Abstract
The E1b55K and E4orf6 proteins of adenovirus type 5 (Ad5) assemble into a complex together with cellular proteins including cullin 5, elongins B and C, and Rbx1. This complex possesses E3 ubiquitin ligase activity and targets cellular proteins for proteasome-mediated degradation. The ligase activity has been suggested to be responsible for all functions of E1b55K/E4orf6, including promoting efficient viral DNA replication, preventing a cellular DNA damage response, and stimulating late viral mRNA nuclear export and late protein synthesis. The known cellular substrates for degradation by E1b55K/E4orf6 are the Mre11/Rad50/Nbs1 DNA repair complex, the tumor suppressor p53, and DNA ligase IV. Here we show that the degradation of individual targets can occur independently of other substrates. Furthermore, we identify separation-of-function mutant forms of E1b55K that can distinguish substrates for binding and degradation. Our results identify distinct regions of E1b55K that are involved in substrate recognition but also imply that there are additional requirements beyond protein association. These mutant proteins will facilitate the determination of the relevance of specific substrates to the functions of E1b55K in promoting infection and inactivating host defenses.
The linear, double-stranded DNA genome of adenovirus serotype 5 (Ad5) is approximately 36 kb and encodes five early transcription units whose proteins perform essential functions for efficient infection. Viruses such as adenovirus employ numerous strategies to contend with host cell factors during the course of establishing a productive infection (10, 68). Examining mutant viruses has implicated E1b and E4 proteins in the modulation of the host cell environment, the promotion of viral DNA replication and viral mRNA export, the prevention of viral genome concatemerization, and the synthesis of late viral proteins (reviewed in references 3, 61, 67, and 68). In adenovirus-infected cells, most of the viral E1b55K protein is present in a complex with E4orf6 (52, 53). The E1b55K/E4orf6 complex has been implicated in the selective modulation of nucleocytoplasmic mRNA during the late phase of virus infection (reviewed in references 18 and 23). This viral complex also promotes efficient viral replication (8, 29, 33, 66) and prevents the viral genome from being concatemerized by host factors (60, 65). The precise mechanisms by which the E1b55K/E4orf6 complex achieves its many functions remain to be elucidated.
E1b55K displays a complex distribution pattern in infected cells and requires E4orf6 for nuclear localization (17, 27, 37, 42, 47) and association with viral replication centers (48). The E1b55K protein possesses a leucine-rich nuclear export signal (NES), which has been implicated in nucleocytoplasmic shuttling via the CRM1-mediated nuclear export receptor (19, 39). Export-deficient mutant forms of E1b55K accumulate in subnuclear aggregates that also contain cellular binding partners (20, 31). In the absence of E4orf6, the expression of E1b55K produces cytoplasmic aggregates (17, 27, 37, 42, 47). In transformed cells, E1b55K from Ad2 or Ad5 accumulates in a large cytoplasmic body (4, 9, 20, 74) with characteristics of an aggresome (38, 42). Many cellular proteins that interact with E1b55K localize at the aggresome in transfected and transformed cells (4, 9, 20, 22, 42, 44, 74). Therefore, protein aggregation has been proposed previously as a potential strategy utilized by E1b55K to inactivate cellular proteins (42) and promote transformation (31).
One tactic commonly employed by viruses to inactivate inhibitory host factors is to induce the specific downregulation or degradation of cellular proteins (25, 55). Degradation can be achieved via the covalent modification of target proteins with polyubiquitin chains by an E3 ubiquitin ligase, followed by recognition and destruction by the proteasome (reviewed in reference 35). The adenoviral E1b55K and E4orf6 proteins associate with cellular proteins to form an E3 ligase complex that contains elongins B and C, cullin 5, and Rbx1 (7, 30, 49). BC-box motifs in E4orf6 have been identified previously as important for binding elongins B and C (5, 15, 43). It has been suggested that all functions of the E1b55K/E4orf6 complex are due to the degradation of cellular proteins by the ubiquitin ligase activity (6, 16, 70). Since E1b55K alone can physically associate with p53 (13, 34, 41, 56, 71, 73), DNA ligase IV (2), and the Mre11/Rad50/Nbs1 (MRN) complex (12), it is believed to mediate substrate recognition, while both E4orf6 and E1b55K are required for proteasome-mediated degradation (12, 13, 49, 50, 56, 69). Not all proteins that interact with E1b55K are downregulated, but cellular proteins so far identified as degradation substrates of the E1b55K/E4orf6 complex include p53 (13, 28, 30, 46, 49-51, 58), the MRN DNA repair complex (12, 60), and the DNA ligase IV protein (2). The degradation of these cellular factors has been demonstrated by inhibition with proteasome inhibitors and by increased turnover in studies of protein half-life (12, 46, 49-51, 58, 60, 69). Exactly which substrates are relevant to specific functions of the E1b55K/E4orf6 complex is unclear. Additionally, the mechanisms for the selection and degradation of distinct proteins remain to be resolved. Understanding how the E1b55K/E4orf6 viral ubiquitin ligase targets specific cellular substrates is crucial to elucidating how these viral proteins perform their myriad of functions.
In the present study, we characterized some of the cellular and viral requirements for the degradation of each target. By examining the steady-state levels of E1b55K/E4orf6 substrates in mutant cell lines, we demonstrated that the downregulation of individual cellular factors can occur independently of the other factors. A series of E1b55K mutant proteins were characterized with respect to localization and the degradation of the known substrates. We identified regions of E1b55K important for cellular localization, and we demonstrated that there are independent requirements for p53, MRN, and DNA ligase IV downregulation. Our results show that affinity for substrates appears to be a major, but not sole, determinant of E1b55K-mediated degradation.
MATERIALS AND METHODS
E1b55K mutagenesis and cloning and cell transfections.
Previously described E1b55K mutant genes were amplified from corresponding viruses (56, 72) with PCR primers RVF (forward primer; 5′-CCGCTCGAGATGGAGCGAAGAAACCCATC-3′) and RVR (reverse primer; 5′-CCATCGATTCAATCTGTATCTTCATCGCTAG-3′) and cloned into the Xho1 and Cla1 sites of a modified version of the pCLNC retroviral vector, as previously described (12). Site-directed mutagenesis of the E1b55K gene in either the pDC516 backbone vector (provided by H. Young) or pCLNC was performed using a QuikChange mutagenesis kit (Stratagene) and the primers listed in Table 1. Mutant proteins were screened as described below, and interesting mutant constructs in the pDC516 vector were PCR amplified using RVF and RVR primers (or a modified RVR primer for the triple phosphorylation site mutant protein [3XPA]; 5′-CCATCGATTCAATCTGCATCTTCATCGGCAG-3′) and subcloned into the Xho1 and Cla1 sites of pCLNC (54). To generate NES mutant constructs in pDC516 (see the legend to Fig. 7B), genes encoding the E1b555K mutant proteins R240A and H354 were PCR amplified and introduced in place of the wild-type (WT) E1b55K gene in pDC516 by using SmaI and SacI sites. E1b gene fragments containing mutations were then subcloned as follows: for the protein with both NES and H373A mutations (NES-H373A), the EcoRI-PstI fragment corresponding to the E1b55K NES was cloned into pDC516 expressing the H373A mutant protein, and for NES-R240A and NES-H354, the PstI-BglII fragment corresponding to the region containing H354 or R240A (in pDC516) was cloned into pDC516 expressing the E1b55K NES. All mutant proteins used in this study were verified by sequencing. Cells were transfected with plasmids by using Lipofectamine 2000 according to the protocol of the manufacturer (Invitrogen).
TABLE 1.
