Abstract
Methionine sulfoxide reductase enzymes MsrA and MsrB have complementary stereospecificies that respectively reduce the S- and R-stereoisomers of methionine sulfoxide (MetSO), and together function as critical antioxidant enzymes. In some pathogenic and metal -reducing bacteria these genes are fused to form a bifunctional methionine sulfoxide reductase (i.e., MsrBA) enzyme. To investigate how gene fusion affects the substrate specificity and catalytic activities of Msr, we have cloned and expressed the MsrBA enzyme from Shewanella oneidensis, a metal-reducing bacterium and fish pathogen. For comparison, we also cloned and expressed the wild-type MsrA enzyme from Shewanella oneidensis and a genetically engineered MsrB protein. MsrBA is able to completely reduce (i.e., repair) MetSO in the calcium regulatory protein calmodulin (CaM); while only partial repair is observed using both MsrA and MsrB enzymes together at 25 °C. A restoration of the normal protein fold is observed coincident with the repair of MetSO in oxidized CaM by MsrBA, as monitored by the time-dependent increases in the anisotropy associated with the rigidly bound multiuse affinity probe 4′5′-bis(1,3,2-dithoarsolan-2yl)fluorescein (FlAsH). Underlying the efficient repair of MetSO in oxidized CaM is the coordinate activity of the two catalytic domains in the MsrBA fusion protein, which results in an order of magnitude rate enhancement in comparison to the individual MsrA or MsrB enzymes alone. The coordinate binding of both domains of MsrBA permits the full repair of all MetSO in CaMox. The common expression of Msr fusion proteins in bacterial pathogens is consistent with an important role for this enzyme activity in the maintenance of protein function necessary for bacterial survival under highly oxidizing conditions associated with pathogenesis or bioremediation.
Methionines are highly susceptible to oxidation to their corresponding methionine sulfoxides (MetSO1) by a range of commonly generated reactive oxygen species (ROS), such as hydrogen peroxide, singlet oxygen, or peroxynitrite (1–3). Methionine sulfoxide reductase (Msr) enzymes recognize MetSO within unfolded sequences of proteins, and bind with high-affinity (i.e., Kd = 70 ± 10 nM) prior to the reduction of MetSO to restore the native Met structure (4–6). In addition, distinct Msr enzymes are critical to the maintenance of cellular Met pools (7), whose oxidation can down-regulate the initiation of protein synthesis.
Two different enzyme classes of Msr, i.e., MsrA and MsrB, are expressed in virtually all organisms (8, 9). While these enzymes have little structural homology, the catalytic sites of MsrA and MsrB possess a mirror image symmetry that promotes the selective reduction of S- and R-diastereomers of MetSO (10–12). High-affinity binding and reduction of free MetSO by either MsrA or MsrB is rapid (kob > 50 sec−1), with subsequent steps involving reformation of the reduced catalytic center, which limits steady-state rates of catalysis (13, 14). However substantial amounts of MetSO are present in a range of different healthy mouse tissues (i.e., >4% of all Met) (2), suggesting that Msr enzymes are insufficient to maintain cellular Met in the fully reduced state. The incomplete repair of MetSO in proteins has been linked to the substrate requirements of Msr enzymes, which do not recognize the majority of MetSO within folded protein structures (5). Rather, sensitive proteins whose structures are disrupted upon oxidation of Met are the major substrates of Msr enzymes, and repair of these regulatory proteins has the potential to modulate cellular metabolism in a redox-dependent manner that is regulated by the NADPH-dependent reduction of thioredoxin by thioredoxin reductase (15).
Msr enzymes are critical to bacterial survival in high oxidative stress environments, and permit the colonization of host organisms by a range of pathogens, including Erwinia chrysanthemi, Escherichia coli, Helicobacter pylori, Mycobacterium smegmatis, Mycobacterium tuberculosi, Mycoplasma genitlium, and Neisseria gonorrhoeae (11, 16–21). Bifunctional Msr enzymes, which reduce both S- and R-diastereomers of MetSO, are present in many pathogenic and metal-reducing bacteria (including Haemophilus influenze, Helicobacter pylori, Vibrio cholerae, Neisseria meningitidis, Bacillus anthracis, and Shewanella oneidensis), and have arisen through gene fusion to create both MsrAB or MsrBA variants (8). These fusion proteins are commonly located on the outer surface of the inner membrane in the periplasm, permitting the efficient redox coupling of cytosolic NADPH to maintain these enzymes in a reduced state necessary for the repair of oxidized proteins (11, 22). It remains unclear whether the formation of bifunctional Msr enzymes through gene fusion has any additional functional significance that may, for example, be related to the catalytic ability to simultaneously bind multiple MetSO within an oxidized protein.
To assess possible increases in the ability of bifunctional Msr enzymes to reduce MetSO in oxidized proteins, we have cloned and expressed the two genes encoding MsrBA and MsrA from Shewanella oneidensis (Figure 1). This soil microbe encodes 42 multiheme cytochromes, which underlie a diverse metabolism that allows growth using solid metal oxides as terminal electron acceptors, resulting in a considerable functional sensitivity to reactive oxygen species (ROS) (23, 24). For comparison, we have constructed and expressed the MsrB encoding region of MsrBA. The abilities of MsrBA, MsrA, and MsrB to repair protein-bound MetSO were assessed using the calcium regulatory protein calmodulin (CaM) that has previously been used as a protein substrate to assess the ability of mammalian forms of both MsrA and MsrB to recognize MetSO in proteins (6, 25). Furthermore, CaM can play an important role in promoting bacterial pathogenesis, suggesting the physiological relevance of its oxidation (26). We find that the bifunctional MsrBA enzyme is able to fully reduce all S- and R-stereoisomers of MetSO in CaMox. In contrast, a mixture of the isolated MsrA and MsrB proteins is unable to catalyze the complete repair of all MetSO, consistent with the inability of Msr enzymes to recognize MetSO within folded proteins (5). Accompanying reduction of MetSO in CaMox by MsrBA is the restoration of the overall protein fold of CaM, which is not observed using either MsrA or MsrB enzyme alone. These results suggest an important role for bifunctional Msr enzymes in the maintenance of protein functions under conditions of acute oxidative stress.
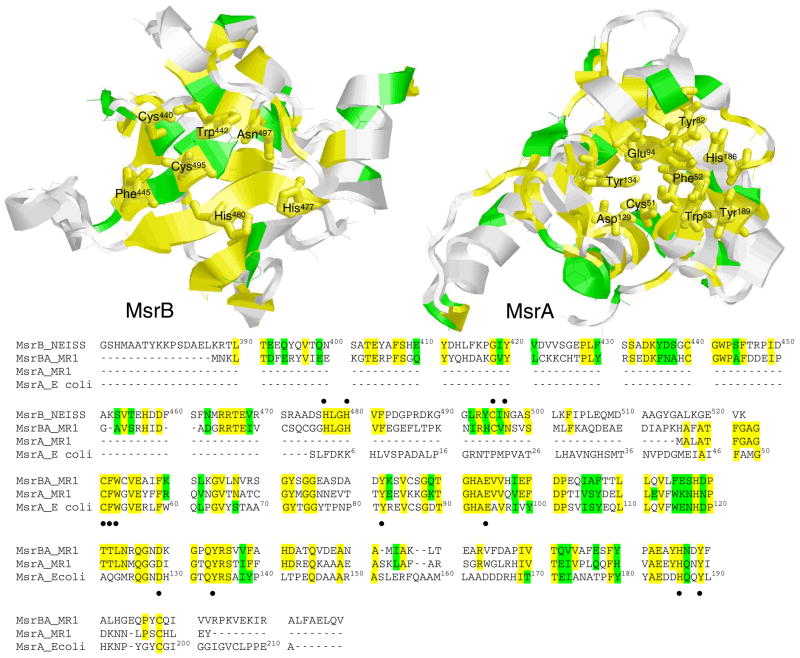
Figure 1. Conserved Active Site Sequences of Msr Proteins.
