-glucopyranosyl-myo-inositol produced by Streptomyces sp. Biosci. Biotechnol. Biochem. 581347-1348. [DOI] [PubMed] [Google Scholar]
- 124.Sareen, D., G. L. Newton, R. C. Fahey, and N. A. Buchmeier. 2003. Mycothiol is essential for growth of Mycobacterium tuberculosis Erdman. J. Bacteriol. 1856736-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sareen, D., M. Steffek, G. L. Newton, and R. C. Fahey. 2002. ATP-dependent l-cysteine:1d-myo-inosityl 2-amino-2-deoxy-α-d-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry 416885-6890. [DOI] [PubMed] [Google Scholar]
- 126.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 4877-84. [DOI] [PubMed] [Google Scholar]
- 127.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 10012989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schneider, M. 2004. A rational approach to maximize success rate in target discovery. Arch. Pharm. (Weinheim) 337625-633. [DOI] [PubMed] [Google Scholar]
- 130.Shen, X. H., C. Y. Jiang, Y. Huang, Z. P. Liu, and S. J. Liu. 2005. Functional identification of novel genes involved in the glutathione-independent gentisate pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 713442-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Singh, A., L. Guidry, K. V. Narasimhulu, D. Mai, J. Trombley, K. E. Redding, G. I. Giles, J. R. Lancaster, Jr., and A. J. Steyn. 2007. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc. Natl. Acad. Sci. USA 10411562-11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Smirnova, G. V., N. G. Muzyka, M. N. Glukhovchenko, and O. N. Oktyabrsky. 2000. Effects of menadione and hydrogen peroxide on glutathione status in growing Escherichia coli. Free Radic. Biol. Med. 281009-1016. [DOI] [PubMed] [Google Scholar]
- 133.Spies, H. S., and D. J. Steenkamp. 1994. Thiols of intracellular pathogens. Identification of ovothiol A in Leishmania donovani and structural analysis of a novel thiol from Mycobacterium bovis. Eur. J. Biochem. 224203-213. [DOI] [PubMed] [Google Scholar]
- 134.Stadler, M., J. Bitzer, A. Mayer-Bartschmid, H. Muller, J. Benet-Buchholz, F. Gantner, H. V. Tichy, P. Reinemer, and K. B. Bacon. 2007. Cinnabaramides A-G: analogues of lactacystin and salinosporamide from a terrestrial streptomycete. J. Nat. Prod. 70246-252. [DOI] [PubMed] [Google Scholar]
- 135.Steenkamp, D. J., and R. N. Vogt. 2004. Preparation and utilization of a reagent for the isolation and purification of low-molecular-mass thiols. Anal. Biochem. 32521-27. [DOI] [PubMed] [Google Scholar]
- 136.Steffek, M., G. L. Newton, Y. Av-Gay, and R. C. Fahey. 2003. Characterization of Mycobacterium tuberculosis mycothiol S-conjugate amidase. Biochemistry 4212067-12076. [DOI] [PubMed] [Google Scholar]
- 137.Stewart, M. J., V. K. Jothivasan, A. S. Rowan, J. Wagg, and C. J. Hamilton. 2008. Mycothiol disulfide reductase: solid phase synthesis and evaluation of alternative substrate analogues. Org. Biomol. Chem. 6385-390. [DOI] [PubMed] [Google Scholar]
- 138.Sundquist, A. R., and R. C. Fahey. 1989. The function of gamma-glutamylcysteine and bis-gamma-glutamylcystine reductase in Halobacterium halobium. J. Biol. Chem. 264719-725. [PubMed] [Google Scholar]
- 139.Truman, A. W., F. Huang, N. M. Llewellyn, and J. B. Spencer. 2007. Characterization of the enzyme BtrD from Bacillus circulans and revision of its functional assignment in the biosynthesis of butirosin. Angew. Chem. Int. Ed. Engl. 461462-1464. [DOI] [PubMed] [Google Scholar]
- 140.Tsen, C. C., and A. L. Tappel. 1958. Catalytic oxidation of glutathione and other sulfhydryl compounds by selenite. J. Biol. Chem. 2331230-1232. [PubMed] [Google Scholar]
- 141.Ung, K. S., and Y. Av-Gay. 2006. Mycothiol-dependent mycobacterial response to oxidative stress. FEBS Lett. 5802712-2716. [DOI] [PubMed] [Google Scholar]
- 142.Unson, M. D., G. L. Newton, C. Davis, and R. C. Fahey. 1998. An immunoassay for the detection and quantitative determination of mycothiol. J. Immunol. Methods 21429-39. [DOI] [PubMed] [Google Scholar]
- 143.Urbaniak, M. D., A. Crossman, T. Chang, T. K. Smith, D. M. van Aalten, and M. A. Ferguson. 2005. The N-acetyl-d-glucosaminylphosphatidylinositol de-N-acetylase of glycosylphosphatidylinositol biosynthesis is a zinc metalloenzyme. J. Biol. Chem. 28022831-22838. [DOI] [PubMed] [Google Scholar]