E1b55K mutagenesis primers
| Mutant protein | Vector backbone | Round of mutagenesis | Primer type | Primer sequence (5′ to 3′)a |
|---|---|---|---|---|
| L83,87,91A (NES mutant protein) | pDC516 | First round | Forward | GGCTGAACTGAGACGCATTGCGACAATTACAGAGG |
| Reverse | CCTCTGTAATTGTCGCAATGCGTCTCAGTTCAGCC | |||
| Second round | Forward | GGCTGAACTGTATCCAGAAGCGAGACGCATTGCG | ||
| Reverse | CGCAATGCGTCTCGCTTCTGGATACAGTTCAGCC | |||
| Third round | Forward | GTACAGGTGGCTGAAGCGTATCCAGAAGCGAGACGC | ||
| Reverse | GCGTCTCGCTTCTGGATACGCTTCAGCCACCTGTAC | |||
| K104R | pDC516 | Forward | GGGCTAAAGGGGGTGAGGAGGCAGCGGGGGG | |
| Reverse | CCCCCCGCTGCCTCCTCACCCCCTTTAG | |||
| C348,351S | pDC516 | Forward | CATAACATGGTAAGTGGCAACAGCGAGGACAGG | |
| Reverse | CCTGTCCTCGCTGTTGCCACTTACCATGTTATG | |||
| C361,366S | pDC516 | First round | Forward | TCAGATGCTGACCTCCTCGGACGGCAA |
| Reverse | TTGCCGTCCGAGGAGGTCAGCATCTGA | |||
| Second round | Forward | TCGGACGGCAACTCTCACCTGCTGAAG | ||
| Reverse | CTTCAGCAGGTGAGAGTTGCCGTCCGA | |||
| H373A | pDC516 | Forward | TGCTGAAGACCATTGCCGTAGCCAGCCACT | |
| Reverse | AGTGGCTGGCTACGGCAATGGTCTTCAGCA | |||
| C454,456S | pDC516 | Forward | CCAGGTGCAGACCCTCCGAGTCTGGCGGTAAACATATTAGG | |
| Reverse | CCTAATATGTTTACCGCCAGACTCGGAGGGTCTGCACCTGG | |||
| S490,491,T495A | pDC516 | First round | Forward | GCTCTAGCGATGAAGATGCAGATTGAGGTACTG |
| Reverse | CAGTACCTCAATCTGCATCTTCATCGCTAGAGC | |||
| Second round | Forward | CGCTGAGTTTGGCGCTGCCGATGAAGATGCAGATTGAGG | ||
| Reverse | CCTCAATCTGCATCTTCATCGGCAGCGCCAAACTCAGCG | |||
| S490,491,T495D | pCLNC | First round | Forward | GGCTCTAGCGATGAAGATGACGATTGAGGTACTGAA |
| Reverse | CAGTACCTCAATCGTCATCTTCATCGCTAGAGC | |||
| Second round | Forward | CGCTGAGTTTGGCGATGACGATGAAGATGACGATTGAGG | ||
| Reverse | CCTCAATCGTCATCTTCATCGTCATCGCCAAACTCAGCG |
Codons changed in the primer sequence are underlined. Primers were used to introduce changes into E1b55K with the backbone vector pDC516 or pCLNC. Constructs encoding interesting mutant proteins were subcloned into retroviral vector pCLNC in order to make cell lines. All mutations were verified by sequencing.
FIG. 7.
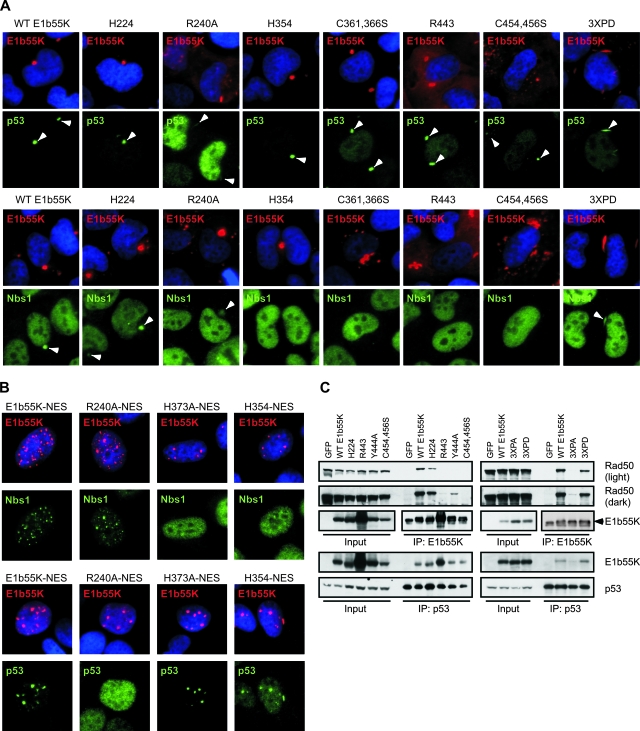
Defects in substrate binding of E1b55K mutant proteins. (A) E1b55K mutant proteins colocalize with cellular substrates in cytoplasmic aggresomes/aggregates. U2OS cells were transfected with E1b55K mutant constructs to induce cytoplasmic aggresome/aggregate formation. Cells were processed for immunofluorescence analysis after 24 to 36 h by using antibodies to the indicated proteins. DAPI staining marks cell nuclei. Arrowheads indicate the colocalization of substrates with E1b55K in cytoplasmic aggregates. (B) E1b55K NES mutant proteins relocalize cellular targets into nuclear aggregates. U2OS cells were transfected with constructs expressing E1b55K proteins with NES mutations. Cells were processed for immunofluorescence analysis after 24 h by using antibodies to the indicated proteins. DAPI staining marks cell nuclei. (C) Lysates from mutant E1b55K cell lines were subjected to immunoprecipitation (IP) with the 2A6 antibody to E1b55K or a p53 antibody as described in Materials and Methods. Approximately half the immunoprecipitate was analyzed by immunoblotting alongside 5% of the input lysate.
Cell lines.
HeLa, U2OS, IMR90, Saos2, and 293 cells were purchased from the American Tissue Culture Collection. Stable cell lines derived from HeLa and U2OS that express WT and mutant E1b55K from retrovirus vectors have been described previously (12). Immortalized A-TLD3 and A-TLD1 cells were described previously (12, 59, 60). NBS cells were provided by P. Concannon (14). FUS9 cells with mutant DNA-dependent protein kinase (DNA-PKcs) (32) were provided by T. Melendy. All cells were maintained as monolayers in Dulbecco modified Eagle's medium (DMEM) supplemented with 10 or 20% fetal bovine serum (FBS) and penicillin-streptomycin, except FUS9 cells, which required a 1:1 ratio of DMEM and Ham's F10 medium supplemented with 10% FBS and penicillin-streptomycin. All cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
Viruses and infections.
The mutant viruses dl3112 (ΔE1b55K ΔE4orf3) and dl110 (ΔE1b55K) have been described previously (1, 57) and were obtained from D. Ornelles and G. Ketner, respectively. The viruses with mutated phosphorylation sites in E1b55K (pm490/1/5A and pm490/1A) have been described previously (62, 63) and were obtained from P. Branton. The H224, H354, and R443 viruses with mutations in E1b55K have been described previously (72) and were obtained from A. Berk. The Y444A (ONYX85) virus has been described previously (56) and was obtained from Y. Shen. The E1 deletion mutant recombinant adenovirus vector expressing E4orf6 (rAdE4orf6) was obtained from P. Branton (50). All viruses were propagated in 293 cells, purified by two sequential rounds of ultracentrifugation in cesium chloride gradients, and stored in 40% glycerol at −20°C. Infections were performed on monolayers of cells in DMEM supplemented with 2% FBS. After 2 h at 37°C, additional serum was added to obtain a total concentration of 10%.
Antibodies, immunoblotting, and immunofluorescence.
Primary antibodies to the indicated proteins were purchased from Novus Biologicals Inc. (Nbs1), Genetex (Mre11 and Rad50), Serotech (DNA ligase IV), Santa Cruz (Ku70 and p53 [antibody C-19]), Research Diagnostics Inc. (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]), and Calbiochem (PCNA and p53 [antibody Ab-6]). The monoclonal B6 antibody to DBP and the monoclonal 2A6 antibody to E1b55K were obtained from A. Levine. The E4orf6 monoclonal antibody Rsa#3 (45) was obtained from D. Ornelles. Secondary antibodies were purchased from Jackson Laboratories or Eurogentec. Immunoblotting and immunofluorescence analyses were performed essentially as described previously (12). For immunofluorescence analyses, cells were fixed with 4% paraformaldehyde before being permeabilized with 0.5% Triton X-100 and processed as described previously (12).
Immunoprecipitations.
U2OS E1b55K-expressing cells were lysed with buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 0.5% NP-40, and 1× protease inhibitors without EDTA (Roche) for 30 min on ice. After cellular debris was spun out, 400 to 500 μg of total protein per reaction (amounts were constant among experiments) was precleared with protein A/G beads (Santa Cruz) for 1 h at 4°C. Twenty-five microliters of the anti-E1b55K antibody (2A6) or the anti-DBP antibody (B6) as a control or 10 μl of the p53 antibody (C-19) was added to the precleared supernatant along with protein A/G beads, and the mixture was rotated overnight at 4°C. Beads were then washed three times for 30 min each in fresh lysis buffer before being boiled in sodium dodecyl sulfate (SDS) loading buffer and run on SDS-polyacrylamide gels for the resolution of interacting proteins by immunoblotting. To detect the interaction of E1b55K mutant proteins with E4orf6, beads were heated at 55°C for 15 min in SDS loading buffer before gel loading.
Analysis of concatemer formation by pulsed-field gel electrophoresis.
Cells were infected with WT Ad5 or the ΔE1b55K ΔE4orf3 mutant dl3112 at a multiplicity of infection (MOI) of 10. Cells were harvested at 30 h postinfection (hpi), and DNA was analyzed for concatemer formation as described previously (60, 65). Briefly, cells were incorporated into agarose plugs that were treated with a proteinase K solution (1.2% SDS, 0.125 M EDTA, and 5 mg of proteinase K/ml) at 50°C overnight and then washed with 50 mM EDTA solution. Plugs were loaded onto a 1.2% high-gelling-temperature agarose gel and subjected to pulsed-field electrophoresis for 16 h. DNA was visualized by staining the gel with Sybr green.