Tertiary structures (top) and sequences (bottom) of Msr proteins highlighting identical (yellow) and conserved (green) amino acids. Side chains implicated in catalysis or substrate recognition are highlighted (12, 49, 50). Structures correspond to MsrB (left) or MsrA (right). Sequences compare amino acids between MsrB from Neisserium gonorrhoeae and MsrA from E. coli with Shewanella oneidensis MR-1 proteins MsrA (SO2337) and MsrBA (SO2588), as determined using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). Structures correspond to 2gt3.pdb for MsrA from E. coli or 1l1d.pdb for MsrB from Neisseria gonorrhoeae (12, 51, 52), and were drawn using the program RASMOL (53).
EXPERIMENTAL PROCEDURES
Materials
Thioredoxin, thioredoxin reductase, L-methionine sulfoxide (L-MetSO), 2-mercaptoethanol (β-ME), triscarboxyethylphosphine-HCl (TCEP), and β-nicotinamide adenine dinucleotide phosphate (NADPH) were purchased from Sigma Chemical Co. (St. Louis). 4′5′-bis(1,3,2-dithoarsolan-2yl)fluorescein (FLAsH-EDT2) was synthesized as previously described (27, 28). All other chemicals were of reagent grade.
Cloning and Expression of Msr Isoforms
Two putative Msr genes, i.e., MsrA (SO2337) and MsrBA (SO2588) (Figure 1), identified in the Shewanella oneidensis MR-1 genome (29), respectively contained 480- and 903-base pairs and their DNA fragments were respectively PCR amplified with MsrA-F (5′-CACCARGGCGTTAGCAAC-TTTCGGT-3′) and MsrA-R (5′-GTACTCGAGCTGACAACTT GGTAA-3′) primers, and MsrBA-F (5′-CACCATGGACAA ACTGACTGATTTTGAA-3′) and MsrBA-R (5′-AACTTGCAGTTCGGCAAATAA ATGC-3′) primers. For comparison, the MsrB region of the MsrBA gene containing the first 128 amino acids in the sequence was PCR amplified using the following forward and reverse primers, i.e., MSRB#2-F (5′-CACCATGAACAAACTGACTGATTTTGA-3′) and MSRB#2R (5′-AGCATGTTTTGGGGCAA TAT-3′). This sequence encodes the entire MsrB gene based on homology with other MsrB sequences in a range of microbes, including Bacillus anthracis, Escherichia coli, Mycobacterium tuberculosis, Salmonella typhimurium, Vibrio cholerae, Yersinia pestis. The amplified DNA fragments were cloned into pENTR/SD/D-TOPO vector (Invitrogen) to contain a V5-His tag sequence (KGGRADPAFLYKVVINSKLEGKPIPNPLLGLDSTRTGHHH HHH) engineered at the C-terminus of the expressed proteins and transformed into Escherichia coli strain TOP10. Positive clones were screened and all DNA sequences were verified by DNA sequencing (Amplicon Express, Pullman, WA).
For overexpression of Msr isoforms in Escherichia coli, individual plasmids were cloned into pETDEST 42 (Invitrogen CAT #12276-010) using LR Clonase (Invitrogen CAT #11791-019) and transformed into Escherichia coli strain BL21Star (DE3). One liter of innoculated culture was grown at 37 °C to an optical density of 0.4 at 600 nm. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 1.0 mM, and growth was continued overnight at 30 °C. Bacteria were harvested by centrifugation, and following resuspension in 10 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 10 mM imidazole, cells were lysed by three passes through a French pressure cell (Glenmills Inc., Clifton, NJ). Lysates were first clarified by centrifugation at 5,000 rpm for 15 min and the resulting supernatant was then centrifuged at 15,000 rpm for an additional 30 min. The clear supernatant was loaded onto a cobalt column (Talon Metal Affinity Resins; BD Biosciences – Clontech; Palo Alto, CA), and following five volumes of washing buffer [10 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 20 mM imidazole]. MsrBA, MsrA, and MsrB recombinant proteins were eluted with 5 ml of elution buffer [10 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 300 mM imidazole]. The N-terminal His-Patch thioredoxin sequence from the MsrB recombinant protein was enzymatically digested using EnterokinaseMax (EKMax; Invitrogen Cat # 190-01). Msr protein concentrations were determined using the Coomassie Plus protein assay (Pierce Inc., Rockford, IL).
Expression and Purification of CaM
DNA encoding a wild type and a mutant CaM containing four cysteines engineered in helix A (at Glu6, Glu7, Ala10, and Glu11) (i.e., C4-CaM) was cloned into pET-15b (Novagen) expression vector, and expressed in BL21 (DE3) Escherichia coli strain, as previously described (30). CaM was purified by chromatography on phenyl sepharose CL-4B (Pharmacia, Piscataway, NJ) (31), and its protein concentration was measured using a micro BCA assay reagent kit (Pierce, Rockford, IL), using desalted wild type CaM as the standard (ε277nm = 3029 M−1 cm−1) (31).
Oxidation of Calmodulin
Following incubation with H2O2 (100 mM) in 10 mM Tris-HCl (pH 7.5) for 24 hrs at 25 °C all nine methionines in CaM (60 μM) were oxidized to their corresponding methionine sulfoxides (25, 32). The concentration of H2O2 was determined by using the published extinction coefficient, ε240 = 39.4 M−1 cm−1 (33). The reaction was stopped by exhaustive dialysis at 4 °C.
FlAsH-labeled CaM
C4-CaM (50 μM) was incubated in 50 mM HEPES (pH 7.5), 140 mM KCl, 1 mM β-ME, and 1 mM TCEP for one hour to reduce any disulfide bonds prior to the addition of FlAsH-EDT2 (50 μM); essentially complete labeling required two hours at room temperature. FlAsH-labeled CaM was separated from unbound FlAsH-EDT2 using a Sephadex G25 size exclusion column (34).
Fluorescence Measurements
Rotational mobilities (anisotropy) of FlAsH-labeled CaM were measured using a SPEX FluoroMax-2 fluorometer (Edison, NJ), where the anisotropy (r) was calculated as the ratio of fluorescence intensities (I) with the polarizers in the vertical (v) or horizontal (h) position:
| (1) |
where g = Ihv/Ihh.
Enzymatic Activities of Msr
The reductase activity of Msr isoforms was measured in 10 mM Tris-HCl buffer (pH 7.5), NADPH (400 μM), thioredoxin (50 μM), and thioredoxin reductase (2 μM) following the decrease in the absorbance of NADPH at 340 nm (ε340 = 6220 M−1 cm−1) at 24 °C, essentially as previously described (6, 35, 36).