- 144.van Ophem, P. W., J. Van Beeumen, and J. A. Duine. 1992. NAD-linked, factor-dependent formaldehyde dehydrogenase or trimeric, zinc-containing, long-chain alcohol dehydrogenase from Amycolatopsis methanolica. Eur. J. Biochem. 206511-518. [DOI] [PubMed] [Google Scholar]
- 145.Ventura, M., C. Canchaya, A. Tauch, G. Chandra, G. F. Fitzgerald, K. F. Chater, and D. van Sinderen. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71495-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vetting, M. W., P. A. Frantom, and J. S. Blanchard. 2008. Structural and enzymatic analysis of MshA from Corynebacterium glutamicum: substrate-assisted catalysis. J. Biol. Chem. 28315834-15844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vetting, M. W., S. L. Roderick, M. Yu, and J. S. Blanchard. 2003. Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases. Protein Sci. 121954-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vetting, M. W., M. Yu, P. M. Rendle, and J. S. Blanchard. 2006. The substrate-induced conformational change of Mycobacterium tuberculosis mycothiol synthase. J. Biol. Chem. 2812795-2802. [DOI] [PubMed] [Google Scholar]
- 149.Vogt, R. N., D. J. Steenkamp, R. Zheng, and J. S. Blanchard. 2003. The metabolism of nitrosothiols in the mycobacteria: identification and characterization of S-nitrosomycothiol reductase. Biochem. J. 374657-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Voskuil, M. I., K. C. Visconti, and G. K. Schoolnik. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinburgh) 84218-227. [DOI] [PubMed] [Google Scholar]
- 152.Walsh, C. 2003. Antibiotics: actions, origins, resistance. ASM Press, Washington, DC.
- 153.Wang, R., Y. J. Yin, F. Wang, M. Li, J. Feng, H. M. Zhang, J. P. Zhang, S. J. Liu, and W. R. Chang. 2007. Crystal structures and site-directed mutagenesis of a mycothiol-dependent enzyme reveal a novel folding and molecular basis for mycothiol-mediated maleylpyruvate isomerization. J. Biol. Chem. 28216288-16294. [DOI] [PubMed] [Google Scholar]
- 154.Watanabe, R., K. Ohishi, Y. Maeda, N. Nakamura, and T. Kinoshita. 1999. Mammalian PIG-L and its yeast homologue Gpi12p are N-acetylglucosaminylphosphatidylinositol de-N-acetylases essential in glycosylphosphatidylinositol biosynthesis. Biochem. J. 339185-192. [PMC free article] [PubMed] [Google Scholar]
- 155.Weber, T., K. Welzel, S. Pelzer, A. Vente, and W. Wohlleben. 2003. Exploiting the genetic potential of polyketide producing streptomycetes. J. Biotechnol. 106221-232. [DOI] [PubMed] [Google Scholar]
- 156.Wheeler, P. R., N. G. Coldham, L. Keating, S. V. Gordon, E. E. Wooff, T. Parish, and R. G. Hewinson. 2005. Functional demonstration of reverse transsulfuration in the Mycobacterium tuberculosis complex reveals that methionine is the preferred sulfur source for pathogenic mycobacteria. J. Biol. Chem. 2808069-8078. [DOI] [PubMed] [Google Scholar]
- 157.Williams, L., F. Fan, J. S. Blanchard, and F. M. Raushel. 2008. Positional isotope exchange analysis of the Mycobacterium smegmatis cysteine ligase (MshC). Biochemistry 474843-4850. [DOI] [PubMed] [Google Scholar]
- 158.Wong, K. K., and J. S. Blanchard. 1989. Human erythrocyte glutathione reductase: pH dependence of kinetic parameters. Biochemistry 283586-3590. [DOI] [PubMed] [Google Scholar]
- 159.Xie, Z., N. Siddiqi, and E. J. Rubin. 2005. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 494778-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yu, T. W., R. Muller, M. Muller, X. Zhang, G. Draeger, C. G. Kim, E. Leistner, and H. G. Floss. 2001. Mutational analysis and reconstituted expression of the biosynthetic genes involved in the formation of 3-amino-5-hydroxybenzoic acid, the starter unit of rifamycin biosynthesis in Amycolatopsis mediterranei S699. J. Biol. Chem. 27612546-12555. [DOI] [PubMed] [Google Scholar]
- 161.Zdanowski, K., P. Doughty, P. Jakimowicz, L. O'Hara, M. J. Buttner, M. S. Paget, and C. Kleanthous. 2006. Assignment of the zinc ligands in RsrA, a redox-sensing ZAS protein from Streptomyces coelicolor. Biochemistry 458294-8300. [DOI] [PubMed] [Google Scholar]