RESULTS
Cellular substrates of the E1b55K/E4orf6 complex are degraded independently of one another.
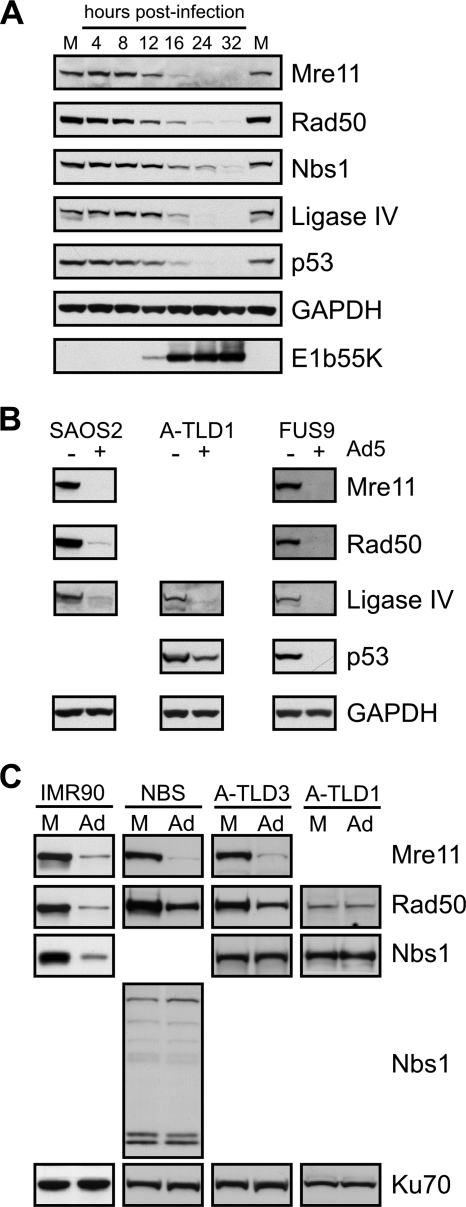
We first analyzed the kinetics of substrate degradation in U2OS cells, which express all currently known targets of the E1b55K/E4orf6 complex. The degradation of cellular proteins was examined by immunoblotting over a time course of infection with WT Ad5 (Fig. 1A). During infection, the E1b55K protein was first detected at 12 hpi. At this time point, the total steady-state levels of the Mre11, Rad50, Nbs1, p53, and DNA ligase IV proteins began to decrease. Nbs1 downregulation was slightly delayed compared to that of the other substrates, suggesting that Nbs1 may not be a direct target. Since many of the cellular DNA repair proteins are part of large protein complexes, we determined whether cellular proteins could be degraded independently of one another by using mutant cell lines (Fig. 1B). Mre11, Rad50, and DNA ligase IV were degraded in Saos2 cells that do not express p53. A-TLD1 cells (59) harbor a mutation in Mre11 that destabilizes the MRN complex. We found that DNA ligase IV and p53 could still be degraded in these cells, although we observed in experiments with reproducible results that p53 levels were not completely diminished. DNA ligase IV and the MRN complex are critical to DNA repair by nonhomologous end joining. We therefore also examined whether DNA-PKcs, another key nonhomologous end-joining factor, was required for the degradation of substrates. Ad5 infection of FUS9 cells, which lack DNA-PKcs (32), still induced the downregulation of all known substrates. We have also previously shown that the ATM and ATR kinases and signaling pathways are not required for the degradation of MRN (12). Together, these data demonstrate that MRN, p53, and DNA ligase IV can undergo virus-induced downregulation independently of one another and other proteins involved in the DNA damage response.
FIG. 1.
Cellular substrates of the E1b55K/E4orf6 complex are degraded independently of one another during adenovirus infection. (A) Degradation of cellular proteins over a time course of Ad5 infection. U2OS cells were infected with WT Ad5 (MOI of 25), and lysates were harvested at the indicated times postinfection for analysis by immunoblotting with specific antibodies. GAPDH served as a loading control. Protein levels were compared to those in mock-infected cells (M). (B) Independent degradation of p53, the MRN complex, and DNA ligase IV. Cell lines with mutant p53 (Saos2), Mre11 (A-TLD1), or DNA-PKcs (FUS9) were infected with WT Ad5 (MOIs of 25, 100, and 25, respectively). Lysates were prepared at 24 to 48 hpi and analyzed by immunoblotting for the indicated proteins. +, present; −, absent. (C) Mre11 may be the degradation target within the MRN complex. IMR90, A-TLD3, A-TLD1, or NBS cells were either mock treated (M) or infected with WT Ad5 (Ad; MOI of 75). Cells were harvested at 24 to 48 hpi, and lysates were analyzed by immunoblotting for the indicated proteins. Ku70 served as a loading control.
To examine the degradation of the MRN complex by E1b55K/E4orf6, we employed mutant NBS and A-TLD cell lines. IMR90 fibroblast cells were infected as a control and showed the degradation of all MRN complex members (Fig. 1C). NBS cells harbor a mutation in the Nbs1 gene resulting in a truncated protein defective in complex formation with Mre11 and, thus, Rad50 (11). The infection of these cells with Ad5 caused the degradation of both Mre11 and Rad50 (Fig. 1C), suggesting that full-length Nbs1 is not required. Smaller proteins recognized by the Nbs1 antibody were not altered by Ad5 infection. A-TLD3 cells contain a missense mutation in Mre11 that diminishes its interaction with Nbs1 (59), while A-TLD1 cells express a prematurely truncated Mre11 protein that is unstable and defective for Rad50 and Nbs1 interaction (59). A-TLD3 cells showed reduced levels of both Mre11 and Rad50 during Ad5 infection (Fig. 1C). Although we had difficulty visualizing the truncated form of Mre11 in A-TLD1 cells, Rad50 was unaltered (Fig. 1C), suggesting that full-length Mre11 is required for the destabilization of Rad50 and Nbs1 during Ad5 infection.
Identification of additional E1b55K mutant proteins with defects in MRN degradation.
Previous studies have described a large number of E1b55K mutant proteins (26, 52, 56, 63, 72), but many of these have not been assessed for the degradation of all the known substrates. We previously identified two E1b55K mutant forms that can distinguish between the degradation targets p53 and MRN (12). In the presence of E4orf6, the R240A mutant protein degrades the MRN complex but not p53, whereas the H354 insertion mutant protein degrades p53 but not MRN. We extended these studies by examining a number of E1b55K mutant viruses (56, 72) with either point mutations or small insertions across the length of E1b55K. A subset of mutations is shown in the schematic in Fig. 2.
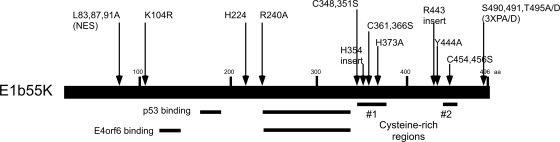
FIG. 2.
Schematic representation of E1b55K and the relative locations of the mutations within the mutant proteins used in this study. The relative locations of p53 binding domains (34, 71) and E4orf6 binding domains (52) are underlined. Two cysteine-containing regions important for the degradation of substrates are also highlighted.
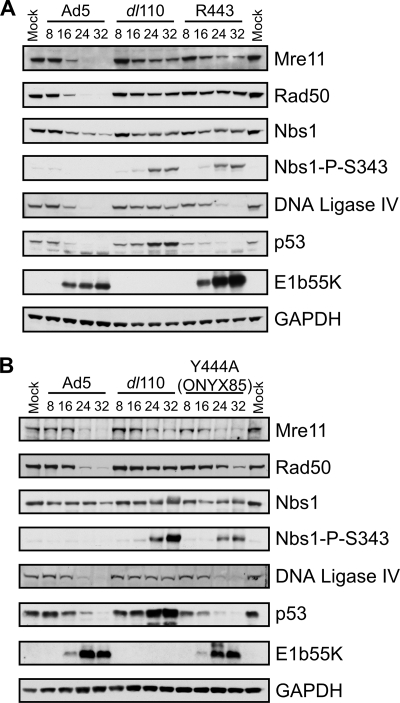
Infections with mutant E1b55K viruses were compared to those with WT Ad5 and the E1b55K deletion virus dl110 with respect to the degradation of p53 and MRN over the time course of infection. In addition to the previously described H354 and R240A viruses, we found separation-of-function phenotypes for E1b55K mutant viruses R443 (which has a 4-amino-acid insertion at residue 443) (72) and Y444A (ONYX85) (56). While both viruses induced the degradation of p53, in agreement with previous reports (56, 58), the kinetics of p53 degradation was delayed for R443. We found that neither the R443 virus nor the Y444A virus exhibited complete downregulation of all three MRN complex members (Fig. 3). The R443 virus also produced more E1b55K than WT Ad5. It is interesting that the insertion creating the R443 mutation also results in a Y444D mutation (72), which may contribute to a compound phenotype. In the course of our survey, we found that the R443A mutation (in virus ONYX84) (56) did not affect the degradation of MRN (data not shown), suggesting that the region around position 443, rather than the actual residue at this position, is important to MRN downregulation.