Mass Spectrometric Analysis
Whole protein ESI spectra were acquired on an AUTOSPEC-Q mass spectrometer equipped with Mark III ESI source, essentially as previously described (5).
RESULTS
Conserved Structures of Msr Protein from Shewanella
We assessed the ability of the two Msr proteins encoded in the genome of Shewanella oneidensis MR-1 (i.e., MsrBA and MsrA) to repair oxidized CaM (CaMox), in which all nine methionines were oxidized to their methionine sulfoxides. Together these proteins function to reduce methionine sulfoxides in Shewanella proteins, and contribute to the ability of this oxidatively sensitive organism to contribute to the bioremediation of soils through its diverse metabolism that permits the reduction of solid metal oxides as terminal electron acceptors (24). The C-terminal sequence of MsrBA (SO2588) is homologous to the protein sequence of MsrA (SO2337), with 61% overall positive sequence homology determined using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). The active site and substrate binding recognition sequences in MsrA are absolutely conserved (Figure 1), permitting a functional comparison between these two enzyme activities to determine the functional implications of the MsrB fusion protein. As there is no homologous MsrB protein encoded in the Shewanella genome, we have cloned and expressed the N-terminal sequence of MsrBA, whose sequence is essentially identical within the active site region and has a 50% overall positive sequence homology to the MsrB protein sequence in Neisseria gonorrhoeae (Figure 1).
Enhanced Repair by MsrBA Reduces All Nine MetSO in CaMox
Taking advantage of large shifts in the mobility of CaM following Met oxidation using SDS-PAGE, we assessed the ability of MsrBA in the presence of the artificial electron donor DTT to repair MetSO in fully oxidized CaM (CaMox), where authentic standards of unoxidized or fully oxidized CaM with apparent molecular masses of 17.9 and 20.6 kDa were used for comparison. Upon incubation of CaMox with MsrBA there is a time-dependent increase in the mobility of CaM on SDS-PAGE, which approaches that of unoxidized CaM after four hours of incubation (Figure 2). These results suggest that MsrBA is able to fully repair the majority of the MetSO in CaMox. In comparison, much smaller changes in CaM mobility are apparent upon incubation of CaMox with MsrA over the same time course, suggesting a diminished capacity of MsrA to recognize and repair MetSO in CaMox.
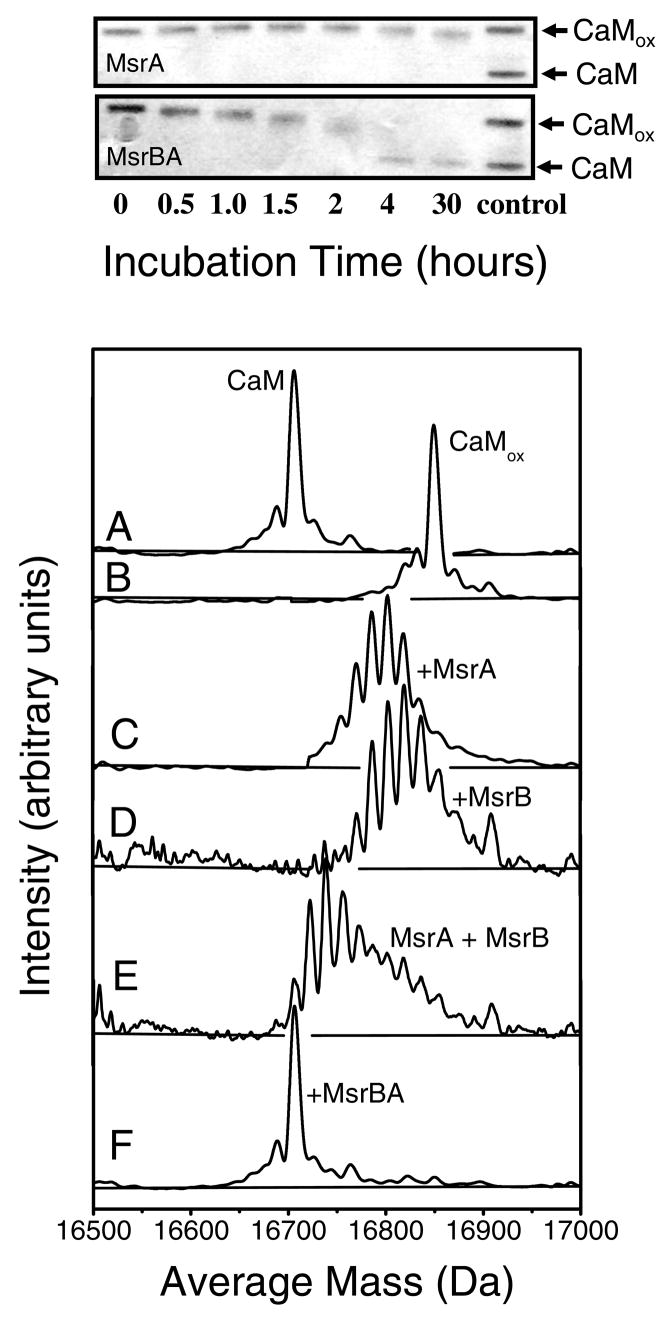
Figure 2. Complete Repair of CaMox by MsrBA.
(Top) SDS-PAGE (14% Tris-glycine) gel showing time-dependent mobility changes of CaMox in the presence of either MsrA or MsrBA. (Bottom) Intact protein ESI-MS spectra of unoxidized CaM (A) and oxidized CaM prior to (B) and following incubation for 12 hours with MsrA (C), MsrB (D), an equimolar mixture of MsrA + MsrB (E), and MsrBA (F). Experimental conditions involved CaM (10 μM) incubated with indicated Msr isoforms (1.0 μM) of either MsrBA, MsrA, or MsrB in 10 mM MOPS (pH 7.5), 50 mM KCl, and 15 mM DTT at 25°C. No MetSO reduction is observed in the absence of added DTT, which acts to reduce the active site of Msr enzymes to permit reduction of MetSO.
Additional resolution regarding the abilities of Msr enzymes to recognize and reduce MetSO in proteins was obtained using intact protein mass spectrometry to assess the extent of Met oxidation in oxidized CaM (CaMox). CaMox has previously been used to assess the ability of both MsrA and MsrB to repair oxidized proteins due to the presence of nine surface exposed methionines (6, 25). Prior to oxidation the average mass of CaM was 16,706.7 Da (Figure 2A), which is consistent with the theoretical mass of 16,706.4 Da. A dehydration artifact associated with the loss of water in the ion trap results in the appearance of a lower molecular mass feature at 16,688.4 Da (37). For these experiments Mets in CaM were quantitatively oxidized to their corresponding MetSO using hydrogen peroxide, resulting in a 143.3 Da shift in the average mass of CaM to 16,850.0 Da which is indicative of the quantitative oxidation of all nine methionines (i.e., 9 Met × 16 Da/oxygen = 144 Da)(Figure 2B).
Upon incubation for 12 hr with MsrBA in the presence of 15 mM DTT all nine MetSO in CaM are reduced to form methionine (Figure 2F), and the resulting mass spectrum is virtually identical to that of unoxidized CaM (Figure 2A). In comparison, a distribution of CaM oxiforms remains that contains up to eight MetSO per protein following incubation of CaMox with an equimolar mixture of MsrA and MsrB (Figure 2E). The retention of large numbers of MetSO in CaM following repair by both MsrA and MsrB is consistent with prior results that indicate the restoration of the protein fold prevents further repair of MetSO (5, 25). Thus, while the complementary specificities of MsrA and MsrB to repair the S- and R-stereoisomers of MetSO result in additional repair in comparison to either enzyme alone (Figures 2C and 2D), the fusion of these enzyme activities to form the bifunctional MsrBA results in a striking enhancement in the extent of CaMox repair.
Our results demonstrate an enhanced ability to reduce S- and R-MetSOs in oxidized proteins by the MsrBA fusion protein in comparison to either MsrA or MsrB enzymes. These letter results are consistent with the observation that the oxidation of Met145 disrupts the tertiary structure of CaM, resulting in an ability of Msr enzymes to recognize MetSO within disordered sequences (5, 15, 38). Reduction of this conformationally sensitive site in CaMox by either MsrA or MsrB will result in a restoration of the protein fold and will diminish the ability of Msr enzymes to recognize and repair other MetSO in the refolded protein. Thus, depending on the order in which individual MetSO are reduced in CaMox the protein will assume its native fold and essentially trap remaining MetSO within the folded structure. In contrast, MsrBA is able to bind and fully repair all MetSO in CaMox, suggesting that the simultaneous binding of both domains facilitates protein repair.
Redox-Dependent Modulation of MsrBA Function
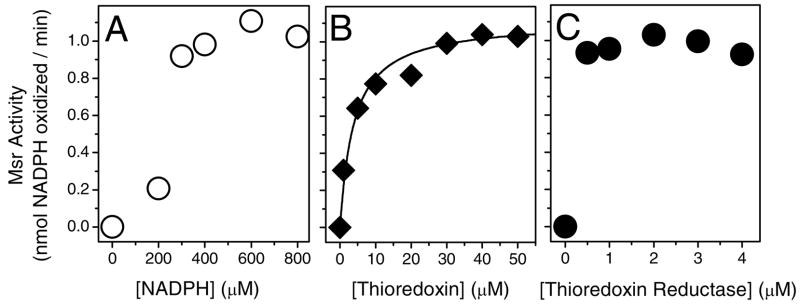
The repair of free MetSO involves the reduction of the active site dithiol, which under cellular conditions is mediated by reduced thioredoxin whose concentration is maintained by the NADPH-dependent activity of thioredoxin reductase (39). To determine whether changes in cellular concentrations of NADPH and reduced thioredoxin affect rates of MsrBA activity, we measured the substrate-dependence of repair as a function of NADPH and thioredoxin concentrations. An equimolar stoichiometry of thioredoxin reductase relative to MsrBA is sufficient to maintain optimal function (Figure 3C), indicating a high-affinity binding interaction that promotes the efficient reduction in the catalytic site of MsrBA. Half-points of maximal catalytic activity occur at 240 ± 10 μM NADPH and 3.3 ± 0.8 μM thioredoxin (Figure 3A and 3B), which are near physiological levels of NADPH and thioredoxin in bacteria (40–43). These latter results indicate that the action of MsrBA is under redox control, such that reduced rates of MetSO repair will occur under conditions of oxidative stress and associated reductions in NADPH and reduced thioredoxin.
Figure 3. Substrate-Dependence of MsrBA Mediated Repair of CaMox.
MsrBA-dependent rates of repair of CaMox as a function of NADPH (A), thioredoxin (B), or thioredoxin reductase (C). Experimental conditions included MsrBA (0.5 μM) and CaMox (30 μM) in 10 mM Tris-HCl (pH 7.5) containing indicated substrate concentrations and saturating concentrations of NADPH (400 μM) (B and C), thioredoxin (50 μM) (A and C) or thioredoxin reductase (2 μM) (A and B) at 24 °C. Line in panel B represents nonlinear least-squares fit to the Michaelis-Menten rate equation, where KM = 3.3 ± 0.8 μM.
Restoration of Protein Fold Upon Repair of CaMox by MsrBA
The oxidation of Met145 in CaM induces a loss of function and results in global structural changes that disrupt the secondary structure and uncouple the opposing domains of CaM, resulting in the nonproductive binding between CaMox and some target proteins (15, 32, 38, 44, 45). This structural change results in the uncoupling between the opposing domains of CaM, which is normally associated with the calcium-dependent activation of CaM. Following binding of the fluorophore 4′5′-bis(1,3,2-dithoarsolan-2yl)fluorescein (FlAsH), the oxidant-induced disruption of the protein fold of CaM can be readily monitored in real time as a reduction in the steady-state anisotropy (30, 44). Similar oxidant-induced changes in protein structure are observed using SDS-PAGE, where oxidation of all nine Mets result in a substantial reduction in the electrophoretic mobility (Figure 4).
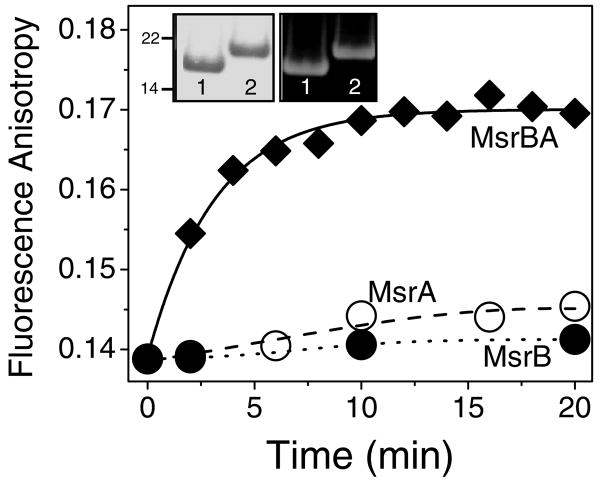
Figure 4. Restoration of CaMox Protein Fold Upon Repair of Met(SO) by MsrBA.
Steady-state anisotropies for FlAsH-labeled CaMox (1.0 μM) in the presence of MsrBA (◆), MsrA (○) and MsrB (●). Reaction mixture consists of NADPH (400 μM), thioredoxin (50 μM), thioredoxin reductase (2 μM), and indicated isoform of Msr (1.0 μM) in 50 mM HEPES (pH 7.5), 140 mM KCl, and 0.2 mM CaCl2 in a total volume of 2 mL. Initial rates associated with MsrBA-dependent increases in anisotropy are (5.9 ± 1.1) × 10−3/min, (0.53 ± 0.12) × 10−3/min, and (0.19 ± 0.02) × 10−3/min for MsrBA, MsrA and MsrB, respectively. Excitation was at 500 nm and emitted light was measured at 530 nm; slit widths were set at 5 nm. Inset: Electrophoretic mobility on SDS-PAGE of FlAsH-labeled CaM (10 μg) prior to (lane 1) and following oxidation of all nine methionines (lane 2) using a 14% Tris-Glycine gel visualized by fluorescence detection (right) or following Coomassie blue staining (left).