FIG. 3.
E1b55K C-terminal mutant proteins with defects in the degradation of the MRN complex. U2OS cells were infected (MOI of 50) with WT Ad5, the ΔE1b55K mutant dl110, or the R443 (A) or Y444A (B) mutant virus, and cells were harvested at the times postinfection indicated above the gels. Lysates were analyzed by immunoblotting with the indicated antibodies, and the results were compared to those from mock-infected cells. GAPDH served as a loading control. Nbs1-P-S343, Nbs1 phosphorylated at S343.
Given that the MRN complex is required for the DNA damage response to mutant adenoviruses (12), we tested whether these mutants induced damage signaling. We observed that the partial degradation of MRN by R443 and Y444A was not sufficient to prevent signaling during viral infection (Fig. 3; also data not shown), as shown by Nbs1 phosphorylation (at S343). This finding suggests that infection with adenovirus mutants unable to neutralize MRN completely can induce DNA damage signaling. Together with our previous observations (12), these data indicate that multiple regions of E1b55K are important for MRN degradation. The complete survey results for mutant proteins are presented in Table 2, together with a summary of observations previously reported in the literature.
TABLE 2.
Summary of E1b55K mutant phenotypesa
| Protein | Substrate degradationb phenotype (during coexpression with E4orf6 alone) for:
|
Substrate degradationc phenotype (during virus infection) for:
|
Substrate bindingd phenotype for:
|
Aggregate colocalizatione phenotype for:
|
Cytoaggregatef phenotype | Nuclear retention by E4orf6g | Concatemer formationh phenotype | Additional reference(s)j | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p53 | MRN | Lig4 | p53 | MRN | Lig4 | p53 | MRN | p53 | MRN | |||||
| WT E1b55K | + | + | + | + | + | + | + | + | + | + | + | + | − | |
| L83,87,89A (NES mutant protein) | ND | + | ND | ± | + | ND | + | + | + | + | NA | + | ND | 20, 31, 36, 39 |
| K104R | ND | + | ND | + | + | ND | ND | + | ND | + | + | + | ND | 21, 36 |
| H224 insertion mutant protein | + | + | + | + | + | + | + | + | + | ± | + | ± | − | 26, 34, 52, 72 |
| R240A | − | + | + | − | + | + | − | + | − | ± | + | + | − | 12, 56 |
| C348,351S | ND | + | ND | ND | ND | ND | ND | ND | ND | ND | + | + | ND | |
| H354 insertion mutant protein | + | − | + | + | − | + | + | − | + | − | + | + | − | 12, 26, 34, 42, 52, 71, 72 |
| C361,366S | − | − | − | − | − | − | ± | − | + | − | + | ± | + | |
| H373A | + | − | + | + | − | + | + | − | + | − | + | + | − | |
| R443 insertion mutant protein | ± | − | + | ± | − | ± | + | − | + | − | +/diffuse | ± | + | 26, 34, 52, 58, 71, 72 |
| Y444A | + | + | + | + | ± | + | + | ± | + | ± | + | ± | − | 56 |
| C454,456S | − | − | − | − | − | − | + | − | + | −i | + | ± | + | 31 |
| S490,491,T495A (3XPA) | − | + | ± | − | − | − | − | − | NA | NA | Diffuse | ± | + | 50, 62 |
| S490,491,T495D (3XPD) | ± | + | + | ND | ND | ND | + | + | + | ± | + | + | − | |
Unless otherwise indicated, symbols and abbreviations are as follows: +, proficient (may not reflect WT E1b55K activity); ±, defective; −, very defective; ND, not determined; NA, not applicable.
Degradation of endogenous proteins by E1b55K mutant forms in the presence of rAdE4orf6 as assessed by Western blotting or immunofluorescence analysis.
Degradation of endogenous proteins assessed in the context of viruses expressing E1b55K mutant proteins or E1b55K mutant cell lines infected with dl3112.
Interaction measured by coimmunoprecipitation experiments.
MRN or p53 colocalization with E1b55K aggregates as assessed by immunofluorescence.
E1b55K staining in the absence of rAdE4orf6 was assayed by using immunofluorescence. +, presence of E1b55K aggregates; diffuse, diffuse cytoplasmic E1b55K. See Fig. 4A and the corresponding legend for data and details.
E1b55K staining in the presence of rAdE4orf6 was assayed by using immunofluorescence. See Fig. 4A and the corresponding legend for data and details.
Concatemer formation by mutant virus dl3112 measured in mutant E1b55K-expressing cells.
Observation from the present study based on E1b55K expression in human cells.
This table summarizes data from this paper as well as the additional references indicated.
Two cysteine-containing regions of E1b55K are important for substrate degradation.
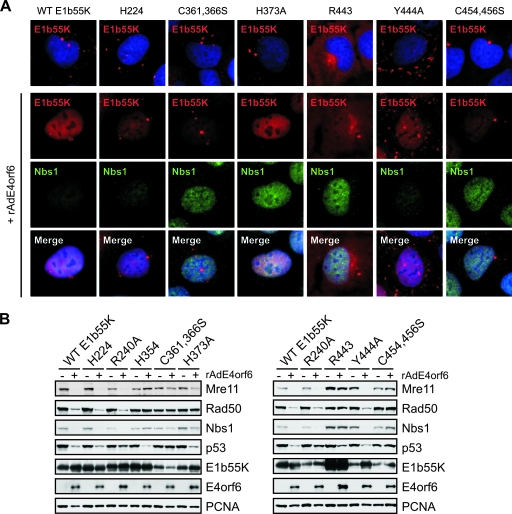
The changes in the E1b55K mutant proteins that we identified as defective for MRN degradation (H354, R443, and Y444A) are located in or near two regions that contain cysteine residues and may resemble zinc fingers (26). To analyze E1b55K further, we cloned the genes for these and other site-specific mutant forms into retroviral vectors (Fig. 2). Stable cell lines expressing individual E1b55K proteins were generated by retrovirus transduction as described previously (12). Mutant proteins were examined by immunofluorescence analysis in the presence and absence of E4orf6 for cellular localization and the degradation of the MRN complex. In the absence of E4orf6, WT E1b55K was localized in cytoplasmic aggregates (Fig. 4A), as described previously (27, 37). In the presence of E4orf6, E1b55K accumulated in a diffuse nuclear pattern with few remaining cytoplasmic foci and this outcome was accompanied by the degradation of Nbs1. The E1b55K mutant proteins exhibited a variety of patterns, which are shown in Fig. 4A and summarized in Table 2. Those with patterns differing from the WT E1b55K pattern included the R443 protein, which displayed predominantly diffuse cytoplasmic staining, and the Y444A protein, which showed more elongated cytoplasmic foci (Fig. 4A). With the exception of the Y444A and C348,351S proteins, the mutant forms with changes near cysteine regions were unable to degrade MRN in the presence of E4orf6 (Fig. 4A and Table 2), regardless of their localization patterns. Many mutant proteins were not retained in the nucleus to the same extent as WT E1b55K in the presence of E4orf6 (Fig. 4A and Table 2). However, this phenotype did not correlate completely with the lack of MRN degradation, since mutant forms H224 and Y444A were still able to degrade MRN (Fig. 4A). This finding suggests that nuclear retention by E4orf6 is not absolutely required for the degradation of cellular substrates.
FIG. 4.
Localization and substrate degradation in cell lines expressing WT and mutant E1b55K proteins. (A) U2OS cell lines stably expressing WT and mutant E1b55K proteins were either mock treated or infected with rAdE4orf6 (MOI of 50) for 24 h. E1b55K and Nbs1 were detected by immunofluorescence. DAPI (4′,6-diamidino-2-phenylindole) staining marks the cell nuclei in the top and bottom panels. (B) Lysates from the infected cells described above were analyzed by immunoblotting with antibodies to the indicated cellular and viral proteins. PCNA served as a cellular loading control. +, present; −, absent.