To measure alterations in the protein fold of CaM, we have used the same FlAsH-labeled CaM mutant that we have previously used to assess oxidant-induced changes in the conformation of CaM (30, 44). Upon incubation of equimolar amounts of CaMox with MsrBA in the presence of saturating amounts of NADPH, thioredoxin, and thioredoxin reductase there is a time-dependent increase in the fluorescence anisotropy of FlAsH-labeled CaM, which after approximately ten minutes increases from 0.139 ± 0.001 to a value that approaches that of unoxidized CaM (i.e., 0.170 ± 0.001) (Figure 4). These measurements demonstrate the full restoration of the protein fold associated with the repair of oxidized calmodulin by MsrBA over a time-scale that is relevant to cellular repair processes. In comparison, much smaller increases in fluorescence anisotropy are observed following the incubation of either MsrA or MsrB with FlAsH-labeled CaM, indicating that the separate enzyme activities are unable to efficiently bind and reduce MetSO in oxidized calmodulin. Only MsrBA has the ability to recognize and repair MetSO within oxidized proteins to restore the protein fold associated with native function. In comparison, limited refolding occurs using either MsrA or MsrB enzymes alone.
Enhanced Rates of CaMox Repair by MsrBA
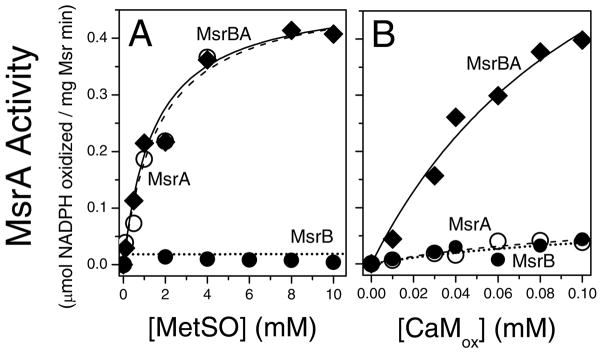
The enhanced catalytic efficiency of MsrBA in comparison to either MsrA or MsrB alone towards CaMox is apparent using an enzyme-linked assay involving measurements of decreases in the absorbance of NADPH to measure initial rates of catalysis (Figure 5B). In the presence of saturating amounts of all substrates and 100 μM CaMox (in which all nine Met are oxidized to their corresponding sulfoxides), MsrBA exhibits the highest initial rate (39.2 mol of NADPH oxidized/mol MsrBA · min) relative to that of MsrA and MsrB (3.3 and 1.9 mol of NADPH oxidized/mol Msr · min, respectively). These results indicate that fused protein MsrBA results in significantly increased activity in comparison to the individual MsrA or MsrB enzymes.
Figure 5. Preferential Recognition and Repair of CaMox.
Catalytic activities of Msr isoforms (0.5 μM) against L-MetSO (A) and CaMox (B) for MsrBA (◆; solid lines), MsrA (○; dashed lines) and MsrB (●; dotted lines). Experimental conditions involved 10 mM Tris-HCl (pH 7.5), NADPH (400 μM), thioredoxin (50 μM), and thioredoxin reductase (2 μM) in a total volume of 0.2 mL at 24 °C. Lines represent fits to the Michaelis-Menten rate equation (see Table 1).
To further assess the specificity of these bacterial Msr enzymes with respect to the ability to recognize and reduce either free or protein-bound MetSO, we compared their substrate-dependencies using both L-MetSO and CaMox (Figure 5). Maximal catalytic rates (i.e., kcat) against CaMox are approximately 20-fold higher for MsrBA in comparison to either MsrA or MsrB alone, indicating that the fusion of these enzyme activities results in large rate enhancements (Figure 5B; Table 1). Similar increases in the catalytic efficiency (kcat/KM) of MsrBA are apparent, which are more than 10-fold larger than that of either MsrA or MsrB. These results indicate that the ability of MsrBA to maintain the MetSO within protein substrates in an accessible partially unfolded state results in an increase in the overall catalytic rates of repair.
Table 1.
Catalytic Rates of MetSO Repair by Msr Enzymes
| Enzyme | Substrate | Vmaxa (μmol NADPH/mg min) | Kma (mM) | kcatb (s−1) | kcat/Kmc (M−1 s−1) |
|---|---|---|---|---|---|
| MsrBA | MetSO | 0.49 ± 0.03 | 1.7 ± 0.4 | 0.31 ± 0.02 | 180 ± 40 |
| CaMox | 0.9 ± 0.2 | 0.12 ± 0.03d | 0.6 ± 0.1 | 5,000 ± 2,000 | |
| MsrA | MetSO | 0.5 ± 0.1 | 2.0 ± 0.8 | 0.19 ± 0.04 | 100 ± 40 |
| CaMox | 0.07 ± 0.02 | 0.07 ± 0.05d | 0.027 ± 0.007 | 400 ± 300 | |
| MsrB | MetSO | ND | ND | ND | ND |
| CaMox | 0.05 ± 0.01 | 0.05 ± 0.02d | 0.01 ± 0.01 | 200 ± 200 |
Data presented were obtained by fitting the experimental data to the Michaelis-Menten equation, , where [S] is concentration of substrate, Km is the Michaelis constant and Vmax is the velocity when the reaction approaches a maximum. Reactions conditions are described in the legend of Figure 5.
kcat = Vmax × molecular weight = turnover number.
Errors were propagated.
Value are apparent and do not take into account the presence of multiple MetSO in oxidized CaM.
In comparison to the large rate enhancements observed for MsrBA against CaMox relative to values observed for MsrA or MsrB, the catalytic rates for MsrBA and MsrA are very similar when MetSO is used as a substrate (Figure 5A). This insensitivity of catalytic rates of repair of free MetSO for MsrBA and MsrA activities is consistent with earlier data using MsrAB isolated from Neisseria, where the steady-state activities against MetSO were also observed to be unaffected by gene fusion (46). In contrast, minimal rates of repair are observed for the genetic construct encoding the MsrB domain, consistent with prior measurements indicating that the turnover number of MsrA is more than 30-fold that of MsrB for free MetSO using physiological substrates (i.e., NADPH, thioredoxin, and thioredoxin reductase) (6).
The similar catalytic efficiencies of MsrBA and MsrA against free MetSO indicates that increases in the catalytic efficiencies association with fusion of both enzyme activities in bifunctional MsrBA enymes against CaMox requires the simultaneous binding of both domains to the protein-bound MetSO substrates. These results are consistent with earlier reports indicating a cooperative binding between mammalian MsrA enzymes and highly oxidized CaM (5), which suggested that induced structural changes within CaMox upon binding one domain of MsrBA enhances the binding of the other domain.