Using the stable E1b55K-expressing cell lines, we also examined E1b55K/E4orf6-mediated degradation of targets by immunoblotting (Fig. 4B). The previously characterized cell lines expressing green fluorescent protein (GFP), WT E1b55K, and mutant proteins R240A and H354 (12) were included as controls. The immunoblotting results confirmed our initial immunofluorescence observations and indicated that, with the exception of Y444A, most of the isolated mutant proteins were defective in MRN degradation (Fig. 4B and Table 2). C361,366S and C454,456S did not appear to degrade either p53 or MRN (Fig. 4B). This phenotype is unlikely to be due to defective E4orf6 binding since these mutant forms exhibited an interaction with E4orf6 similar to that of H224 (Fig. 4A; data not shown), a mutant protein competent to degrade all substrates but previously reported to exhibit reduced interaction with E4orf6 (52). H373A behaved similarly to H354 (12) in cellular localization and the degradation of p53 but not MRN (Fig. 4). R443 also appeared to be a separation-of-function mutant protein upon immunoblotting, although considering the levels of E1b55K, p53 degradation was not as robust as that by the WT. Together with our previous data (12), the results for these mutant proteins illustrate that the requirements for E1b55K-mediated degradation of MRN are distinct from those for the degradation of p53, and they identify two cysteine regions as critical for the proper localization of E1b55K and MRN degradation.
The phosphorylation of E1b55K is required for correct cellular localization and the degradation of substrates.
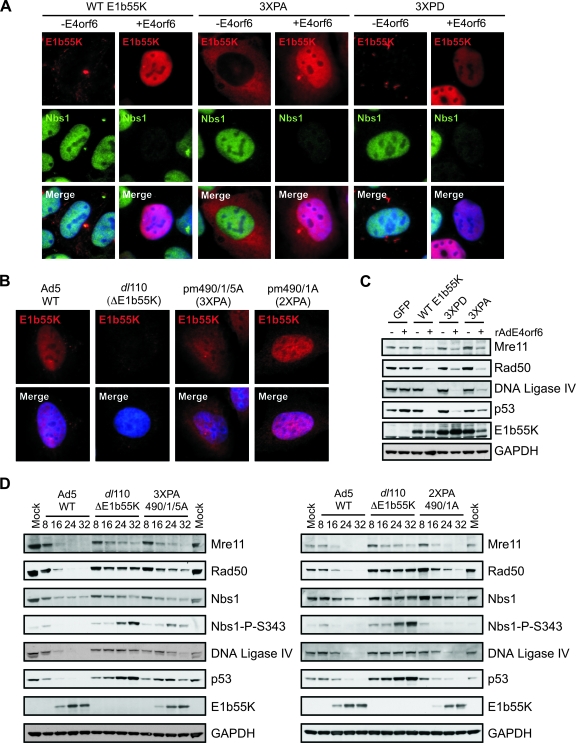
Multiple posttranslational modifications of E1b55K have been reported, including SUMO-1 conjugation at K104 (21, 36, 40) and phosphorylation at the C terminus (62, 63). We wanted to address whether these modifications affected E1b55K localization or substrate degradation. E1b55K proteins harboring mutations at the SUMO modification site (K104) (21) and the known phosphorylation sites in the C terminus (S490, S491, and T495) (62, 63) were expressed from plasmids and first assessed for localization in the absence and presence of E4orf6 (Fig. 5A). The mutation of the sumoylation site did not affect E1b55K localization in the absence of E4orf6 (Table 2 and data not shown). However, the triple phosphorylation site mutant protein, 3XPA, displayed a pattern drastically different from that of WT E1b55K. Instead of localizing in characteristic cytoplasmic aggregates, 3XPA was found diffusely spread throughout the cytoplasm, somewhat similar to R443 (Fig. 5A). In some cells, one or two very small E1b55K foci in the cytoplasm were noted, but no larger aggregates were detected. In the presence of E4orf6 expressed after transfection (Fig. 5A) or during infection (Fig. 5B) with the 3XPA mutant virus, pm490/1/5A (62), we observed increased nuclear staining, although a large amount of E1b55K also remained in the cytoplasm. The 2XPA double phosphorylation site mutant virus, pm490/1A (62), displayed a more predominantly nuclear E1b55K staining pattern (Fig. 5B). To address whether phosphorylation was required for the proper localization of E1b55K, we constructed a phosphomimic mutant protein in which the amino acids at positions 490, 491, and 495 were changed to aspartic acid (3XPD). After transient transfection with the construct expressing this mutant protein, we observed a cellular localization pattern more similar to that of WT E1b55K (Fig. 5A). However, the cytoplasmic aggregates of 3XPD appeared slightly more elongated than those of WT E1b55K, somewhat similar to those of the Y444A protein. These results suggest that the phosphorylation of E1b55K is critical to proper cellular localization.
FIG. 5.
The phosphorylation of E1b55K is required for correct cellular localization and substrate degradation. (A) U2OS cells were transfected with constructs expressing WT E1b55K, E1b55K mutant protein 3XPA, or E1b55K phosphomimic 3XPD. After 16 h, cells were superinfected with rAdE4orf6 (MOI of 20) for 24 h before being processed for immunofluorescence analysis with the indicated antibodies. Cell nuclei are stained with DAPI in the merged images. −E4orf6, without E4orf6; +E4orf6, with E4orf6. (B) U2OS cells were infected with WT Ad5, the ΔE1b55K mutant dl110, or E1b55K mutant virus pm490/1/5A (expressing 3XPA) or pm490/1A (expressing 2XPA) for approximately 24 h. Cells were fixed and analyzed for E1b55K localization by using immunofluorescence. DAPI indicates cell nuclei in the merged images. (C) Cell lines expressing E1b55K were infected with rAdE4orf6 (MOI of 50) for 24 h. Cells were harvested, and lysates were analyzed by immunoblotting with the indicated antibodies. +, present; −, absent. (D) U2OS cells were infected with WT Ad5, the ΔE1b55K mutant dl110, or E1b55K phosphomutant virus pm490/1/5A (expressing 3XPA; left panel) or pm490/1A (expressing 2XPA; right panel), all at an MOI of 50. Cells were harvested at the times postinfection indicated above the gels and processed for immunoblotting with the indicated antibodies. GAPDH served as a cellular loading control. Nbs1-P-S343, Nbs1 phosphorylated at S343.
We then examined how the SUMO and phosphorylation site mutations affected substrate downregulation. Immunofluorescence analyses revealed that the mutant proteins were competent to degrade MRN in the presence of E4orf6 (Fig. 5A and Table 2). To analyze further the requirement of E1b55K phosphorylation for degradation, we generated stable U2OS cell lines expressing the 3XPA and 3XPD proteins and compared them to the WT E1b55K-expressing cell line. The localization pattern in these stable cell lines was similar to that seen after transient transfection, although less nuclear staining in the presence of E4orf6 was noted (data not shown). The degradation of cellular targets in these cell lines was assessed with E4orf6 by immunoblotting. The MRN complex was degraded in all three E1b55K-expressing cell lines (Fig. 5C), confirming the immunofluorescence data (Fig. 5A). The 3XPA protein was defective in p53 downregulation, while the phosphomimic 3XPD partially rescued degradation (Fig. 5C). To confirm that these phenotypes occurred in the context of virus infection, we infected U2OS cells with either the 3XPA or 2XPA mutant virus (pm490/1/5A or pm490/1A, respectively) and analyzed degradation over time (Fig. 5D). The 2XPA virus was competent to downregulate substrates and prevent damage signaling, consistent with previously reported p53 degradation (50). In contrast, we found that the 3XPA virus was defective for the degradation of cellular targets in general (Fig. 5D) and damage signaling was observed as indicated by the phosphorylation of Nbs1. Our data suggest that the phosphorylation of the residues mutated in 3XPA is required for efficient degradation in the context of virus infection.
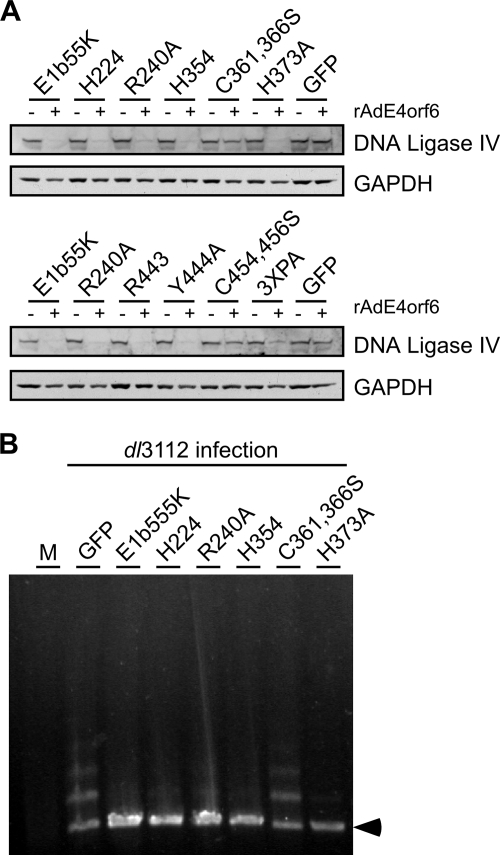
E1b55K separation-of-function mutant proteins still degrade DNA ligase IV to prevent concatemer formation.
In addition to MRN and p53, DNA ligase IV was recently described as a degradation substrate of E1b55K/E4orf6 (2). We tested the E1b55K mutant cell lines for the downregulation of this target in the presence of E4orf6. As was observed for MRN and p53 degradation, the proteins C361,366S and C454,456S were defective for the degradation of DNA ligase IV (Fig. 6A). The 3XPA mutant protein displayed reduced DNA ligase IV downregulation (Fig. 5C and D and 6A). Interestingly, we found that the separation-of-function mutant proteins could all degrade DNA ligase IV, regardless of p53 or MRN specificity (Fig. 6A). This pattern was also true for most of the E1b55K mutant viruses (Fig. 3 and Table 2), although the R443 virus had a slight defect (Fig. 3A). These data indicate that the requirements of E1b55K for DNA ligase IV degradation are distinct from those for both p53 and MRN degradation.
FIG. 6.
E1b55K separation-of-function mutant proteins still degrade DNA ligase IV to prevent concatemer formation. (A) U2OS-based cell lines stably expressing WT and mutant E1b55K proteins were either mock treated or infected with rAdE4orf6 (MOI of 50) for 24 h. Lysates were analyzed by immunoblotting for DNA ligase IV and GAPDH (loading control). +, present; −, absent. (B) E1b55K mutant proteins that cannot degrade MRN still retain the ability to prevent concatemer formation. U2OS cells expressing E1b55K mutant forms were infected with dl3112 at an MOI of 10 for 30 h, and viral DNA was analyzed by pulsed-field gel electrophoresis as described in Materials and Methods. Controls included a GFP-expressing U2OS cell line that was either mock treated (M) or infected with dl3112 to produce concatemers. An arrowhead indicates the position of the linear viral genome.
The concatemerization of mutant viral genomes requires both the MRN complex and DNA ligase IV (60). Therefore, E1b55K proteins unable to degrade MRN may still be able to prevent the concatemerization of a mutant virus if DNA ligase IV is downregulated. We examined whether concatemers formed during the infection of E1b55K mutant cell lines (Fig. 6B). Since the E4orf3 protein can independently prevent concatemers (60), we utilized the dl3112 mutant virus, which has both E1b55K and E4orf3 deleted (1, 57). As expected, mutant viral genomes were concatemerized in GFP-expressing control cells but not in cells expressing WT E1b55K protein. We also found that concatemers were inhibited in mutant cell lines proficient in the degradation of DNA ligase IV alone (those expressing H354 or H373A) or in cell lines in which both MRN and DNA ligase IV were degraded (those expressing H224, R240A, Y444A, or 3XPD) (Fig. 6B and Table 2). There was no correlation between p53 degradation and concatemers (Table 2). Additionally, the cell lines expressing C361,366S and C454,456S, which were unable to downregulate any substrates, could not prevent viral genome concatemerization (Fig. 6B and Table 2). In these experiments, we found that mutant proteins R443 and 3XPA could not fully complement dl3112 to prevent concatemer formation and degrade MRN or DNA ligase IV sufficiently (Table 2). These data confirm the requirement of DNA ligase IV for concatemer formation (60). They also demonstrate that the downregulation of DNA ligase IV is sufficient to prevent the joining of viral genomes even when MRN is not degraded.
E1b55K separation-of-function mutant proteins have defects in substrate binding.
We next examined how mutations in E1b55K affect interactions with cellular degradation substrates. The expression of E1b55K in cells can induce a large, perinuclear accumulation of the protein with characteristics of an aggresome (42). Many cellular proteins interacting with E1b55K have been observed to colocalize with E1b55K aggresomes (4, 22, 42, 44, 74). We therefore examined the ability of E1b55K mutant proteins to concentrate MRN and p53 into cytoplasmic aggregates (Fig. 7A). We found that mutant proteins which degraded MRN (H224, R240A, Y444A, and 3XPD) could all relocalize Nbs1 to cytoplasmic aggregates, although some mutant forms were less efficient than the WT E1b55K protein (Fig. 7A and Table 2). Interestingly, we found that all E1b55K mutant forms concentrated p53 into cytoplasmic aggregates to some extent (Fig. 7A and Table 2). Surprisingly, despite their inability to downregulate the p53 substrate, even the C361,366S and C454,456S proteins relocalized p53. R240A, however, was extremely defective in this activity (Fig. 4B), consistent with its diminished capacity to degrade p53 (12, 56).
To confirm our aggresome immunofluorescence results, we generated mutant E1b55K constructs that also contained a mutation (L83,87,91A) in the NES (39). These E1b55K proteins were expressed by transfection in U2OS cells and analyzed by immunofluorescence (Fig. 7B). Cells expressing E1b55K with a mutated NES displayed nuclear aggregates of E1b55K staining, as previously reported (20). The endogenous cellular Nbs1 and p53 proteins appeared to colocalize with E1b55K foci, suggesting possible interaction (Fig. 7B). When our mutations were combined with the NES mutation, all proteins tested formed distinct nuclear foci, but there were differences in their abilities to redistribute cellular proteins. Consistent with the degradation data, NES-R240A relocalized Nbs1 efficiently but not p53, whereas NES-H373A and NES-H354 recruited p53 efficiently but not Nbs1. Together with the aggresome immunofluorescence data, these results suggest that most mutant proteins with defects in degradation also have defects in binding substrates. However, the cysteine mutant forms (C361,366S and C454,4456S) may have additional defects related to p53 degradation, since they can still bind and relocalize this substrate.
To validate the immunofluorescence results, we performed coimmunoprecipitation experiments with E1b55K mutant proteins in the absence of E4orf6 (Fig. 7C and Table 2). E1b55K was immunoprecipitated from lysates of U2OS E1b55K mutant cells with the 2A6 antibody and analyzed for MRN and p53 interaction. Because it was difficult to resolve p53 from the background immunoglobulin band in E1b55K immunoprecipitates, we performed a reverse immunoprecipitation against p53 and probed for E1b55K interaction (Fig. 7C, bottom panels). Most mutant forms that failed to degrade either MRN or p53 also failed to interact with that particular substrate (Fig. 7C and Table 2), confirming the immunofluorescence results. In the absence of E4orf6, MRN binding to Y444A and 3XPA was significantly weaker than that to WT E1b55K. This interaction, however, was sufficient to induce the degradation of MRN in the presence of E4orf6 (Fig. 4 and 5). This finding suggests that E4orf6 may enhance E1b55K interaction with MRN by relocalizing E1b55K to the nucleus. We also found that the cysteine mutant proteins C361,366S and C454,456S were defective for MRN interaction but still retained the ability to bind p53, although not to the same extent as WT E1b55K (Table 2). Unfortunately, we were unable to examine endogenous DNA ligase IV by using immunofluorescence and could not detect an interaction between E1b55K and endogenous DNA ligase IV in our immunoprecipitation experiments (data not shown). Together, our data suggest that the E1b55K separation-of-function mutant proteins described here are defective in degradation due to diminished substrate binding. The ability of the cysteine mutant forms to bind p53 in the absence of downregulation suggests that there are additional E1b55K requirements for substrate degradation.
DISCUSSION
In this study, we examined requirements for adenovirus-mediated degradation of the known cellular substrates of the E1b55K/E4orf6 ubiquitin ligase complex: the MRN proteins, p53, and DNA ligase IV. We first addressed whether the degradation of each of these cellular factors required other proteins in the DNA damage response. While our previous use of E1b55K mutant proteins suggested that MRN degradation and p53 degradation are separable events (12), it was not clear whether the prior absence of cellular factors would affect the degradation of the other substrates. Using mutant cell lines, we found that adenovirus downregulates each substrate independently of the other substrates and does not require other DNA damage response proteins (Fig. 1B). Additionally, after examining adenovirus infections in A-TLD and NBS mutant cell lines, we suggest that Mre11 is the degradation target of E1b55K/E4orf6 (Fig. 1C). The downregulation of Rad50 and Nbs1, therefore, may be an indirect consequence of complex destabilization, as noted in other studies using RNA interference against particular members of the MRN complex in the absence of virus infection (64, 77). It is possible, however, that Rad50 is the target but that the protein is not recognized by E1b55K in the absence of Mre11 due to conformational changes. Further analysis of protein-protein interactions will be required to resolve the target.
We analyzed a large collection of E1b55K mutant forms in order to define further the viral requirements for degradation. We found that the downregulation of the MRN complex is separable from p53 and DNA ligase IV degradation, as discerned by using a number of E1b55K mutant proteins. The results for these mutant proteins indicate that at least two C-terminal cysteine-containing regions are critical to MRN degradation (Fig. 2 and Table 2). These regions include a proposed zinc finger motif (23) and a transcriptional repression domain (71, 73). In the context of viral infection, mutants with defects in MRN degradation cannot completely prevent DNA damage signaling (Fig. 3 and 5D) (12). However, some of these mutants can still prevent viral genome concatemerization. This effect appears to occur through the degradation of DNA ligase IV (Fig. 6), although we cannot exclude targeting of additional, unidentified substrates. Many of the mutant proteins analyzed could degrade DNA ligase IV independently of their p53 and MRN phenotypes, which suggests that there are distinct E1b55K requirements for DNA ligase IV destabilization.