DISCUSSION
Using both intact protein mass spectrometry and fluorescence spectroscopy to respectively measure reduction of MetSO and restoration of the structural coupling between the opposing domains of CaMox, we have demonstrated that the fusion of complementary enzyme activities associated with the repair of R- and S-stereoisomers of MetSO in MsrBA results in the full repair of all MetSO within CaMox and the restoration of the native fold (Figures 2 and 4). MsrBA acts to stabilize an unfolded state through the coordinated binding of either MsrA or MsrB domains to CaMox (Figure 6), permitting the full reduction of all MetSO in CaMox within ten minutes (Figure 4), a time scale that is relevant to bacterial host colonization. A consequence of the stabilization of the unfolded state is an increased catalytic rate due to the increased local concentration of MetSO within unstructured sequences, which permit access of the oxidized side-chain of MetSO into the Msr active site for repair (5). In comparison, an incomplete repair of either S- or R-MetSO in CaMox is observed for the individual MsrA or MsrB enzymes, where the rebinding of the Msr enzyme following dissociation competes with protein refolding and the associated retention of MetSO and reductions in catalytic rates. These latter results are in agreement with prior measurements for homologous Msr proteins from bovine sources under these experimental conditions (6, 25), indicating that the inability of mixtures of MsrA and MsrB enzymes to fully reduce MetSO in comparison to MsrBA is not the result of sequence differences between these homologous enzymes. Likewise, limited restoration of the overall structure of CaMox occurs using either MsrA or MsrB enzymes alone, indicating that the separate enzyme activities are unable to efficiently bind and reduce all MetSO within CaMox. These measurements demonstrate that the enhanced ability of the MsrBA fusion protein to reduce MetSO in oxidized proteins in comparison to the individual MsrA or MsrB enzymes arises as a result of the coordinate action of the MsrA and MsrB elements that maintain CaMox in a unfolded state necessary for the recognition and repair of MetSO.
Figure 6. Coordinated Binding at Multiple MetSO in CaMox by MsrBA Facilitates Repair.
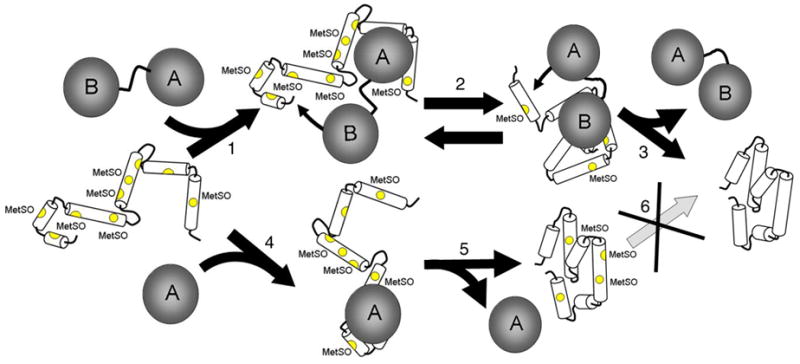
Depiction of proposed role of MsrBA fusion protein (gray connected circles denoted A and B) in binding and stabilizing unfolded state of oxidized CaM (white cylinders)(step 1), permitting reduction of all nine MetSO (yellow) to their native Met structure through the coordinate binding of both active sites A and B in MsrBA (top) at multiple MetSO sites (step 2), where diffusional steps are minimized due to the role of the complemetary protein domain in anchoring MsrBA to the oxidized protein. Underlying thecapacity of MsrBA to fully repair CaMox is an ability to stabilize the oxidized and partially unfolded CaM to promote recognition of exposed MetSO by the Msr enzymes (Figure 4). The ability to maintain the MetSO within protein substrates in an accessible partially unfolded state results in an increased catalytic rates of repair (Figure 5). Following MetSO reduction, MsrBA dissociates, releasing the fully repaired protein (step 3). In contrast, while binding of MsrA (bottom) or MsrB (not depicted) to fully oxidized CaM (step 4) readily occurs, neither MsrA or MsrB alone can fully repair all S- or R-MetSO in CaMox under these experimental conditions, where there is substantial protein refolding at 25 °C (step 5) that competes with the ability of the individual Msr enzymes to recognize and bind MetSO in CaMox (step 5) (5, 6, 25, 32). Following protein refolding, there is no additional reduction of MetSO (step 6 is blocked), resulting in the retention of MetSO as occurs during biological aging (2).
The retention of S-MetSO in CaMox following enzymatic repair by MsrA is consistent with the spatial requirements around the active site cysteine, as steric limitations prevent the catalytic sulfur atom from interacting with MetSO residues within helices (5). Thus, upon reduction of a critical MetSO in CaMox (i.e., Met145) the restoration of the protein fold results in the retention of MetSO residues within secondary structures that are inaccessible to the active sites of Msr enzymes (5, 25, 32, 38). Substantial amounts of both S- and R-MetSO are retained following the simultaneous incubation of MsrB and MsrA with CaMox, where a stochastic distribution of CaM oxiforms are observed that retain up to eight MetSO per CaM (Figure 2E). The simultaneous binding of both catalytic domains of MsrBA prevents the refolding of CaMox (irrespective of the reduction of MetSO145) and promotes the complete repair of all MetSO in oxidized proteins (i.e., CaMox) (Figure 2F). These results indicate that the simultaneous binding of both active sites in MsrBA maintains CaMox in a partially unfolded state that exposes MetSO within conformationally disordered sequences for Msr enzyme recognition and repair (5)(Figure 6). Consistent with the latter suggestion that the inability of individual MsrA or MsrB enzymes to fully repair MetSO in CaMox is due to protein refolding, prior results have demonstrated the full repair of all MetSO by MsrA and MsrB upon disruption of the structure of CaMox at higher temperatures (47). In total, these results indicate that the simultaneous binding of both domains in MsrBA contributes to an ability of this bifunctional Msr enzyme to efficiently and completely reduce MetSO in CaMox. This enhanced repair activity against oxidized proteins represents an advantage under acute conditions of oxidative stress associated with microbial colonization, whe n high levels of protein oxidation are likely to occur (11, 16, 48).
Prior measurements have established the physiological importance of the correct localization of MsrAB to the outer membrane in both Neisseria gonorrhoeae and Helicobacter pylori, whose periplasmic localization promotes colonization under conditions of oxidative stress (11, 16). Coupling MsrAB to a thioredoxin domain (i.e., NT) at the N-terminus of MsrAB in Neisseria efficiently reduces the active site in both catalytic domains in the absence of added thioredoxin (22). Cytosolic reducing potential (i.e., NADPH) is coupled to the MsrAB systems through the integral membrane protein DspD, which promotes the reduction of the active sites in both MsrA and MsrB domains of MsrAB (22). These results suggested that enzyme fusion in bifunctional Msr enzymes functions primarily as an organizing mechanism to effectively couple the cytosolic reducing equivalents to active MsrA and MsrB sites in the periplasm. Consistent with this suggestion, gene fusion results in no change in the catalytic repair of free MetSO for MsrAB in comparison to MsrA and MsrB alone (46). Our results suggest that gene fusion of MsrA and MsrB to form MsrBA in Shewanella plays an additional role in promoting the rapid and complete repair of MetSO in oxidized proteins, which is expected to promote bacterial colonization in oxidizing environments.
Acknowledgments
This work was supported by grants from the National Institutes of Health (AG12993) and U. S. Department of Energy, Office of Science Genomics:GTL project. Pacific Northwest National Laboratory is operated for the DOE by Battelle Memorial Institute under contract DE-AC06-76RLO 1830.
We wish to thank Todd D. Williams for assistance in collecting the mass spectra and Diana J. Bigelow and H. Steven Wiley for insightful discussions.