Further analysis of the panel of E1b55K mutant proteins also identified cysteine mutant forms (C361,366S and C454,456S) with multiple defects that may affect degradation. In our studies, these mutant proteins were unable to downregulate any of the known cellular substrates (Fig. 4 and 6). We found that these cysteine mutant forms were defective in their association with MRN, but not with p53 (Fig. 7). This result is consistent with the location of the mutations outside the previously defined p53 interaction domain in E1b55K (71). In contrast to our findings, Hartl et al. recently observed that the C454,456S protein degraded exogenously expressed p53 and mislocalized Rad50 but not Mre11 in transformed baby rat kidney cells (31). The discrepancies may reflect different functions of E1b55K in transformed rodent cells versus human cells and the fact that we examined degradation specifically for endogenous proteins. From our data, it appears that the cysteine mutant proteins have an additional defect related to p53 degradation. The cysteine amino acids mutated in these E1b55K proteins may be important for modifying degradation substrates with ubiquitin or another marker required for ubiquitination and degradation. Unfortunately, we could not detect E1b55K interactions with endogenous DNA ligase IV, so it is unclear why the cysteine mutant proteins do not downregulate this substrate. Further insights will come from studies that delineate regions in E1b55K important for binding and degrading DNA ligase IV.
We examined a number of mutant proteins to address whether posttranslational modifications of E1b55K are important for the degradation of substrates. We found that the sumoylation site in E1b55K could be modified without impairing MRN degradation (Table 2), in agreement with a previous report (36). Phosphorylation sites in the C terminus appeared to be important for both the degradation of cellular substrates and the correct cellular localization of E1b55K (Fig. 5 and 6). Consistent with our data, a similar Ad12 E1b55K mutant protein (S476,477A) was also found previously to be defective in the formation of cytoplasmic aggregates (76). Phosphorylation may alter protein conformation, thereby affecting intramolecular E1b55K interactions or association with partner proteins. Along these lines, we found by coimmunoprecipitation that the interaction between 3XPA and p53 was severely defective or unstable (Table 2), consistent with the inability of this mutant protein to prevent p53-mediated transactivation and promote cellular transformation (62). The phosphorylation of all three amino acids may be required, however, since 2XPA still degraded p53 (Fig. 5D) (50) and other cellular substrates (Fig. 5D) and retained the ability to bind p53 (63). The interaction between 3XPA and MRN was significantly weaker than that between WT E1b55K and MRN (Fig. 7), although it was sufficient to induce MRN degradation in the presence of rAdE4orf6 (Fig. 5A and C). The phosphorylated amino acids in E1b55K lie within consensus casein kinase I and casein kinase II motifs (62, 63). However, it is still unclear whether these kinases are responsible for the phosphorylation of E1b55K in cells. The identification of kinases that modify E1b55K will provide more insight into the effects of phosphorylation on localization, structure, and protein-protein interactions important to function. It is interesting that the R443 protein also displayed a diffuse cytoplasmic staining pattern in U2OS stable cell lines (Fig. 4A), although it still formed some small, perinuclear foci. Therefore, the C terminus of E1b55K may be important to its structure and localization. While we do not know whether phosphorylation is affected for this mutant protein, R443 retained stable p53 binding (Fig. 7) (34, 71), unlike 3XPA.
Our analysis of E1b55K mutant proteins illustrated differences in degradation by proteins coexpressed with E4orf6 alone compared to degradation in the context of virus infection. We noticed a particular difference with proteins mutated in the C terminus (Y444A, R443, and 3XPA), which showed greater defects than the other proteins in the degradation of substrates during viral infection (compare Fig. 3 and 5D to Fig. 4B). Higher levels of E4orf6 expressed from the recombinant adenovirus than in the context of virus infection may partly contribute to these observations. The results of experiments with E1b55K mutant proteins and E4orf6 alone are useful in delineating the absolute requirements for degradation. For example, the phosphorylation of E1b55K may not be absolutely required for MRN degradation but may contribute more to the degradation of p53 and DNA ligase IV (Fig. 5C). Interactions between viral proteins are likely to be far more complex in the context of virus infection, and therefore, it will be informative to incorporate some of the E1b55K mutant proteins analyzed here into the virus to characterize further their phenotypes. It will also be interesting to examine degradation in primary human cells, in order to determine whether the same requirements hold true.
Although the degradation of p53, MRN, and DNA ligase IV induced by E1b55K/E4orf6 appears to be proteasome mediated, only p53 has been shown to be a direct ubiquitination target (30, 49). It will be important to determine whether the ubiquitination of the MRN complex and DNA ligase IV by the E1b55K/E4orf6 ubiquitin ligase is directly involved in degradation. This is likely to be the case, since the downregulation of all these substrates is prevented by the expression of transdominant cullin 5 (43) and by small interfering RNA knockdown of cullin 5 (R. A. Schwartz and M. D. Weitzman, unpublished data; 2, 43, 70). Other proteins that interact with E1b55K have been identified by genetic, biochemical, and proteomic studies (22, 24, 30, 44, 75); however, not all of these interacting proteins are targets for downregulation. It will be interesting to ascertain what determines the ubiquitination and degradation of E1b55K-associated proteins. Understanding the selective targeting of degradation substrates by E1b55K mutant proteins will help illuminate which cellular proteins are involved in viral replication, late protein production, and cell transformation.
Acknowledgments
We thank A. Berk, P. Branton, P. Concannon, G. Ketner, A. Levine, T. Melendy, D. Ornelles, J. Petrini, Y. Shen, and H. Young for generous gifts of reagents. We thank Daniel Linfesty and Darwin Lee for technical assistance. We thank members of the Weitzman lab for discussions and critical reading of the manuscript. We acknowledge the James B. Pendleton Charitable Trust for providing the Pendleton Microscopy Facility.
This work was supported by NIH grant CA97093 (M.D.W.) and by gifts from the Joe W. and Dorothy Dorsett Brown Foundation and the Lebensfeld Foundation. R.A.S. was supported in part by a scholarship from the ARCS Foundation.
Footnotes
Published ahead of print on 9 July 2008.
REFERENCES
- 1.Babiss, L. E., and H. S. Ginsberg. 1984. Adenovirus type 5 early region 1b gene product is required for efficient shutoff of host protein synthesis. J. Virol. 50202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 817034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berk, A. J. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 247673-7685. [DOI] [PubMed] [Google Scholar]
- 4.Blair Zajdel, M. E., and G. E. Blair. 1988. The intracellular distribution of the transformation-associated protein p53 in adenovirus-transformed rodent cells. Oncogene 2579-584. [PubMed] [Google Scholar]
- 5.Blanchette, P., C. Y. Cheng, Q. Yan, G. Ketner, D. A. Ornelles, T. Dobner, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 249619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchette, P., K. Kindsmuller, P. Groitl, F. Dallaire, T. Speiseder, P. E. Branton, and T. Dobner. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 822642-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boivin, D., M. R. Morrison, R. C. Marcellus, E. Querido, and P. E. Branton. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 731245-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, C. R., S. J. Doxsey, E. White, and W. J. Welch. 1994. Both viral (adenovirus E1B) and cellular (hsp 70, p53) components interact with centrosomes. J. Cell. Physiol. 16047-60. [DOI] [PubMed] [Google Scholar]
- 10.Burgert, H. G., Z. Ruzsics, S. Obermeier, A. Hilgendorf, M. Windheim, and A. Elsing. 2002. Subversion of host defense mechanisms by adenoviruses. Curr. Top. Microbiol. Immunol. 269273-318. [DOI] [PubMed] [Google Scholar]
- 11.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates III, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93477-486. [DOI] [PubMed] [Google Scholar]
- 12.Carson, C. T., R. A. Schwartz, T. H. Stracker, C. E. Lilley, D. V. Lee, and M. D. Weitzman. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 226610-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cathomen, T., and M. D. Weitzman. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 7411407-11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerosaletti, K. M., A. Desai-Mehta, T. C. Yeo, M. Kraakman-Van Der Zwet, M. Z. Zdzienicka, and P. Concannon. 2000. Retroviral expression of the NBS1 gene in cultured Nijmegen breakage syndrome cells restores normal radiation sensitivity and nuclear focus formation. Mutagenesis 15281-286. [DOI] [PubMed] [Google Scholar]
- 15.Cheng, C. Y., P. Blanchette, and P. E. Branton. 2007. The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion. Virology 36436-44. [DOI] [PubMed] [Google Scholar]
- 16.Corbin-Lickfett, K. A., and E. Bridge. 2003. Adenovirus E4-34kDa requires active proteasomes to promote late gene expression. Virology 315234-244. [DOI] [PubMed] [Google Scholar]