Abbreviations
- β-ME
2-mercaptoethanol
- CaM
calmodulin
- CaMox
CaM in which all nine Mets are oxidized to MetSO
- ESI-MS
electrospray ionization mass spectrometry
- FLAsH-EDT2
4′5′-bis(1,3,2-dithoarsolan-2yl)fluorescein
- MetSO
methionine sulfoxide
- MS
mass spectrometry
- Msr
methionine sulfoxide reductase
- ROS
reactive oxygen species
- TCEP
triscarboxyethylphosphine
References
- 1.Hoshi T, Heinemann SH. Regulation of cell function by methionine oxidation and reduction. J Physiol (Lond) 2001;531:1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stadtman ER, Van Remmen H, Richardson A, Wehr NB, Levine RL. Methionine oxidation and aging. Biochim Biophys Acta. 2005;1703:135–140. doi: 10.1016/j.bbapap.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Schoneich C. Methionine oxidation by reactive oxygen species: reaction mechanisms and relevance to Alzheimer’s disease. Biochim Biophys Acta. 2005;1703:111–119. doi: 10.1016/j.bbapap.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Boschi-Muller S, Olry A, Antoine M, Branlant G. The enzymology and biochemistry of methionine sulfoxide reductases. Biochimica et Biophysica Acta (BBA) -Proteins & Proteomics. 2005;1703:231–238. doi: 10.1016/j.bbapap.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Xiong Y, Chen B, Smallwood HS, Urbauer RJ, Markille LM, Galeva N, Williams TD, Squier TC. High-affinity and cooperative binding of oxidized calmodulin by methionine sulfoxide reductase. Biochemistry. 2006;45:14642–14654. doi: 10.1021/bi0612465. [DOI] [PubMed] [Google Scholar]
- 6.Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, Derrick PJ, Barras F. Repair of Oxidized Proteins. Identification of a new methionine sulfoxide reductase. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 7.Lin Z, Johnson LC, Weissbach H, Brot N, Lively MO, Lowther WT. Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function. Proc Natl Acad Sci U S A. 2007;104:9597–9602. doi: 10.1073/pnas.0703774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delaye L, Becerra A, Orgel L, Lazcano A. Molecular evolution of peptide methionine sulfoxide reductases (MsrA and MsrB): on the early development of a mechanism that protects against oxidative damage. J Mol Evol. 2007;64:15–32. doi: 10.1007/s00239-005-0281-2. [DOI] [PubMed] [Google Scholar]
- 9.Ezraty B, Aussel L, Barras F. Methionine sulfoxide reductases in prokaryotes. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics. 2005;1703:221–229. doi: 10.1016/j.bbapap.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Sharov VS, Ferrington DA, Squier TC, Schoneich C. Diastereoselective reduction of protein-bound methionine sulfoxide by methionine sulfoxide reductase. FEBS Lett. 1999;455:247–250. doi: 10.1016/s0014-5793(99)00888-1. [DOI] [PubMed] [Google Scholar]
- 11.Skaar EP, Tobiason DM, Quick J, Judd RC, Weissbach H, Etienne F, Brot N, Seifert HS. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc Natl Acad Sci U S A. 2002;99:10108–10113. doi: 10.1073/pnas.152334799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowther WT, Weissbach H, Etienne F, Brot N, Matthews BW. The mirrored methionine sulfoxide reductases of Neisseria gonorrhoeae pilB. Nat Struct Biol. 2002;9:348–352. doi: 10.1038/nsb783. [DOI] [PubMed] [Google Scholar]
- 13.Antoine M, Boschi-Muller S, Branlant G. Kinetic Characterization of the Chemical Steps Involved in the Catalytic Mechanism of Methionine Sulfoxide Reductase A from Neisseria meningitidis. J Biol Chem. 2003;278:45352–45357. doi: 10.1074/jbc.M307471200. [DOI] [PubMed] [Google Scholar]
- 14.Olry A, Boschi-Muller S, Branlant G. Kinetic characterization of the catalytic mechanism of methionine sulfoxide reductase B from Neisseria meningitidis. Biochemistry. 2004;43:11616–11622. doi: 10.1021/bi049306z. [DOI] [PubMed] [Google Scholar]
- 15.Bigelow DJ, Squier TC. Redox modulation of cellular signaling and metabolism through reversible oxidation of methionine sensors in calcium regulatory proteins. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics. 2005;1703:121–134. doi: 10.1016/j.bbapap.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Alamuri P, Maier RJ. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004;53:1397–1406. doi: 10.1111/j.1365-2958.2004.04190.x. [DOI] [PubMed] [Google Scholar]
- 17.Hassouni ME, Chambost JP, Expert D, Van Gijsegem F, Barras F. The minimal gene set member msrA, encoding peptide methionine sulfoxide reductase, is a virulence determinant of the plant pathogen Erwinia chrysanthemi. Proc Natl Acad Sci U S A. 1999;96:887–892. doi: 10.1073/pnas.96.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhandayuthapani S, Blaylock MW, Bebear CM, Rasmussen WG, Baseman JB. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J Bacteriol. 2001;183:5645–5650. doi: 10.1128/JB.183.19.5645-5650.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, Nathan C. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci U S A. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douglas T, Daniel DS, Parida BK, Jagannath C, Dhandayuthapani S. Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J Bacteriol. 2004;186:3590–3598. doi: 10.1128/JB.186.11.3590-3598.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moskovitz J, Flescher E, Berlett BS, Azare J, Poston JM, Stadtman ER. Overexpression of peptide-methionine sulfoxide reductase in Saccharomyces cerevisiae and human T cells provides them with high resistance to oxidative stress. Proc Natl Acad Sci U S A. 1998;95:14071–14075. doi: 10.1073/pnas.95.24.14071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brot N, Collet JF, Johnson LC, Jonsson TJ, Weissbach H, Lowther WT. The thioredoxin domain of Neisseria gonorrhoeae PilB can use electrons from DsbD to reduce downstream methionine sulfoxide reductases. J Biol Chem. 2006;281:32668–32675. doi: 10.1074/jbc.M604971200. [DOI] [PubMed] [Google Scholar]
- 23.Ghosal D, Omelchenko MV, Gaidamakova EK, Matrosova VY, Vasilenko A, Venkateswaran A, Zhai M, Kostandarithes HM, Brim H, Makarova KS, Wackett LP, Fredrickson JK, Daly MJ. How radiation kills cells: survival of Deinococcus radiodurans and Shewanella oneidensis under oxidative stress. FEMS Microbiol Rev. 2005;29:361–375. doi: 10.1016/j.femsre.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Shi L, Squier TC, Zachara JM, Fredrickson JK. Respiration of metal (hydr)oxides by Shewanella and Geobacter: A key role for multiheme c-type cytochromes. Molecular Microbiology. 2007 doi: 10.1111/j.1365-2958.2007.05783.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun H, Gao J, Ferrington DA, Biesiada H, Williams TD, Squier TC. Repair of oxidized calmodulin by methionine sulfoxide reductase restores ability to activate the plasma membrane Ca-ATPase. Biochemistry. 1999;38:105–112. doi: 10.1021/bi981295k. [DOI] [PubMed] [Google Scholar]
- 26.Turk BE. Manipulation of host signalling pathways by anthrax toxins. Biochem J. 2007;402:405–417. doi: 10.1042/BJ20061891. [DOI] [PubMed] [Google Scholar]
- 27.Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 28.Thorn KS, Naber N, Matuska M, Vale RD, Cooke R. A novel method of affinity-purifying proteins using a bis-arsenical fluorescein. Protein Sci. 2000;9:213–217. doi: 10.1110/ps.9.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, Clayton RA, Meyer T, Tsapin A, Scott J, Beanan M, Brinkac L, Daugherty S, DeBoy RT, Dodson RJ, Durkin AS, Haft DH, Kolonay JF, Madupu R, Peterson JD, Umayam LA, White O, Wolf AM, Vamathevan J, Weidman J, Impraim M, Lee K, Berry K, Lee C, Mueller J, Khouri H, Gill J, Utterback TR, McDonald LA, Feldblyum TV, Smith HO, Venter JC, Nealson KH, Fraser CM. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotechnol. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 30.Chen B, Mayer MU, Markillie LM, Stenoien DL, Squier TC. Dynamic motion of helix A in the amino-terminal domain of calmodulin is stabilized upon calcium activation. Biochemistry. 2005;44:905–914. doi: 10.1021/bi048332u. [DOI] [PubMed] [Google Scholar]
- 31.Strasburg GM, Hogan M, Birmachu W, Thomas DD, Louis CF. Site-specific derivatives of wheat germ calmodulin. Interactions with troponin and sarcoplasmic reticulum. J Biol Chem. 1988;263:542–548. [PubMed] [Google Scholar]
- 32.Ferrington DA, Sun H, Murray KK, Costa J, Williams TD, Bigelow DJ, Squier TC. Selective degradation of oxidized calmodulin by the 20 S proteasome. J Biol Chem. 2001;276:937–943. doi: 10.1074/jbc.M005356200. [DOI] [PubMed] [Google Scholar]
- 33.Nelson DP, Kiesow LA. Enthalpy of decomposition of hydrogen peroxide by catalase at 25 degrees C (with molar extinction coefficients of H 2 O 2 solutions in the UV) Anal Biochem. 1972;49:474–478. doi: 10.1016/0003-2697(72)90451-4. [DOI] [PubMed] [Google Scholar]
- 34.Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 35.Rutberg B, Hoch JA. Citric acid cycle: gene-enzyme relationships in Bacillus subtilis. J Bacteriol. 1970;104:826–833. doi: 10.1128/jb.104.2.826-833.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boschi-Muller S, Azza S, Branlant G. E. coli methionine sulfoxide reductase with a truncated N terminus or C terminus, or both, retains the ability to reduce methionine sulfoxide. Protein Sci. 2001;10:2272–2279. doi: 10.1110/ps.10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, Yin DH, Yao Y, Sun H, Qin Z, Schoneich C, Williams TD, Squier TC. Loss of conformational stability in calmodulin upon methionine oxidation. Biophys J. 1998;74:1115–1134. doi: 10.1016/S0006-3495(98)77830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sacksteder CA, Whittier JE, Xiong Y, Li J, Galeva N, Jacoby ME, Purvine S, Williams TD, Rechsteiner MC, Bigelow DJ, Squier TC. Tertiary Structural Rearrangements upon Oxidation of Methionine145 in Calmodulin Promotes Targeted Proteasomal Degradation. Biophys J. 2006:1675–1245. doi: 10.1529/biophysj.106.086033. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams CH., Jr Thioredoxin-thioredoxin reductase--a system that has come of age. Eur J Biochem. 2000;267:6101. doi: 10.1046/j.1432-1327.2000.01700.x. [DOI] [PubMed] [Google Scholar]
- 40.Louie TM, Yang H, Karnchanaphanurach P, Xie XS, Xun L. FAD is a preferred substrate and an inhibitor of Escherichia coli general NAD(P)H:flavin oxidoreductase. J Biol Chem. 2002;277:39450–39455. doi: 10.1074/jbc.M206339200. [DOI] [PubMed] [Google Scholar]
- 41.Roy SO, Packard TT. NADP-Isocitrate dehydrogenase from Pseudomonas nautica: kinetic constant determination and carbon limitation effects on the pool of intracellular substrates. Appl Environ Microbiol. 1998;64:4958–4964. doi: 10.1128/aem.64.12.4958-4964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Keulen G, Girbal L, van den Bergh ER, Dijkhuizen L, Meijer WG. The LysR-type transcriptional regulator CbbR controlling autotrophic CO2 fixation by Xanthobacter flavus is an NADPH sensor. J Bacteriol. 1998;180:1411–1417. doi: 10.1128/jb.180.6.1411-1417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson WH, Yang X, Choi YE, Jones DP, Kehrer JP. Thioredoxin and its role in toxicology. Toxicol Sci. 2004;78:3–14. doi: 10.1093/toxsci/kfh050. [DOI] [PubMed] [Google Scholar]
- 44.Chen B, Mayer MU, Squier TC. Structural uncoupling between opposing domains of oxidized calmodulin underlies the enhanced binding affinity and inhibition of the plasma membrane Ca-ATPase. Biochemistry. 2005;44:4737–4747. doi: 10.1021/bi0474113. [DOI] [PubMed] [Google Scholar]
- 45.Gao J, Yao Y, Squier TC. Oxidatively modified calmodulin binds to the plasma membrane Ca-ATPase in a nonproductive and conformationally disordered complex. Biophys J. 2001;80:1791–1801. doi: 10.1016/S0006-3495(01)76149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olry A, Boschi-Muller S, Marraud M, Sanglier-Cianferani S, Van Dorsselear A, Branlant G. Characterization of the methionine sulfoxide reductase activities of PILB, a probable virulence factor from Neisseria meningitidis. J Biol Chem. 2002;277:12016–12022. doi: 10.1074/jbc.M112350200. [DOI] [PubMed] [Google Scholar]
- 47.Vougier S, Mary J, Dautin N, Vinh J, Friguet B, Ladant D. Essential Role of Methionine Residues in Calmodulin Binding to Bordetella pertussis Adenylate Cyclase, as Probed by Selective Oxidation and Repair by the Peptide Methionine Sulfoxide Reductases. J Biol Chem. 2004;279:30210–30218. doi: 10.1074/jbc.M400604200. [DOI] [PubMed] [Google Scholar]
- 48.Gunesekere IC, Kahler CM, Ryan CS, Snyder LA, Saunders NJ, Rood JI, Davies JK. Ecf, an alternative sigma factor from Neisseria gonorrhoeae, controls expression of msrAB, which encodes methionine sulfoxide reductase. J Bacteriol. 2006;188:3463–3469. doi: 10.1128/JB.188.10.3463-3469.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antoine M, Gand A, Boschi-Muller S, Branlant G. Characterization of the amino acids from Neisseria meningitidis MsrA involved in the chemical catalysis of the methionine sulfoxide reduction step. J Biol Chem. 2006;281:39062–39070. doi: 10.1074/jbc.M608844200. [DOI] [PubMed] [Google Scholar]
- 50.Gand A, Antoine M, Boschi-Muller S, Branlant G. Characterization of the amino-acids involved in substrate specificity of methionine sulfoxide reductase A. J Biol Chem. 2007;282:20484–20491. doi: 10.1074/jbc.M702350200. [DOI] [PubMed] [Google Scholar]
- 51.Coudevylle N, Antoine M, Bouguet-Bonnet S, Mutzenhardt P, Boschi-Muller S, Branlant G, Cung MT. Solution structure and backbone dynamics of the reduced form and an oxidized form of E. coli methionine sulfoxide reductase A (MsrA): structural insight of the MsrA catalytic cycle. J Mol Biol. 2007;366:193–206. doi: 10.1016/j.jmb.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 52.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]