- 17.Dobbelstein, M., J. Roth, W. T. Kimberly, A. J. Levine, and T. Shenk. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 164276-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobner, T., and J. Kzhyshkowska. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 25925-54. [DOI] [PubMed] [Google Scholar]
- 19.Dosch, T., F. Horn, G. Schneider, F. Kratzer, T. Dobner, J. Hauber, and R. H. Stauber. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 755677-5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endter, C., B. Hartl, T. Spruss, J. Hauber, and T. Dobner. 2005. Blockage of CRM1-dependent nuclear export of the adenovirus type 5 early region 1B 55-kDa protein augments oncogenic transformation of primary rat cells. Oncogene 2455-64. [DOI] [PubMed] [Google Scholar]
- 21.Endter, C., J. Kzhyshkowska, R. Stauber, and T. Dobner. 2001. SUMO-1 modification required for transformation by adenovirus type 5 early region 1B 55-kDa oncoprotein. Proc. Natl. Acad. Sci. USA 9811312-11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fleisig, H. B., N. I. Orazio, H. Liang, A. F. Tyler, H. P. Adams, M. D. Weitzman, and L. Nagarajan. 2007. Adenoviral E1B55K oncoprotein sequesters candidate leukemia suppressor sequence-specific single-stranded DNA-binding protein 2 into aggresomes. Oncogene 264797-4805. [DOI] [PubMed] [Google Scholar]
- 23.Flint, S. J., and R. A. Gonzalez. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272287-330. [DOI] [PubMed] [Google Scholar]
- 24.Gabler, S., H. Schutt, P. Groitl, H. Wolf, T. Shenk, and T. Dobner. 1998. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 727960-7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao, G., and H. Luo. 2006. The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 845-14. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez, R. A., and S. J. Flint. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 764507-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodrum, F. D., T. Shenk, and D. A. Ornelles. 1996. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J. Virol. 706323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grand, R. J., M. L. Grant, and P. H. Gallimore. 1994. Enhanced expression of p53 in human cells infected with mutant adenoviruses. Virology 203229-240. [DOI] [PubMed] [Google Scholar]
- 29.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 769194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartl, B., T. Zeller, P. Blanchette, E. Kremmer, and T. Dobner. 2008. Adenovirus type 5 early region 1B 55-kDa oncoprotein can promote cell transformation by a mechanism independent from blocking p53-activated transcription. Oncogene 273673-3684. [DOI] [PubMed] [Google Scholar]
- 32.Hoppe, B. S., R. B. Jensen, and C. U. Kirchgessner. 2000. Complementation of the radiosensitive M059J cell line. Radiat. Res. 153125-130. [DOI] [PubMed] [Google Scholar]
- 33.Huang, M.-H., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 632605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao, C. C., P. R. Yew, and A. J. Berk. 1990. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology 179806-814. [DOI] [PubMed] [Google Scholar]
- 35.Kerscher, O., R. Felberbaum, and M. Hochstrasser. 2006. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22159-180. [DOI] [PubMed] [Google Scholar]
- 36.Kindsmuller, K., P. Groitl, B. Hartl, P. Blanchette, J. Hauber, and T. Dobner. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. USA 1046684-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konig, C., J. Roth, and M. Dobbelstein. 1999. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B-55-kilodalton protein. J. Virol. 732253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10524-530. [DOI] [PubMed] [Google Scholar]
- 39.Kratzer, F., O. Rosorius, P. Heger, N. Hirschmann, T. Dobner, J. Hauber, and R. H. Stauber. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene 19850-857. [DOI] [PubMed] [Google Scholar]
- 40.Lethbridge, K. J., G. E. Scott, and K. N. Leppard. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J. Gen. Virol. 84259-268. [DOI] [PubMed] [Google Scholar]
- 41.Lin, J., J. Chen, B. Elenbaas, and A. J. Levine. 1994. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 81235-1246. [DOI] [PubMed] [Google Scholar]
- 42.Liu, Y., A. Shevchenko, A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 7914004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo, K., E. Ehrlich, Z. Xiao, W. Zhang, G. Ketner, and X. F. Yu. 2007. Adenovirus E4orf6 assembles with Cullin5-ElonginB-ElonginC E3 ubiquitin ligase through an HIV/SIV Vif-like BC-box to regulate p53. FASEB J. 211742-1750. [DOI] [PubMed] [Google Scholar]
- 44.Maheswaran, S., C. Englert, S. B. Lee, R. M. Ezzel, J. Settleman, and D. A. Haber. 1998. E1B 55K sequesters WT1 along with p53 within a cytoplasmic body in adenovirus-transformed kidney cells. Oncogene 162041-2050. [DOI] [PubMed] [Google Scholar]
- 45.Marton, M. J., S. B. Baim, D. A. Ornelles, and T. Shenk. 1990. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J. Virol. 642345-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 9311295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orlando, J. S., and D. A. Ornelles. 1999. An arginine-faced amphipathic alpha helix is required for adenovirus type 5 E4orf6 protein function. J. Virol. 734600-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 153104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 713788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roth, J., C. Konig, S. Wienzek, S. Weigel, S. Ristea, and M. Dobbelstein. 1998. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J. Virol. 728510-8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rubenwolf, S., H. Schutt, M. Nevels, H. Wolf, and T. Dobner. 1997. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 711115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarnow, P., P. Hearing, C. W. Anderson, D. N. Halbert, T. Shenk, and A. J. Levine. 1984. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J. Virol. 49692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88593-602. [DOI] [PubMed] [Google Scholar]
- 55.Shackelford, J., and J. S. Pagano. 2005. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 41139-156. [DOI] [PubMed] [Google Scholar]
- 56.Shen, Y., G. Kitzes, J. A. Nye, A. Fattaey, and T. Hermiston. 2001. Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1B-55K protein. J. Virol. 754297-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shepard, R. N., and D. A. Ornelles. 2004. Diverse roles for E4orf3 at late times of infection revealed in an E1B 55-kilodalton protein mutant background. J. Virol. 789924-9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16349-357. [DOI] [PubMed] [Google Scholar]
- 59.Stewart, G. S., R. S. Maser, T. Stankovic, D. A. Bressan, M. I. Kaplan, N. G. Jaspers, A. Raams, P. J. Byrd, J. H. Petrini, and A. M. Taylor. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99577-587. [DOI] [PubMed] [Google Scholar]
- 60.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418348-352. [DOI] [PubMed] [Google Scholar]
- 61.Tauber, B., and T. Dobner. 2001. Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene 2781-23. [DOI] [PubMed] [Google Scholar]
- 62.Teodoro, J. G., and P. E. Branton. 1997. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J. Virol. 713620-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teodoro, J. G., T. Halliday, S. G. Whalen, D. Takayesu, F. L. Graham, and P. E. Branton. 1994. Phosphorylation at the carboxy terminus of the 55-kilodalton adenovirus type 5 E1B protein regulates transforming activity. J. Virol. 68776-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vo, A. T., F. Zhu, X. Wu, F. Yuan, Y. Gao, L. Gu, G. M. Li, T. H. Lee, and C. Her. 2005. hMRE11 deficiency leads to microsatellite instability and defective DNA mismatch repair. EMBO Rep. 6438-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. USA 91153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weinberg, D. H., and G. Ketner. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weitzman, M. D. 2005. Functions of the adenovirus E4 proteins and their impact on viral vectors. Front. Biosci. 101106-1117. [DOI] [PubMed] [Google Scholar]
- 68.Weitzman, M. D., and D. A. Ornelles. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 247686-7696. [DOI] [PubMed] [Google Scholar]
- 69.Wienzek, S., J. Roth, and M. Dobbelstein. 2000. E1B 55-kilodalton oncoproteins of adenovirus types 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J. Virol. 74193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 35782-85. [DOI] [PubMed] [Google Scholar]
- 72.Yew, P. R., C. C. Kao, and A. J. Berk. 1990. Dissection of functional domains in the adenovirus 2 early 1B 55K polypeptide by suppressor-linker insertional mutagenesis. Virology 179795-805. [DOI] [PubMed] [Google Scholar]
- 73.Yew, P. R., X. Liu, and A. J. Berk. 1994. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 8190-202. [DOI] [PubMed] [Google Scholar]
- 74.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 14244-58. [DOI] [PubMed] [Google Scholar]
- 75.Zhao, L. Y., A. L. Colosimo, Y. Liu, Y. Wan, and D. Liao. 2003. Adenovirus E1B 55-kilodalton oncoprotein binds to Daxx and eliminates enhancement of p53-dependent transcription by Daxx. J. Virol. 7711809-11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao, L. Y., and D. Liao. 2003. Sequestration of p53 in the cytoplasm by adenovirus type 12 E1B 55-kilodalton oncoprotein is required for inhibition of p53-mediated apoptosis. J. Virol. 7713171-13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong, H., A. Bryson, M. Eckersdorff, and D. O. Ferguson. 2005. Rad50 depletion impacts upon ATR-dependent DNA damage responses. Hum. Mol. Genet. 142685-2693. [DOI] [PubMed] [Google Scholar]